Abstract
The small viral channel Kcv is a Kir-like K+ channel of only 94 amino acids. With this simple structure, the tetramer of Kcv represents the pore module of all complex K+ channels. To examine the structural contribution of the transmembrane domains (TMDs) to channel function, we performed Ala scanning mutagenesis of the two domains and tested the functionality of the mutants in a yeast complementation assay. The data reveal, in combination with computational models, that the upper halves of both TMDs, which face toward the external medium, are rather rigid, whereas the inner parts are more flexible. The rigidity of the outer TMD is conferred by a number of essential aromatic amino acids that face the membrane and probably anchor this domain in the bilayer. The inner TMD is intimately connected with the rigid part of the outer TMD via π···π interactions between a pair of aromatic amino acids. This structural principle is conserved within the viral K+ channels and also present in Kir2.2, implying a general importance of this architecture for K+ channel function.
Keywords: Membrane Biophysics, Membrane Proteins, Potassium Channels, Protein Structure, Protein-Protein Interactions
Introduction
K+ channels are transmembrane proteins, which catalyze the selective and regulated flux of potassium ions across membranes. A breakthrough in understanding of structure/function correlates in these important proteins occurred with the high-resolution structures determined for several K+ channels (1–4). Many functional properties such as selectivity and gating of K+ channels are now understood on the basis of the specific architecture of these channel proteins. Still, many aspects of function and regulation cannot be explained only on the basis of the protein structure. The performance of the protein also depends on the lipid environment and on the organization of the protein in this environment. Indeed, there is increasing evidence that many different properties of K+ channels are depending on the interaction between the protein and the surrounding host membranes (5–7). As a consequence of this dependence, many amphiphilic drugs, which target ion channels, have dual effects, one effect being directly related to an interaction with the protein and a secondary effect being mediated by modification of the bilayer properties (8).
A good model system for understanding the interplay of membrane proteins with the membrane is provided by the viral potassium channel Kcv (9, 10). This Kir-like potassium channel with two transmembrane domains is, with only 94 amino acids, very small. The protein is almost completely embedded in the membrane. Only a small, 15 amino acid-long domain at the N terminus and a short turret domain between the two transmembrane domains stick out of the membrane into aqueous solution (11). It can be assumed that in this arrangement any protein/membrane interaction strongly reflects back on function.
Unbiased information on the structural significance of the two transmembrane domains (TMDs)2 and their importance for channel function can be obtained by an alanine scan of the relevant domains. In this approach, each amino acid in a primary sequence is individually replaced by the amino acid alanine, and the effect of this mutation is tested in a functional assay. In this way, it is possible to eliminate all side chain interactions, except for the Cβ atom, without altering the main chain conformation or the insertion of steric effects (12–14). Alanine is a common natural amino acid in all kinds of secondary structures, including TMDs (12). For these reasons, alanine is the amino acid of choice in the mutagenesis scan. Hence, the replacement of a residue with alanine should reveal the contribution of the replaced residue to the overall stability and fold of the protein (15).
In the present study, we use alanine scanning mutagenesis of the two transmembrane domains of Kcv in conjunction with a structural Kcv model (11, 16). The data provide a comprehensive picture of the functional architecture of the channel. The upper part of the outer transmembrane domain is anchored via aromatic side chains with the membrane. A mutual interaction between a pair of amino acids on the two TMDs attaches the inner TMD to the outer TMD and in this way also immobilizes the upper part of TMD2. The application of an elastic network model supports the structural significance of a mutual interaction of these amino acids (17, 18).
EXPERIMENTAL PROCEDURES
For the yeast complementation assay, the Kcv gene was cloned into pYES2 (Invitrogen) into the EcoRI and XhoI site or as a chimera with EGFP into the BamHI and XhoI site of the plasmid (19). The QuikChange site-directed mutagenesis method (Stratagene) was used to insert the point mutations. The point mutations were designed on the primer level or randomized on the primer level by using the wobble base codon NNK (where N is AGCT and K is GT), which codes for all possible amino acids. The resulting constructs were checked by DNA sequencing.
Yeast complementation experiments were done with the yeast strain SGY1528 (Mat a ade 2–1 can 1–100 his 3–11,15 leu 2–3,112 trp 1–1 ura 3–1 trk 1::HIS3 trk 2::TRP1) (19). This strain, kindly provided by Dr. Minor (University of California San Francisco), lacks an endogenous potassium uptake system and is not able to grow on media ≤ 10 mm potassium. For the complementation assay, yeasts were grown in parallel under selective (1 mm and 0.5 mm KCl agar plates) and non-selective (100 mm KCl agar plates) conditions for 3 days at 30 °C.
To analyze the spatial distribution of amino acids in the channel protein, the MD model of Kcv based on the tetrameric form of the KirBac1.1 (PDB code 1P7B) x-ray template structure was used (11). The programs PyMOL and MOLCAD (20) were used for studying the three-dimensional structure and the spatial relationships of the amino acid residues.
Elastic network models (18) have already been successfully applied to determine vibrational dynamics and kinetically hot residues, avoiding time-consuming molecular dynamics simulations (21). Such network models use a coarse-grained representation for amino acids by substituting the complex amino acid by a bead on the respective Cα position. Interactions between residues are modeled as harmonic springs for contacts within a cutoff distance. Anisotropic network models (17) additionally invoke anisotropy of positional fluctuations to assess the directions of collective motions. Eigenmodes and eigenfrequencies are derived from the so-called Hessian matrix by diagonalization techniques. The approach to quantify the impact of thought-experiment-style mutations, e.g. altering intramolecular forces up to the full deletion of contacts, is described elsewhere (22). We expanded this approach by computing overlaps of corresponding eigenvectors belonging to eigenvalues greater than zero. This allows us to qualitatively assess the effect the presence or absence of an amino acid pair interaction has on the functional mechanics of the protein.
RESULTS
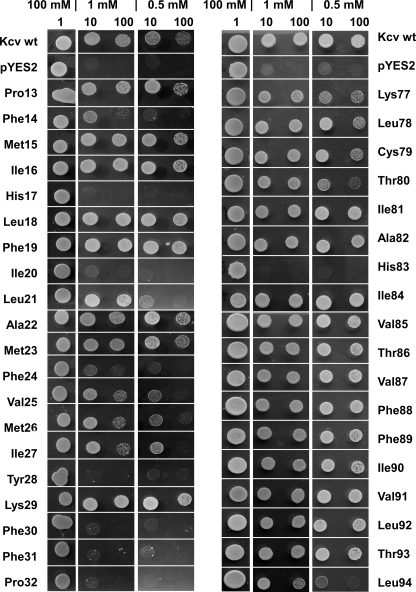
To detect functionally important side chains in the amino acid composition of the transmembrane domains of Kcv, the respective amino acids were replaced one by one by alanine. The two alanines (Ala-22 and Ala-82), which are already present in the TMDs of the wild-type channel (Fig. 1A), were replaced by glycine. All channel mutants were expressed individually in a yeast strain, which lacks a functional K+ uptake system. These yeasts are only able to survive in medium with a high K+ concentration. They do not grow on a selective low K+ medium unless they are expressing a heterologous K+ uptake system (19, 23). Fig. 2 shows representative data from a growth assay of the yeast mutants expressing only the vector (pYES2), the Kcv-WT channel, or its mutants. All yeasts transfected with either of the constructs were able to grow on high (100 mm) K+ medium, meaning that the expression of the mutations was deleterious for the cells. The two top rows show that the yeast strain transfected with the vector is, as expected, not able to grow under selective conditions with only 1 mm or 0.5 mm K+. Growth under selective conditions can be rescued by expressing the Kcv-WT channel. Hence, an active viral K+ channel in the plasma membrane provides a sufficient K+ influx for yeast growth under selective pressure.
FIGURE 1.
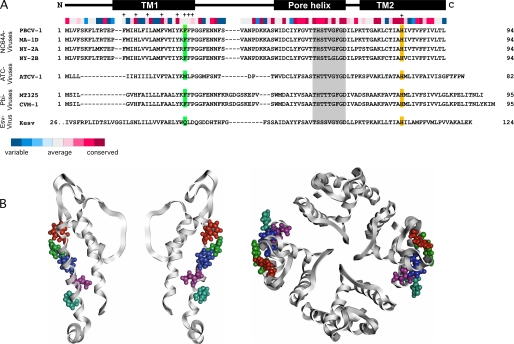
Alignment of different viral K+ channels and structure of Kcv. A, several algae viruses (the names are given on the left) that infect different hosts code for K+ channels (second row). The channel proteins are variable between the viruses but can also differ in isolates from the same virus, e.g. in NC64A viruses. The alignment highlights the position of the canonical K+ channel selectivity filter (in gray) as well as two amino acids corresponding to Phe-30 and His-83 in Kcv from virus Paramecium bursaria chlorella virus 1 (PBCV-1). The structural organization of Kcv from PBCV-1 with the position of the two transmembrane domains (TMD1 and TMD2) as well as the pore helix are illustrated in the top panel (A) and are shown explicitly in the simulation model (B). The degree of amino acid conservation between the viral K+ channels was estimated according to Ref. 46 and is given as a color-coded bar below the structure sketch. Different tones of blue indicate variable amino acids, whereas red tones indicate conserved amino acids. B, position of selected amino acid residues in the first transmembrane domain of Kcv. Two opposing subunits of Kcv are shown as ribbons in the left panel in side view and all four subunits in top view (right panel). Critical amino acids in TMD1, which were detected by the alanine-scanning mutagenesis, namely Phe-31 (red), Tyr-28 (green), Phe-24 (blue), Ile-20 (magenta), and His-17 (cyan), are highlighted. Both perspectives show that all critical residues are facing toward the lipid bilayer.
FIGURE 2.
Yeast complementation assay of Kcv mutants. The amino acids of the first transmembrane domain (left panel) and of the second transmembrane domain (right panel) were individually mutated to alanine. The two endogenous alanines were replaced by Gly. When the K+ uptake deficient yeast strain SGY1528 was transfected with the vector only (pYES2), it was only able to grow on medium with high (100 mm) K+. Transfection of the yeast strain with the WT Kcv channel allowed growth on selective media with low (1 mm or 0.5 mm) K+. Mutations in the transmembrane domains resulted in variable growth under selective conditions. The yeasts transfected with Kcv-WT, Kcv mutants, and with empty vector were plotted in different dilutions: 1 (undiluted), 1:10, and 1:100.
A scrutiny of the yeasts transfected with Kcv mutants shows that many mutants also grew under the selective conditions (Fig. 2). This implies that a large number of amino acid positions in the TMDs tolerate a mutation into alanine without loss of function. The data also highlight several important amino acids that are crucial for channel function in yeast. The substitutions of nearly all phenylalanines (Phe-14, Phe-24, Phe-30, and Phe-31), histidine (His-17), isoleucine (Ile-20), tyrosine (Tyr-28), and proline (Pro-32) in the first outer transmembrane (TMD1) domain showed the most dramatic effects. In these cases, the substitution results in a near or complete loss of channel function.
The picture is different in the second, inner transmembrane domain (TMD2). In this domain, alanine scanning revealed that nearly all of the amino acid side chains were not essential for channel function. Only the substitution of histidine (His-83) caused a total loss of function, whereas substitution of leucine (Leu-94) leads only to a significantly reduced function of yeast growth (Fig. 2).
To understand the spatial organization of the sensitive amino acids in the context of the three-dimensional structure of the channel, we localized their positions in the simulation model of Kcv (11). Fig. 1B shows the Kcv structure with the respective amino acids highlighted. It seems that the relevant amino acids of TMD1 nearly all present their side chains to the membrane. Fig. 1A illustrates that nearly all of the sensitive amino acids shown in Fig. 2, with the exception of Phe-31 and Ile-20, are also conserved (24–26) within the sequences of all viral potassium channels. This emphasizes that the overall architecture of the channel is maintained throughout this family of channels.
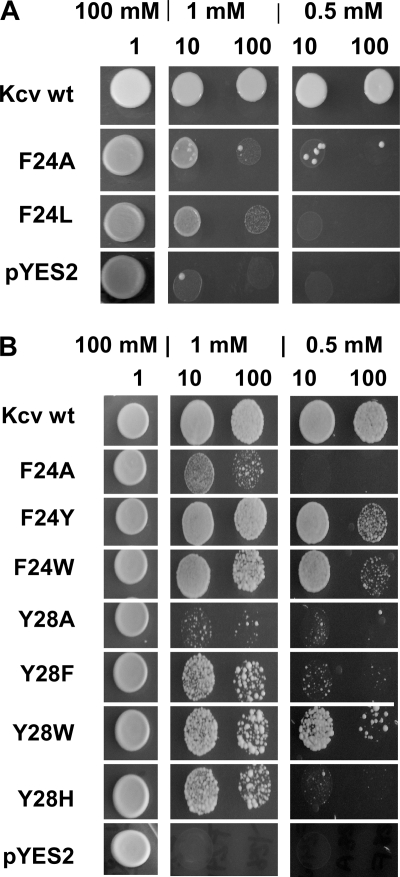
The conservation and orientation together with the aromatic side chain character suggests that these amino acids are involved in an anchoring of TMD1 in the lipid. An alternative explanation for the loss of function in the mutants could be the reduced hydrophobicity that is introduced with the alanine. To test the importance of a hydrophobic flavor of the critical amino acids, one of them, Phe-24, was mutated to Leu. This eliminates the aromatic side chain but preserves the hydrophobic character of this amino acid. A test of the mutant in the yeast system revealed that the substitution of Phe-24 with leucine failed to recover channel function (Fig. 3A). Hence, the hydrophobic nature of the amino acid in this position is not the only requirement.
FIGURE 3.
Yeast complementation assay of aromatic amino acids in TMD1. A, a replacement of the hydrophobic amino acid Phe-24 by the hydrophobic amino acid Leu did not rescue yeast growth on selective media. B, substitution of Phe-24 and Tyr-28 by other amino acids with an aromatic side chain rescued yeast growth on selective media. The complementation assay was done as described in Fig. 2.
To test the alternative hypothesis, namely the importance of the aromatic side chains, the amino acids Phe-24 and Tyr-28 were replaced by tryptophan or tyrosine for position 24 or with tryptophan, histidine, or phenylalanine for position 28, respectively. The results of the yeast complementation assay in Fig. 3B imply that the presence of an aromatic side chain in these positions is indeed sufficient for channel function. All aromatic substitutions were able to rescue channel function with varying degrees. The results of these experiments stress that proper channel function requires the presence of aromatic amino acids in TMD1, which are oriented toward the membrane for anchoring of the protein in the bilayer.
In further experiments we also tested the significance of another aromatic amino acid, His-17, in TMD1. In this case we replaced His-17 with a randomization approach, meaning that in principle any of the 20 possible amino acids could occur at this position. Twelve of the 60 tested yeast colonies transformed with the randomized plasmids showed growth on selective media with 0.5 mm KCl. After eliminating false positives and WT-like channels, we found that a mutant in which histidine was substituted by tryptophan was able to rescue channel function. The results of these experiments imply that the amino acid tryptophan with its aromatic side chain can replace the aromatic histidine in a functional channel. The results of these experiments are in agreement with the findings of Fig. 1B and underscore the importance of the aromatic side chains for anchoring of the protein in the membrane. Hence, the essential features of the critical amino acids are exposure and geometric orientation of the side chains toward the lipid bilayer. This kind of anchoring of transmembrane domains seems to be a general feature and has been described for several transmembrane domains (27–29).
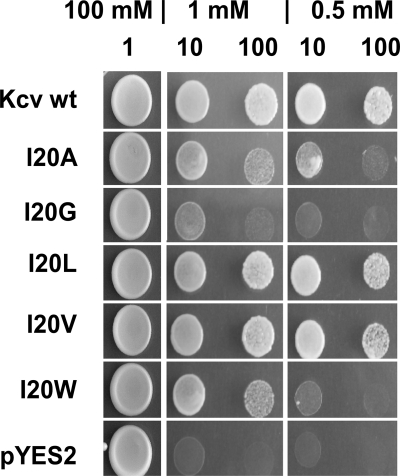
Among the critical amino acids in TMD1, Ile-20 is an exception in that it does not carry an aromatic side chain. Yeasts expressing a channel with an Ala substitution are barely growing (Fig. 2). Previous studies already revealed that this position in TMD1 influences functional properties of the Kcv channel via long-distance interactions with the pore (30). This mutant is, for example, more sensitive to cesium than the wild-type channel in voltage clamp measurements in Xenopus oocytes (30). Because of this critical role of Ile-20, further mutations were made to test the influence of the position on channel function and behavior. Fig. 4 shows the complementation assay of a series of different mutants. The data support a critical role of this position in channel function in the sense that channel performance is sensitive to the amino acid flavor in this position. The Kcv-I20V mutant is, as expected from previous experiments (30), functional. It is able to rescue yeast growth on selection media with 0.5 mm and 1 mm K+. However, the exchange with Trp or Gly modifies channel function. The substitution of Ile to Trp leads to a mutant that can still support yeast growth on 1 mm K+ selective media but not on 0.5 mm KCl. The second mutant, Kcv-I20G, completely fails to rescue yeast growth under selective conditions.
FIGURE 4.
Structural significance of Ile-20 in TMD1. Yeast complementation assay of different mutants of Kcv in which Ile-20 was replaced by alanine, glycine, leucine, valine, or tryptophan. Position 20 was sensitive to the flavor of the amino acid side chain character. The complementation assay was done as described in Fig. 2.
An important feature that is highlighted with the help of the MD model is the interaction of the two TMDs of Kcv. Although TMD2 is very tolerant to exchanges in amino acids, the substitution of His-83 caused a total loss of function (Fig. 2). The reason for the critical role of this amino acid becomes apparent from the MD model, which shows that this residue is in close contact with another essential amino acid in the TMD1, namely Phe-30. The structure reveals that Phe-30 and His-83 are interacting via a π-stack over a distance of only 3.3–3.4 Å (Fig. 5A). Such a stack of π-electron-rich residues occurs if two or more aromatic molecules are oriented in a parallel manner over a distance of about 3.3 Å. London dispersion forces, which dominate these π···π interactions, play important roles for correct folding and stability of many proteins (31) and for mutual interactions between helices in many membrane proteins (32, 33). Evidence for the contribution of these aromatic side chain interactions were already found for acid-sensing ion channels, where the π-π-stacking between the extracellular and transmembrane domain is essential for proton gating (34, 35). Also, the architecture of synthetic ion channels relies on this kind of π-π-stacking interactions (36). It is hence possible that the aromatic-aromatic interaction between Phe-30 and His-83 is an essential factor for structure and function in the Kcv channel.
FIGURE 5.
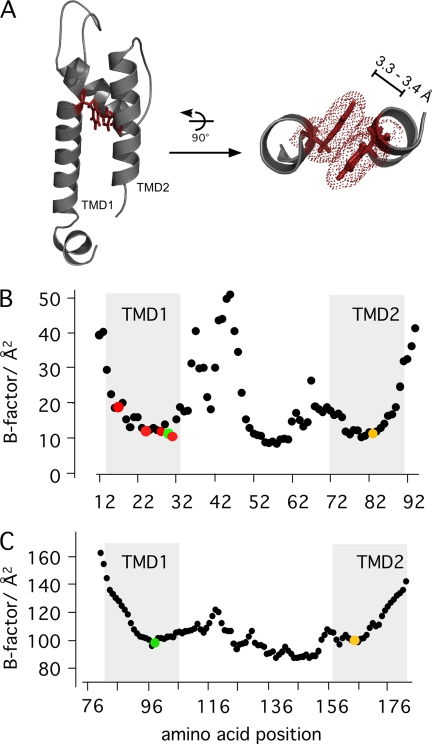
Flexibility of TMDs and aromatic-aromatic interaction between TMD1 and TMD2. A, the distance between Phe-30 in the TMD1 and His-83 in the TMD2 is about 3.3–3.4 Å. This is the typical spacing for π···π interactions (π-stack). Therefore, the two aromatic residues can contribute via this helix-helix interaction to larger protein stability. Cα B-factor distribution of Kcv (B) and in Kir2.2 (C) (4). B, the distribution of B-factors in Kcv as a function of the amino acids. The position of the transmembrane domains is indicated by a gray background. Critical amino acids are highlighted in color: His-83 in yellow and Phe-30 in green. The membrane facing aromatic amino acids His-17, Phe-24, Tyr-28, and Phe-31 are shown in red. C, the B-factors of Kir2.2 (PDB code 3JYC). The position of the interhelical contacts are shown in green (Phe-97) and yellow (Gln-165).
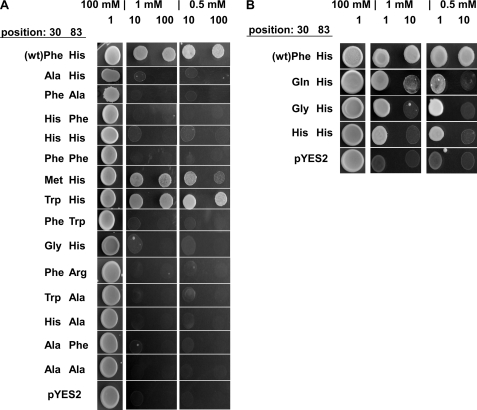
The presumed interaction between Phe and His in Kcv is not a frequently occurring pairing in membrane proteins for interhelical contact points (37). An alignment of viral potassium channels (Fig. 1A), however, shows that the His in TMD2 is highly conserved throughout all viral potassium channels. Even the distantly related potassium channel Kesv of the Ectocarpus siliculosus virus (EsV-1) (38) is conserved at this position. The complementary amino acid in TMD1, e.g. Phe-30 in Kcv, is only semi-conserved (Fig. 1A). In Kesv, we find a glutamine in this position, i.e. an amino acid that is uncharged and aliphatic. All other viral ion channels contain a phenylalanine or methionine in this position. In the case of methionine, it seems possible that the long and extended thiomethyl group is able to stabilize the channel structure through C-H···π interactions, where methionine is the aliphatic C-H donor and histidine is the aromatic π-acceptor (39). To test the importance of π···π or C-H···π interactions for channel structure and function, different substitutions of positions Phe-30 and His-83 with Ala, Gly, Phe, His, Met, Trp, and Gln were made. Note that the F30Q mutant mimics the amino acid pair in the Kesv channel (Fig. 1A). Fig. 6A shows that only two of the 14 substitutions were able to rescue yeast growth on selection media. The substitutions, which generate a functional channel, agree with the idea of a π···π interaction in this site. One of the two positive substitutions of Phe-30 is tryptophan, i.e. an amino acid with an aromatic side chain. As already expected from the alignments (Fig. 1A), Phe-30 could also be replaced by methionine.
FIGURE 6.
Yeast complementation assay to test the nature of a possible interaction between Phe-30 and His-83. Replacements of Phe-30 and His-83 by other amino acids showed that His is obligatory in position 83. The Phe could be replaced by a few amino acids. A, with a low concentration of yeast, a replacement of Phe-30 by Met or Trp conferred growth on selective media with 1 or 0.5 mm K+. B, at an undiluted yeast concentration it was also possible to detect yeast growth on selective media when Phe-30 was replaced by Gln, Gly, or His. The complementation assay was done as described in Fig. 2.
C-H···π interactions, like the Met-His interaction, can contribute to the stability of proteins with the overall stabilization energy of about 0.5 to 1.0 kcal mol−1 per interaction. They are therefore an important factor for protein stability (39). Also, almost all tryptophan residues in proteins are involved in C-H···π or π···π interactions (39, 40). The Trp-His interaction can stabilize a structure by about 1.0 to 4 kcal mol−1 (41). This is in the same range as the Met-His interaction. Hence, it is quite reasonable that the substitution of either methionine or tryptophan can rescue channel function in yeast. This finding highlights the concept of a functional conservation in proteins. This means that the conservation in a protein is not on the level of the specific amino acid but rather on the maintenance of a structural interaction. In the present case, the important factor is the interaction between the two TMDs, which can be brought about either by a π···π interaction or, alternatively, also by the weaker C-H···π-interaction (42).
Additional data show that three other mutations, namely F30H, F30Q, and F30G rescued yeast growth on selective media. However, in these cases, yeast growth could only be detected at high, undiluted yeast concentration on the selective plates (Fig. 6B). The data imply that these channel mutants are in principle also functional. They may provide a lower conductance for K+ uptake and hence a reduced rescue efficiency. Important to note is that the amino acids Gln and His in position 30 are potential partners for a side chain pairing with His-83 (43, 44). Only the mutant F30G does not follow this pattern.
Collectively, the results show that the two positions Phe-30 and His-83 are essential for proper channel function. The structural importance is given by the formation of intra-subunit interactions between the two TMDs. Furthermore, these positions are highly conserved throughout the viral K+ channels and are susceptible to changes. Moreover, the results emphasize an even higher complexity because there is no reciprocity between the two positions, as the mutual exchange of Phe and His leads to a non-functional mutant (Fig. 6A). Additionally, these results demonstrate that the MD model of Kcv (11) reproduces the real conditions in the protein quite well. Delicate interactions such as the π-π-stacking would not have occurred in an inappropriate model that differs from the more realistic one, for instance by a small helix rotation.
The information from the alanine scanning mutagenesis is together with the structural model of Kcv able to provide details on the three-dimensional model of the Kcv channel. More information can be gained by analyzing thermal B-factors originating from the MD simulations (11) that measure the average atomic fluctuations around the equilibrium position and therefore the flexibility of each amino acid in the Kcv protein. Fig. 5B shows Cα atom B-factors that indicate a pronounced dichotomy in both TMDs. Both membrane-spanning domains exhibit a high degree of flexibility toward the cytosolic side. The upper halves, which face the extracellular side, are far less flexible. The present data imply that the more or less rigid nature of TMD1 is achieved by an anchoring of this part of the protein in the membrane. Most of the aforementioned critical aromatic amino acids that face the membrane are located in this rigid part of TMD1 (Fig. 5B). TMD2 on the other hand does not have such a direct anchor in the membrane. The rigid character of the upper part of TMD2 can therefore be best explained by the contact between the two TMDs via the π···π interactions between Phe-30 and His-83. This hypothesis is supported by the fact that the mobility of TMD2 indeed increases downstream of His-83, e.g. the point at which the two transmembrane domains are in intimate contact (Fig. 5B). The fact that any disturbance of the architecture that affects the rigidity of the TMDs results in a loss of channel function implies a certain importance for channel function. It is reasonable to speculate that the maintenance of the rigid nature of the TMDs in this part of the protein is connected to the function of the selectivity filter. The delicate structure of the selectivity filter, which has to maintain its structure to select between cations, probably requires this suspension by the two TMDs.
The relevance of the π···π-interactions between the two TMDs for channel structure and stability is further supported by an application of the theory of anisotropic network models to the Kcv model (11). By using a homogeneous parameterization of the network model, we performed a purely structural-topological sensitivity analysis of the channel: we artificially deleted the mutual contact of amino acids 30–83 within each subunit at the same time. This mutation has no experimental analog and can therefore be performed in silico only. To assess the effect of this deletion on the mechanics of the potassium channel, we computed the difference between the wild-type and mutant covariance matrix using the Frobenius norm. The covariance matrix contains information on correlated motions of all the residues in the tetramer. To avoid obscuration effects that may emerge by summing up the absolute differences of the whole matrix, we defined distinct channel elements and thus, by extension, different subparts of the above-mentioned covariance matrix, which describe local mechanics for relevant parts of the channel (11). The Frobenius norm of a mutant with respect to the wild type increases whenever a mutation has a significant impact on the mechanics of the region under consideration. We detected here a major influence of the contact deletion on both TMDs and the turret region (data not shown).
Another measure of the biomechanical effect that is invariant under rotation is the overlap distance. It is derived for all mechanical modes of the channel. The similarity of corresponding eigenvectors denoting distinct fluctuations of each residue of the wild-type and mutant system can be quantified by the angle between the vectors. We defined the overlap distance dij as dij = 1-cos(αij), with αij being the angle between each of the two eigenvectors (i in the wild type and j in the mutant). A pairing with similar fluctuation vectors shows a distance close to zero, whereas a distance close to 1 indicates large differences between the eigenvectors. In the present case, we identified corresponding modes from the wild type and mutant by using the frequencies of the mechanical motions.
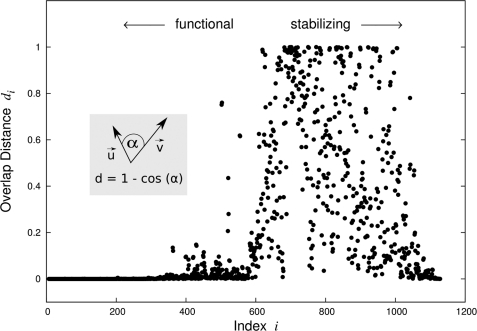
The final results for mechano-dynamical similarity of modes of motions upon mutation of the 30–83 contacts are depicted in Fig. 7. Clearly, large deviations (dij) between wild type and mutant eigenvectors are present for large indices only. Small indices denote small frequencies and represent functional motions of the channel. Such global mechanics are mainly because of function, whereas high-frequency motions, which are populated with only a small probability, describe localized motions having effects mainly on the structure and stability of the protein (45). The data show that the contact between residues 30 and 83 has a major impact on local dynamics as expressed by high-frequency modes and, hence, influences the structure rather than the functionality of the Kcv channel.
FIGURE 7.
Overlap distance for eigenvectors of the wild type and mutant (Δ 30–83) Kcv. With αij denoting the angle between two eigenvectors, ui (wild-type) and vi (mutant), the overlap distance is computed as dij = 1-cos(αij). Overlap distances are shown for corresponding eigenvectors computed for the wild-type and the mutant system using anisotropic network models. In the mutant Kcv, the contact between residues 30 and 83 is artificially deleted within all chains simultaneously. Small indices (slow motions) agree with a high confidence between the wild type and mutant. The respective eigenvectors describe global motions of the molecule that are relevant for biological function(s). High-frequency motions (large indices), however, describe localized dynamics acting on single atoms crucial for the stability of the protein (46). We clearly see that most if not all changes that are caused by the mutation Δ 30–83 stem from the structural modes and thus cause stability issues.
DISCUSSION
The present data support a model for the functional architecture of the small viral K+ channels in which the upper part of the outer transmembrane domain must be anchored via aromatic side chains with the surrounding membrane. Such an anchoring is typical for many membrane proteins (27–29). Mutual interaction of an amino acid pair attaches the inner TMD of the channels to the outer TMD and immobilizes in this way also the upper part of the former TMD. An elastic network model of the Kcv channel underscores the structural significance of this amino acid pairing and reveals that its disruption results in modified protein stability. The experimental data further imply that the respective interaction between the two TMDs supports optimal function of the channel, although it does not seem to be essential. Loss of channel function was induced by the disruption of the pair-wise interaction in most of the mutants that were tested in the rescue assay. The fact that mutants that maintained the stronger π···π interaction or the weaker C-H···π interaction between the TMDs still support proper rescue efficiency implies that the amino acid pairing motive that connects the two TMDs is beneficial for channel function. Interesting to note is that the F30Q mutant, which mimics the amino acid sequence of the Kesv channel, is able to rescue, albeit with a low efficiency, growth of yeast mutants. This observation implies that a general motive of interactions between the two transmembrane domains is indeed conserved within the small viral K+ channels. One mutant, F30G, in which Gly-30 does not interact with His-83, is also able to rescue yeast growth with a low efficiency. The results of these experiments suggest that a close interaction between positions 30 and 83 is not obligatory for basic channel function. The structural connection between the two TMDs may, in the case of the F30G mutant with the small and flexible Gly, be taken over by other interactions between the TMDs, e.g. between Phe-31 and His-83.
The overall architecture and dynamics we find here for the TMDs in Kcvs is not restricted to the small viral K+ channel. The same bipartite organization of the two TMDs, which is characteristic for Kcvs (Fig. 5B), is also present in the Kir2.2 channel. In the latter channel (4), the B-factors imply that those parts of the TMDs that face the external medium are fairly rigid (Fig. 5C). Like in Kcv, the parts of the Kir2.2 TMDs, which are directed toward the cytosol, are more flexible (compare Fig. 5, B and C). A scrutiny of its crystal structure shows that the amino acid pair Phe-97 and Gln-165 can in Kir2.2 generate the contact between the two TMDs and hence a mutual fixation of the two helices in Kir2.2. The XH··π interaction between the two amino acids over a distance of 3 Å is in the same range as the π-stacking interaction in Kcv. Moreover, the two critical amino acids of Kir2.2 are located as in Kcv in the rigid parts of the TMDs at the transition to the flexible parts (Fig. 5C). The mutual interaction between the two amino acids together with their location is consistent with the idea that they generate a close contact between the two transmembrane domains in Kir2.2. The finding of a similar architectural principle in the Kcv and Kir channels implies that the interhelical contact between the two TMDs and the rigid character of the outer part of the TMDs are important features for channel function.
Acknowledgments
We thank Profs. Adam Bertl (Technische Universität Darmstadt) and Dan Minor (University of California San Francisco) for help with the yeast rescue assay and Prof. Mike Blatt (University of Glasgow) for help with the manuscript. Computer time on the IBM Regatta system (Hessischer Hochleistungsrechner) was provided at the Hochschulrechenzentrum Darmstadt.
This work was supported in part by the Deutsche Forschungsgemeinschaft (to G. T. (TH558/19-1), the Adolf-Messer-Stiftung (to S. M. K.), the Fonds der Chemischen Industrie (to S. M. K.) and K. H.) and the European Drug Initiative on Channels and Transporters (EDICT) project EU FP7 (201924) (to A. M.).
- TMD
- transmembrane domain
- TMD1
- first, outer transmembrane domain
- TMD2
- second, inner transmembrane domain
- MD
- molecular dynamic.
REFERENCES
- 1. Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. (1998) Science 280, 69–76 [DOI] [PubMed] [Google Scholar]
- 2. Jiang Y., Lee A., Chen J., Cadene M., Chait B. T., MacKinnon R. (2002) Nature 417, 515–522 [DOI] [PubMed] [Google Scholar]
- 3. Kuo A., Gulbis J. M., Antcliff J. F., Rahman T., Lowe E. D., Zimmer J., Cuthbertson J., Ashcroft F. M., Ezaki T., Doyle D. A. (2003) Science 300, 1922–1926 [DOI] [PubMed] [Google Scholar]
- 4. Tao X., Avalos J. L., Chen J., MacKinnon R. (2009) Science 326, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marius P., Zagnoni M., Sandison M. E., East J. M., Morgan H., Lee A. G. (2008) Biophys. J. 94, 1689–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Valiyaveetil F. I., Zhou Y., MacKinnon R. (2002) Biochemistry 41, 10771–10777 [DOI] [PubMed] [Google Scholar]
- 7. Barrera F. N., Renart M. L., Poveda J. A., de Kruijff B., Killian J. A., González-Ros J. M. (2008) Biochemistry 47, 2123–2133 [DOI] [PubMed] [Google Scholar]
- 8. Lundbaek J. A. (2008) J. Gen. Physiol. 131, 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Plugge B., Gazzarrini S., Nelson M., Cerana R., Van Etten J. L., Derst C., DiFrancesco D., Moroni A., Thiel G. (2000) Science 287, 1641–1644 [DOI] [PubMed] [Google Scholar]
- 10. Thiel G., Baumeister D., Schroeder I., Kast S. M., Van Etten J. L., Moroni A. (2011) Biochim. Biophys. Acta 1808, 580–588 [DOI] [PubMed] [Google Scholar]
- 11. Tayefeh S., Kloss T., Kreim M., Gebhardt M., Baumeister D., Hertel B., Richter C., Schwalbe H., Moroni A., Thiel G., Kast S. M. (2009) Biophys. J. 96, 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cunningham B. C., Wells J. A. (1989) Science 244, 1081–1085 [DOI] [PubMed] [Google Scholar]
- 13. Holst B., Zoffmann S., Elling C. E., Hjorth S. A., Schwartz T. W. (1998) Mol. Pharmacol. 53, 166–175 [DOI] [PubMed] [Google Scholar]
- 14. Di Cera E. (1998) Chem. Rev. 98, 1565–1591 [DOI] [PubMed] [Google Scholar]
- 15. Clackson T., Wells J. A. (1995) Science 267, 383–386 [DOI] [PubMed] [Google Scholar]
- 16. Tayefeh S., Kloss T., Thiel G., Hertel B., Moroni A., Kast S. M. (2007) Biochemistry 46, 4826–4839 [DOI] [PubMed] [Google Scholar]
- 17. Atilgan A. R., Durell S. R., Jernigan R. L., Demirel M. C., Keskin O., Bahar I. (2001) Biophys. J. 80, 505–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tozzini V. (2010) Q. Rev. Biophys. 43, 333–371 [DOI] [PubMed] [Google Scholar]
- 19. Minor D. L., Jr., Masseling S. J., Jan Y. N., Jan L. Y. (1999) Cell 96, 879–891 [DOI] [PubMed] [Google Scholar]
- 20. Brickmann J., Goetze T., Heiden W., Moeckel G., Reiling S., Vollhardt H., Zachmann C. D. (1995) in Data Visualization in Molecular Science: Tools for Insight and Innovation (Bowie J. E. ed) pp. 83–97, Addison-Wesley Reading, MA [Google Scholar]
- 21. Hamacher K., McCammon J. A. (2006) J. Chem. Theory Comput. 2, 873–878 [DOI] [PubMed] [Google Scholar]
- 22. Hamacher K. (2008) Gene 422, 30–36 [DOI] [PubMed] [Google Scholar]
- 23. Balss J., Papatheodorou P., Mehmel M., Baumeister D., Hertel B., Delaroque N., Chatelain F. C., Minor D. L., Jr., Van Etten J. L., Rassow J., Moroni A., Thiel G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12313–12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. (2005) Nucleic Acids Res. 33, W299-W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Glaser F., Pupko T., Paz I., Bell R. E., Bechor-Shental D., Martz E., Ben-Tal N. (2003) Bioinformatics 19, 163–164 [DOI] [PubMed] [Google Scholar]
- 27. Yau W. M., Wimley W. C., Gawrisch K., White S. H. (1998) Biochemistry 37, 14713–14718 [DOI] [PubMed] [Google Scholar]
- 28. Killian J. A. (2003) FEBS Lett. 555, 134–138 [DOI] [PubMed] [Google Scholar]
- 29. De Planque M. R., Killian J. A. (2003) Mol. Membr. Biol. 20, 271–284 [DOI] [PubMed] [Google Scholar]
- 30. Gazzarrini S., Kang M., Van Etten J. L., Tayefeh S., Kast S. M., DiFrancesco D., Thiel G., Moroni A. (2004) J. Biol. Chem. 279, 28443–28449 [DOI] [PubMed] [Google Scholar]
- 31. Shoemaker K. R., Fairman R., Schultz D. A., Robertson A. D., York E. J., Stewart J. M., Baldwin R. L. (1990) Biopolymers 29, 1–11 [DOI] [PubMed] [Google Scholar]
- 32. Popot J. L., Engelman D. M. (1990) Biochemistry 29, 4031–4037 [DOI] [PubMed] [Google Scholar]
- 33. Bowie J. U. (2005) Nature 438, 581–589 [DOI] [PubMed] [Google Scholar]
- 34. Li T., Yang Y., Canessa C. M. (2009) J. Biol. Chem. 284, 4689–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang H., Yu Y., Li W.G., Yu F., Cao H., Xu T.L., Jiang H. (2009) PLoS Biol. 7, e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bhosale S., Sisson A. L., Sakai N., Matile S. (2006) Org. Biomol. Chem. 4, 3031–3039 [DOI] [PubMed] [Google Scholar]
- 37. Adamian L., Liang J. (2001) J. Mol. Biol. 311, 891–907 [DOI] [PubMed] [Google Scholar]
- 38. Delaroque N., Müller D. G., Bothe G., Pohl T., Knippers R., Boland W. (2001) Virology 287, 112–132 [DOI] [PubMed] [Google Scholar]
- 39. Brandl M., Weiss M. S., Jabs A., Sühnel J., Hilgenfeld R. (2001) J. Mol. Biol. 307, 357–377 [DOI] [PubMed] [Google Scholar]
- 40. Wang L., Sun N., Terzyan S., Zhang X., Benson D. R. (2006) Biochemistry 45, 13750–13759 [DOI] [PubMed] [Google Scholar]
- 41. Fernández-Recio J., Vázquez A., Civera C., Sevilla P., Sancho J. (1997) J. Mol. Biol. 267, 184–197 [DOI] [PubMed] [Google Scholar]
- 42. Nishio M., Hirota M., Umezawa Y. (1998) The CH/π Interaction: Evidence, Nature and Consequences, pp. 175–202, Wiley-VCH, New York [Google Scholar]
- 43. Bhattacharyya R., Saha R. P., Samanta U., Chakrabarti P. (2003) J. Prot. Res. 2, 255–263 [DOI] [PubMed] [Google Scholar]
- 44. Heyda J., Mason P. E., Jungwirth P. (2010) J. Phys. Chem. B 114, 8744–8749 [DOI] [PubMed] [Google Scholar]
- 45. Bahar I., Atilgan A. R., Demirel M. C., Erman B. (1998) Phys. Rev. Lett. 80, 2733–2736 [Google Scholar]
- 46. Berezin C., Glaser F., Rosenberg J., Paz I., Pupko T., Fariselli P., Casadio R., Ben-Tal N. (2004) Bioinformatics 20, 1322–1324 [DOI] [PubMed] [Google Scholar]