Abstract
In oncology, simultaneous inhibition of epidermal growth factor receptor (EGFR) and HER2 by monoclonal antibodies (mAbs) is an efficient therapeutic strategy but the underlying mechanisms are not fully understood. Here, we describe a time-resolved fluorescence resonance energy transfer (TR-FRET) method to quantify EGFR/HER2 heterodimers on cell surface to shed some light on the mechanism of such therapies. First, we tested this antibody-based TR-FRET assay in NIH/3T3 cell lines that express EGFR and/or HER2 and in various tumor cell lines. Then, we used the antibody-based TR-FRET assay to evaluate in vitro the effect of different targeted therapies on EGFR/HER2 heterodimers in the ovarian carcinoma cell line SKOV-3. A simultaneous incubation with Cetuximab (anti-EGFR) and Trastuzumab (anti-HER2) disturbed EGFR/HER2 heterodimers resulting in a 72% reduction. Cetuximab, Trastuzumab or Pertuzumab (anti-HER2) alone induced a 48, 44, or 24% reduction, respectively. In contrast, the tyrosine kinase inhibitors Erlotinib and Lapatinib had very little effect on EGFR/HER2 dimers concentration. In vivo, the combination of Cetuximab and Trastuzumab showed a better therapeutic effect (median survival and percentage of tumor-free mice) than the single mAbs. These results suggest a correlation between the extent of the mAb-induced EGFR/HER2 heterodimer reduction and the efficacy of such mAbs in targeted therapies. In conclusion, quantifying EGFR/HER2 heterodimers using our antibody-based TR-FRET assay may represent a useful method to predict the efficacy and explain the mechanisms of action of therapeutic mAbs, in addition to other commonly used techniques that focus on antibody-dependent cellular cytotoxicity, phosphorylation, and cell proliferation.
Keywords: Antibodies, Anticancer Drug, Cancer Therapy, Receptor Regulation, Receptor Tyrosine Kinase, HTRF, Therapeutic Antibodies, Tyrosine Kinase Inhibitors
Introduction
The human ErbB family of receptor tyrosine kinase comprises four members: epidermal growth factor receptor (EGFR),2 HER2 (c-erbB-2), HER3 (c-erbB-3), and HER4 (c-erbB-4). These large glycoproteins contain an extracellular ligand binding domain, a transmembrane region and an intracellular receptor tyrosine kinase (RTK) domain. The extracellular portion consists of four subdomains referred to as domains I-IV (1). Except for HER2, structural changes in solution (from tethered to untethered conformation) that allow the exposure of the dimerization arm are induced following activation by a subset of potential ligands, including EGF (2, 3). Therefore, HERs can form homodimers, heterodimers and possibly higher order oligomers. HER2, which naturally adopt the untethered conformation, is described as the preferred heterodimerization partner with other HERs (4). The three-dimensional structures of the extracellular domains of each family member have been determined and widely support the mode of ligand-mediated dimerization activation of the EGFR signaling pathway (5).
These receptors are expressed in epithelial, mesenchymal, and neuronal tissues and play fundamental roles in cell proliferation, differentiation, adhesion, survival, migration (6), and in the molecular pathogenesis of cancer. Many modifications of HER expression contribute to generate malignant transformation, such as HER2 overexpression in breast cancer. HERs are, therefore, a key therapeutic target in many types of cancer, including breast and colorectal cancers. Clinically, the prognostic significance of each HER receptor is controversial, but generally, HER overexpression is correlated with poor prognosis (7, 8). Specifically, EGFR and HER2 co-expression is reported to occur in the most aggressive carcinomas (9). This association might be explained by the fact that EGFR/HER2 dimers can reduce EGFR endocytosis (10), decrease the dissociation between EGF and EGFR (11), increase EGFR recycling (12) and favor the mobility of cancer cells (13). Finally, HER heterodimers have been reported to be implicated in cancer cell resistance to various drugs by increasing phosphorylation of HER2 (14).
Targeted therapies against HER receptors are now one of the most attractive areas of oncology research. Two major classes of HER-targeted treatments are currently available: anti-HER monoclonal antibodies (mAbs), which target the extracellular domain (15, 16), and HER-specific tyrosine kinase inhibitors (TKIs) (16–18), which target the tyrosine kinase activity of these receptors. Most TKIs are ATP competitors and consequently inactivate the signal transduction pathways that mediate EGFR functions. The mAb mechanisms of action are multifactor and not completely understood. Their effect can be indirect through recruitment of the immune system mediated by the fragment crystallizable region of the antibody (promoting antibody-dependent cellular cytotoxicity, ADCC), or direct on the receptor by, for instance, preventing ligand binding (like in the case of the anti-EGFR mAb Cetuximab). The direct blockade of the binding site inhibits phosphorylation of the tyrosine kinase domain and therefore activation of the downstream signaling pathways (19). However, some other potential effects of anti-HER mAbs should be explored like their ability to modify HER internalization and degradation or to perturb the concentration of homo- and heterodimers.
Although, the expression levels of the different HERs can be used as biomarkers for the choice of anti-HER therapy, the expression of an individual receptor is not always a sufficient indicator of the HER status or of the response to therapy of a tumor (20). For example, patients with colorectal (21, 22) or lung cancer (23, 24) without EGFR overexpression (monitored by immunohistochemistry) might respond to anti-EGFR therapies (such Cetuximab or Gefitinib), while other patients with EGFR over-expression do not. Among breast cancer patients, only 40% of those selected for Trastuzumab treatment on the basis of HER2 overexpression respond to this therapy. Therefore, new biomarkers should be considered, such the global level of HER receptors, HER gene mutations, or dimerization modifications. This raises the question of the choice of the most suitable technology to perform such analyses.
HER dimerization has always been evaluated using biochemical approaches (electrophoretic separation of cross-linked dimers or co-immunoprecipitation). Recently, several new quantitative technologies have been developed (multiplex microbead immunoassay approach (25), VeraTag proximity assay (26, 27), proximity ligation assay (28), flow cytometric FRET (29), and time-resolved FRET (TR-FRET) or HTRF (30)). Except TR-FRET (or HTRF), none of those available in an academic laboratory allows the detection of dimers in intact cells with high specificity and sensitivity. We thus decided to adapt the antibody-based TR-FRET approach using terbium cryptate as long-lived fluorescence donor and the d2 dye as an acceptor to detect EGFR/HER2 heterodimers in various tumor cell lines with different EGFR and HER2 expression levels. Then, the effect of different targeted therapies on EGFR/HER2 dimers was evaluated in an ovarian carcinoma cell line (SKOV-3) and compared with their efficacy in xenografted nude mice.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The anti-EGFR mAbs Cetuximab and m425 were purchased from Merck KGaA (Darmstadt, Germany) and Merck AG (Frankfurt, Germany), respectively. The anti-EGFR mAb m225 was purified from the HB8508 hybridoma from the American Type Culture Collection (ATCC). Trastuzumab, Pertuzumab and the TKI Erlotinib were obtained from Roche Pharma AG (Grenzach-Wyhlen, Germany). Lapatinib was from GlaxoSmithKline (Hertfordshire, UK). The anti-HER2 FRP5 and FSP77 mAbs were kindly provided by Nancy Hynes (Basel, Switzerland) (31). For Western blotting, anti-EGFR, -HER2, -phospho-EGFR, -AKT, and -phospho-AKT antibodies were purchased from Cell Signaling Technology (Ozyme, Saint-Quentin, France). Antibodies against GAPDH and phosphorylated HER2 were purchased from Millipore (Molsheim, France). The irrelevant Px antibody was used for control experiments. Px is an IgG1 mAb that has been purified from the mouse myeloma MOPC21 (32). More details about references, targeted epitopes, and cross-reactivity of the anti-HER antibodies are reported in supplemental Table S1. Human EGF and 10% formalin (HT50–1-1) were from Sigma-Aldrich. Lumi4® Tb is a trademark of Lumiphore Inc.
Cell Lines
The human pancreatic carcinoma (BxPC-3, MiaPaCa-2), ovarian carcinoma (SKOV-3), breast cancer (BT474, SKBR-3), and epidermoid vulva carcinoma (A431) cell lines were from ATCC. The Capan-I (human pancreatic carcinoma cells) and NIH/3T3 (mouse embryonic fibroblasts) lines were kindly provided by L. Buscail (INSERM-U858, Toulouse, France) and by S. Schmidt (CRBM-UMR 7537, Montpellier, France), respectively. BxPC-3, BT474, and SKBR-3 cells were cultured in RPMI (Roswell Park Memorial Institute) 1640 medium (Invitrogen, Fisher Scientific, Illkirch, France); MiaPaCa-2, SKOV-3, A431, and NIH/3T3 cells in DMEM (Dulbecco's modified Eagle's medium) (Invitrogen). Media were supplemented as recommended by ATCC, usually with 10% fetal calf serum (FCS) (Life Technologies).
Plasmids, Viruses, and NIH/3T3-HERs Cell Lines
The Murine Stem Cell Virus (MSCV) retroviral vectors (Clontech, Ozyme) contain the hygromycin (pMSCV-hygro) or the puromycin N-acetyl transferase (pMSCV-puro) resistance gene. The fragments containing the EGFR transcript variant 1 (NM_005228.2) and HER2 transcript variant 1 (NM_004448.2) were isolated from pCMV6-XL4-EGFR and pCMV6-XL4-ERBB2 (Origene, CliniSciences, Montrouge, France) and subcloned in pMSCV-puro and pMSCV-hygro, respectively. pMSCV-puro-EGFR or pMSCV-hygro-HER2 were then transfected into the amphotropic packaging cell line AmphoPack-293 (Clontech). After 5 h, medium was replaced by fresh medium, and cells were cultured for 48 h. Supernatants containing replication-defective virus particles were then collected, centrifuged and filtered through 0.45 μm filters. 2 × 106 NIH/3T3 cells/150-mm plate were grown for 8 h and then incubated with the viral supernatants in complete DMEM (1:4) supplemented with 8 μg/ml polybrene (Sigma-Aldrich) for 16 h. Culture medium was then replaced by fresh medium and antibiotic selection (400 μg/ml hygromycin or 10 μg/ml puromycin) was started 2 days later. After 7 days of selection, NIH/3T3-R1 (infected with MSCV-puro-EGFR) and NIH/3T3-R2 (infected with MSCV-hygro-HER2) cells were labeled with anti-EGFR (m225) or anti-HER2 (FSP77) antibodies and coupled with a secondary anti-mouse FITC-labeled antibody to be cloned using selective fluorescence-activated cell sorting (FACS) with a FACSAria apparatus (Becton Dickinson Biosciences, Le Pont-De-Claix, France). To develop the NIH/3T3-R1R2 cell lines, the same strategy was followed after transfecting NIH/3T3-R2 cells with pMSCV-puro-EGFR.
Co-immunoprecipitation and Western Blotting
NIH/3T3, NIH/3T3-R1, NIH/3T3-R2, NIH/3T3-R1R2, and SKOV-3 cells were serum starved for 16 h and then incubated or not with 100 ng/ml EGF for 10 min. Cells were washed and lysed with lysis buffer (20 mm Tris-HCl pH 7.5, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EDTA, 1% Triton, 10% glycerol, 0.1 mm phenylmethylsulfonyl fluoride, 100 mm sodium fluoride, 1 mm sodium orthovanadate (Sigma-Aldrich), and one tablet of complete protease inhibitor mixture (Roche Diagnostics, Meylan, France)) for 30 min. The insoluble fraction was then eliminated by centrifugation and the protein concentration of the cell lysates was determined using the Bradford assay. Protein lysates were then immunoprecipitated (1 mg total proteins) or directly mixed with Laemmli buffer (1–20 μg total proteins depending on the target and cell line) and heated at 95 °C for 5 min. HER2 was immunoprecipitated using 80 μl of protein A- and protein G-coupled magnetic microbeads (Bio-Adembeads PAG, Ademtech, Pessac, France) that had been preincubated with 8 μg of FSP77 (anti-HER2 mAb) overnight and washed to remove unbound FSP77. Samples were incubated with microbead-bound FSP77 by gentle mixing at room temperature for 1 h. Microbeads were then washed six times with lysis buffer to remove unbound material. Laemmli buffer was added and samples were heated at 95 °C for 5 min. After electrophoresis on 7% SDS-PAGE under reducing conditions, proteins were transferred to polyvinylidene difluoride membranes (Millipore, Molsheim, France) that were saturated in PBS containing 0.1% Tween 20 and 5% nonfat dry milk and then incubated with antibodies against EGFR, HER2 and their phosphorylated forms. After washing and incubation with anti-mouse secondary antibodies (Sigma-Adrich), blots were visualized using a chemiluminescent substrate (Western Lightning Plus-ECL, Perkin Elmer, Courtaboeuf, France).
Flow Cytometry
Cell surface EGFR and HER2 expression was analyzed by FACS using the mouse anti-EGFR (m225) and anti-HER2 (FSP77) mAbs. After washing, an anti-mouse FITC-conjugated mAb (Sigma-Aldrich) was added to detect the primary antibodies. EGFR and HER2 were quantified with the Quantitative Immunofluorescence Indirect assay (QIFI kit, DAKO, Copenhagen, Denmark). This kit contains a series of calibration beads with well-defined quantities of mouse mAbs. These beads mimic cells with different antigen density, which have been labeled with primary mouse IgG mAb. Briefly, cells were incubated with m225 and FSP77 at saturating concentrations, and after washing, they were incubated with the same secondary antibody used for the calibration beads. Results were expressed as antibody binding capacity (ABC) of the analyzed cells. Samples were analyzed with a Coulter® Epics XL-MCLTM Flow Cytometer (Beckman Coulter, Roissy, France) by recording a minimum of 5,000 events to analyze EGFR/HER2 expression, and 10,000 events to quantify EGFR or HER2.
Quantitative EGF Binding
EGF binding in the different cell lines was determined by competition assay between unlabeled and labeled human EGF. Cells were plated at 5 × 104 per well in 96-well sterile, black microplates (Greiner Bio-One, Frickenhausen, Germany) in DMEM medium (without phenol red) supplemented with 10% FCS and after 24 h they were washed with KREBS buffer (146 mm NaCl, 4 mm KCl, 0.5 mm MgCl2, 1 mm CaCl2, 10 mm HEPES, 1 g/liter glucose, and 1 g/liter bovine serum albumin, Sigma-Aldrich) and incubated with 4 nm EGF labeled with Lumi4® Tb cryptate (Cisbio Bioassays, Bagnol sur Cèze, France) and increasing concentrations of unlabeled EGF (Sigma-Aldrich) in KREBS with 0.02% azide (Sigma-Aldrich) at room temperature in the dark for 90 min. After two washes with KREBS, the fluorescence of Lumi4 Tb cryptate was measured in TRF mode (60 μs delay, 400 μs integration) at 620 nm and 337 nm excitation using a Pherastar FS instrument (BMG Labtech, Champigny-sur-Marne, France) to quantify the binding of labeled EGF. IC50 results represent the concentration of unlabeled EGF required to inhibit 50% of the binding of labeled EGF.
Time-resolved Fluorescence Resonance Energy Transfer (TR-FRET) Assay
This assay was performed using the anti-EGFR mAb m425 and the anti-HER2 mAb FRP5 labeled with Lumi4 Tb cryptate (donor) and d2 dye (acceptor) (Cisbio Bioassays). These mAbs were chosen because their targeting epitopes are different from those of the therapeutic mAbs used in this study (see supplemental Table S1). Cells were plated at 105 per well in 96-well sterile, black microplates in DMEM (without phenol red) supplemented with 10% FCS and after 24 h they were washed with KREBS buffer, fixed in 10% formalin for 2 min and washed once with KREBS buffer. After incubation with the labeled mAbs (5 nm each in KREBS buffer) at 37 °C for 6 h, cells were washed four times with KREBS buffer. The fluorescence of Lumi4 Tb and d2 were measured respectively at 620 and 665 nm (60 μs delay, 400 μs integration) upon 337 nm excitation using a Pherastar FS instrument. The fluorescence of serial dilutions of Lumi4 Tb-labeled antibodies in KREBS buffer was simultaneously measured in the same microtiter plate, and the 665 nm emission was plotted against the 620 nm emission. The resulting curve was used to compute the 665 nm contribution from terbium (F665Tb) using the 620 nm emission (F620) of the samples. The TR-FRET signal was expressed as ΔF665(%) = 100 × ΔF665/F665Tb, with ΔF665 = F665c − F665Tb. The 665 nm and 620 nm emissions from the samples were corrected for background as F665c = F665sample − F665background and F620c = F620sample − F620background. The F665background and F620background values were obtained by measuring the fluorescence of a plate containing only reading buffer. The TR-FRET signal expressed as ΔF665(%) represents relative amount of EGFR/HER2 dimers normalized to the level of HER2. This normalization allows avoiding an artificial decrease of EGFR/HER2 dimer signal due to reduced HER2 level. The 620 nm time resolved fluorescence emission is correlated with HER2 concentration when the anti-HER2 mAb is labeled with Lumi4 Tb cryptate and the anti-EGFR mAb with d2. At the same time, the fluorescence of d2 is measured at 670 nm upon 620 nm excitation to quantify EGFR.
Because EGFR and HER2 expression levels are very different according to the studied cell line, the quantification of EGFR/HER2 dimers in the various cell lines required the optimization of the labeled antibody concentrations. The relevant EC50 (i.e. the concentration needed to bind half of d2-m425 in A431 cells that highly express EGFR and half Lumi4 Tb- FRP5 in SKBR-3 cells that strongly express HER2), were obtained from a dose-response curve in which the fluorescence emission arising from the bound labeled antibody was plotted against the initial concentration of labeled antibody. Then the TR-FRET experiments were performed using twice the concentrations corresponding to the EC50. Thus, 3.2 × 105 cells were incubated with 16 nm of d2-m425 and 32 nm of Lumi4 Tb-FRP5 in 2 ml tubes at 37 °C overnight. Then, cells were stained with 10 μg/ml Hoechst 33342 (Invitrogen) at room temperature for 10 min, washed three times and each sample was dispensed into 96-well black microtiter plate in triplicate. Hoechst fluorescence (DNA concentration) was measured at 460 nm upon excitation at 335 nm. The TR-FRET signal representing EGFR/HER2 level was expressed as ΔF665 normalized to the DNA concentration. This normalization allowed us to avoid unspecific differences of signal due to variations in cell numbers due to the experimental handling (particularly the washes). For each sample, controls were obtained by performing the same experiments without cells.
Xenografts and Treatment Procedure
All in vivo experiments were performed in compliance with the national regulations and ethical guidelines for the use of laboratory animals in an accredited establishment (Agreement No. C34-172-27). 6-week-old female athymic mice, purchased from Harlan (Le Malcourlet, France), were injected subcutaneously in the right flank with 5 × 106 SKOV-3 cells. Tumor-bearing mice were randomized in different treatment groups when the tumors reached a minimum of 50 mm3. Mice were treated with Pertuzumab (2 or 10 mg/kg), Trastuzumab (10 mg/kg), Lapatinib (100 or 300 mg/kg) or a combination of Trastuzumab + Cetuximab (ratio 1:1; 2 or 10 mg/kg of each mAb) for 4 weeks. Lapatinib was administrated daily with a feeding tube and antibodies were given intraperitonally twice a week. Tumor dimensions and body weight were measured twice weekly and volumes calculated as follow: D1 × D2 × D3/2. Mice were sacrificed when tumors reached a volume larger than 1500 mm3. Kaplan-Meier survival estimates were calculated from the date of the xenograft to the date of the event of interest (i.e. a tumor volume of 1500 mm3) and compared using the Log-rank test.
Data Analysis
FACS data were represented using the WinMDI software (Joseph Trotter). Data from the TR-FRET and EGF binding experiments were represented using the Prism GraphPad software (San Diego, CA).
Statistical Analysis
Statistical analysis was performed using STATA 11.0 (StataCorp. 2009. Stata: Release 11. Statistical Software. College Station, TX: StataCorp LP.) (xenograft experiments) and Prism GraphPad (TR-FRET experiments).
RESULTS
Characterization of the NIH/3T3-HERs Cell Lines
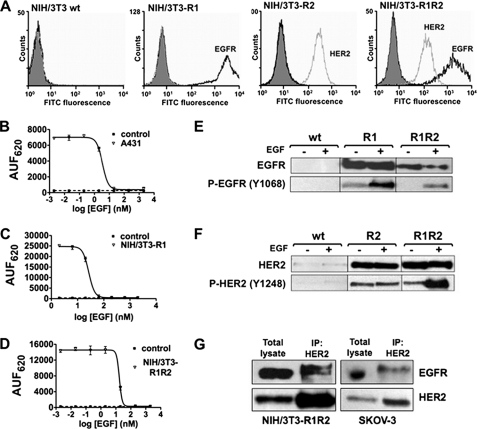
First, the ectopic expression of human EGFR (NIH/3T3-R1 cells) and HER2 (NIH/3T3-R2 cells) or both (NIH/3T3-R1R2 cells) in these cell lines was confirmed by FACS using saturating concentrations of the mAbs m225 (anti-EGFR) and FSP77 (anti-HER2) (Fig. 1A). Although the parental mouse NIH/3T3 cells constitutively express murine EGFR and HER2 at very low level (33), they cannot be detected using antibodies specific for the human receptors. The receptor level was then quantified in the three cell lines using the QIFI assay (Table 1). HER2 expression was similar in NIH/3T3-R2 and -R1R2 cells (about 185 × 103 receptors/cell) and EGFR level in NIH/3T3-R1 and -R1R2 cells was estimated by extrapolation to be about 2,000 × 103 receptors/cell. Binding of EGF to EGFR was verified in A431 cells (positive control) (Fig. 1B) and in the NIH/3T3-R1 and NIH/3T3-R1R2 cell lines (Fig. 1, C and D) using a competition assay between unlabeled and Lumi4 Tb-labeled human EGF. In all cell types, initial binding of labeled EGF was observed, demonstrating that the human EGFR in NIH/3T3-R1 and NIH/3T3-R1R2 cells are still able to normal ligand binding. Moreover, the dose-dependent competition curve obtained by addition of increasing concentration of unlabeled EGF in the three cell lines suggests a specific recognition of EGFR by EGF in these cells. The IC50 were 1.74 nm in A431, 13.28 nm in NIH/3T3-R1, and 7.6 nm in NIH/3T3-R1R2 cells. These values are in agreement with the EGFR levels previously determined in these cell lines (Table 1). However, they also suggest that EGFR level might be higher than the values obtained by extrapolation of the FACS data. The ability of EGFR and HER2 in NIH/3T3-R1, -R2 and -R1R2 cells to be phosphorylated by following EGF binding was assessed by Western blot analysis (Fig. 1, E and F). Basal EGFR phosphorylation was detected in NIH/3T3-R1 cells, but not in NIH/3T3-R1R2 cells. However, after stimulation with EGF, EGFR phosphorylation was increased in both cell lines. HER2 basal phosphorylation level increased upon stimulation with EGF in NIH/3T3-R1R2 cells, but not in NIH/3T3-R2 cells. This result was expected because NIH/3T3-R2 cells do not express EGFR, and EGF cannot stimulate directly HER2, but only in the presence of EGFR (as observed in NIH/3T3-R1R2 cells). Finally, co-immunoprecipitation experiments demonstrated that EGFR and HER2 can also form heterodimers in NIH/3T3-R1R2 cells like in the SKOV-3 cell line, in which both receptors are overexpressed (positive control) (Fig. 1G). In conclusion, the NIH/3T3-HERs cell lines have functional receptors, which can bind to their ligands, can be phosphorylated and form dimers.
FIGURE 1.
Characterization of the NIH/3T3-HERs cell lines. A, flow cytometry analysis of cell surface EGFR (black) and HER2 (gray) expression using the anti-EGFR antibody m225 and the anti-HER2 antibody FSP77 coupled with a FITC-labeled secondary antibody in parental NIH/3T3 (wt), -R1 (EGFR overexpression), -R2 (HER2 overexpression) and -R1R2 (EGFR and HER2) cells. Controls (gray area) were cells incubated only with the FITC-labeled secondary antibody. B–D, EGF binding in A431 cells (positive control) (B), NIH/3T3-R1 (C), and NIH/3T3-R1R2 (D) cells. Cells were incubated with 4 nm EGF labeled with Lumi4 Tb cryptate and increasing concentrations of unlabeled EGF to compete for binding. Unlabeled EGF induced a dose-dependent inhibition of fluorescence emitted at 620 nm. Data are expressed as arbitrary units of fluorescence at 620 nm (AUF620) and represent the mean ± S.E. of triplicates. E and F, Western blot analysis in parental NIH/3T3 (wt), NIH/3T3-R1, NIH/3T3-R2 and NIH/3T3-R1R2 cells. After stimulation with 100 ng/ml EGF, cells were lysed and protein extracts used to determine the expression and the phosphorylation level of EGFR and HER2 with antibodies against total EGFR (E) or HER2 (F) and against phosphorylated EGFR (E) or HER2 (F). G, presence of EGFR/HER2 dimers in NIH/3T3-R1R2 and SKOV-3 cells. Whole cell lysates from EGF-stimulated cells were immunoprecipitated (IP) using the FSP77 anti-HER2 monoclonal antibody and probed with anti-EGFR and -HER2 polyclonal antibodies to detect HER2 and the amount of co-precipitated EGFR (as a dimerization partner).
TABLE 1.
Quantification of EGFR and HER2 levels in various cell lines using the QIFI kit and anti-EGFR (m225) or anti-HER2 (FSP77) antibodies
The calibration curve was drawn using data obtained from control beads. The antigen density was expressed as the ABC in molecules/cell.
| Cell lines | EGFR ABC | HER2 ABC |
|---|---|---|
| 103 molecules/cell | 103 molecules/cell | |
| NIH/3T3-R1 | 1903 ± 266a | Undetectable |
| NIH/3T3-R2 | Undetectable | 187.2 ± 21.1 |
| NIH/3T3-R1R2 | 2188 ± 239a | 183.9 ± 24.3 |
| BT474 | 10.5 ± 2.1 | 478.0 ± 75.8 |
| A431 | 823.9 ± 46.5a | 35.7 ± 3.1 |
| SKOV-3 | 140.7 ± 10.1 | 329.7 ± 36.2 |
| BXPC-3 | 203.3 ± 14.0 | 10.9 ± 1.7 |
| Capan-1 | 26.7 ± 4.6 | 15.6 ± 2.4 |
| MiaPaCa-2 | 60.8 ± 9.9 | 21.5 ± 3.7 |
| SKBR-3 | 41.6 ± 12.1 | 1766 ± 243a |
a Values with an asterisk were extrapolated from the QIFI kit range, between 2,000 and 518,000 molecules/cell.
EGFR/HER2 Heterodimers Can Be Quantified with our Antibody-based TR-FRET Assay
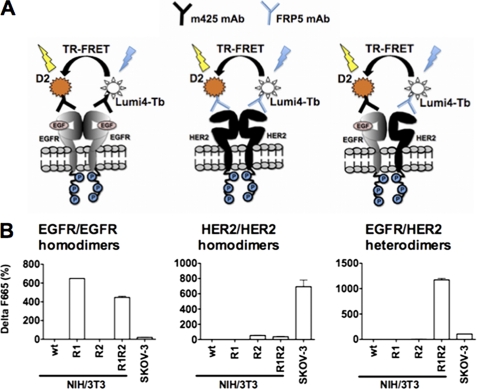
These cell lines were then used to test an antibody-based TR-FRET assay for detecting and quantifying EGFR/HER2 heterodimers using d2-m425 (anti-EGFR antibody labeled with the acceptor fluorophore) and Lumi4 Tb-FRP5 (anti-HER2 antibody labeled with the donor fluorophore). The same principle was used also to detect EGFR/EGFR homodimers with m425 labeled with d2 and Lumi4 Tb and HER2/HER2 homodimers with FRP5 labeled with d2 and Lumi4 Tb (Fig. 2A). In case of dimerization, the proximity between donor and acceptor allows an energy transfer between the two fluorophores. The fluorescence emitted will thus be used to quantify the dimer concentration; however in the case of homodimers this value will represent only 50% of their actual concentration due to the formation also of homodimers that contain two donors or two acceptors and are, therefore, undetectable. To verify the ability of this method to detect specifically EGFR/HER2 heterodimers, parental NIH/3T3 (wt), NIH/3T3-R1, -R2, - R1R2, and SKOV-3 cells were used (Fig. 2B). EGFR/HER2 heterodimers could be detected and quantified only in NIH/3T3-R1R2 cells (ΔF665(%) = 1200) and in SKOV-3 cells (ΔF665(%) = 100). The lower heterodimer concentration in SKOV-3 cells could be due to the much higher quantity of HER2/HER2 homodimers formed in these cells than in NIH/3T3-R1R2 cells (Fig. 2B, middle panel). No background signal was detected with d2-m425 in NIH/3T3-R1 or with Lumi4 Tb-FRP5 alone in NIH/3T3-R2 cells. Dimers were not detected in NIH/3T3 cells as the TR-FRET experiments were performed with human receptors specific antibodies.
FIGURE 2.
Detection of HER homo- and heterodimers in NIH/3T3-R1R2 and SKOV-3 cells. A, schematic of the antibody-based TR-FRET assays to detect EGFR/EGFR or HER2/HER2 homodimers and EGFR/HER2 heterodimers using m425 (anti-EGFR) and/or FRP5 (anti-HER2) mAbs conjugated with terbium cryptate (Lumi4 Tb) or the d2 dye. B, detection of HER dimers in parental NIH/3T3 (wt: no detectable EGFR or HER2 expression), NIH/3T3-R1 (only EGFR), -R2 (only HER2) and -R1R2 and SKOV-3 cells (both EGFR and HER2) using the antibody-based TR-FRET assay. Cells were incubated with labeled antibodies overnight and the fluorescence signal was directly measured, without washes. Data are representative of three independent experiments.
Correlation between EGFR and/or HER2 Expression Levels and EGFR/HER2 Heterodimer Concentration
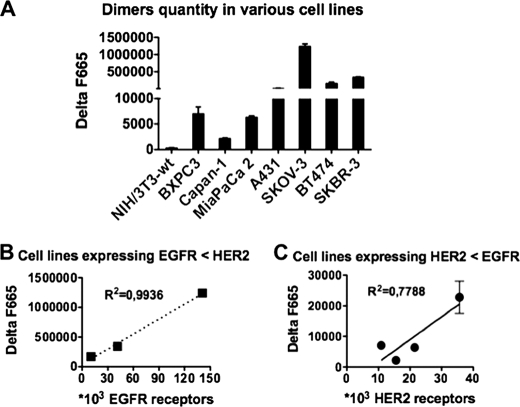
Because the different cancer cell lines used for this work (BT474, A431, SKOV-3, BXPC-3, Capan I, MiaPaCa-2, and SKBR-3) presented very variable EGFR and HER2 expression levels (as determined by QIFI assay and Table 1), EGFR/HER2 dimers in these cell lines were quantified using our antibody-based TR-FRET assay in the presence of excess of labeled mAbs and fluorescence was measured only after several extensive washes to eliminate unbound antibodies. EGFR/HER2 dimers were detected in all cell lines (Fig. 3A), but their concentration was lower in cell lines with lower EGFR or HER2 expression than in cells that express higher levels of one of the two receptors. To determine whether the concentration of EGFR/HER2 dimers was correlated with EGFR or HER2 expression level, correlation curves were drawn. In SKOV-3, BT474, and SKBR-3 cells (higher HER2 expression) the quantity of EGFR/HER2 dimers was significantly correlated with EGFR expression level (Fig. 3B). This kind of correlation was still observed, but was less direct, in cell lines that express more EGFR than HER2, such Capan-I, BXPC-3, MiaPaCa-2, and A431 (Fig. 3C).
FIGURE 3.
Quantification of EGFR/HER2 dimers in different cancer cell lines using the antibody-based TR-FRET assay. A, 3.2 × 105 cells were incubated at 37 °C with d2-labeled m425 (anti-EGFR) Lumi4 Tb-labeled FRP5 (anti-HER2) antibodies overnight. Cells were then stained with 10 μg/ml of Hoechst 33342 and washed three times with KREBS buffer before detection of the fluorescence signals (see “Experimental Procedures” for a complete description of the protocol used). The ΔF665 value represents the EGFR/HER2 dimer concentration normalized to the DNA quantity (Hoechst fluorescence). Data are the mean ± S.E. of three independent experiments performed in triplicate. B, correlation curve between EGFR/HER2 dimer quantity and EGFR expression (determined previously using the quantitative immunofluorescence indirect assay, see Table 1) in cancer cell lines with higher HER2 than EGFR expression level. C, correlation curve between EGFR/HER2 dimer quantity and HER2 expression in cancer cell lines with higher EGFR than HER2 expression level.
Therapeutic anti-EGFR and -HER2 mAbs, but Not TKIs, Can Modulate the Level of EGFR/HER2 Dimers in SKOV-3 Cells
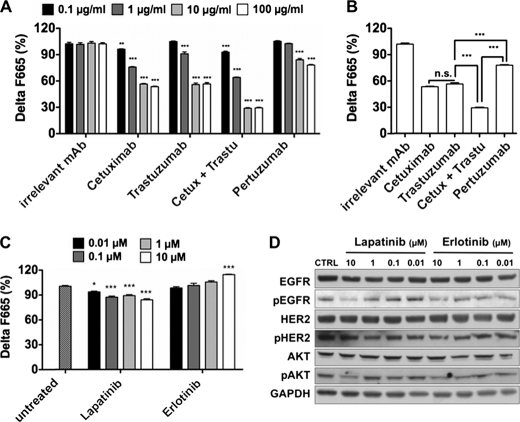
We previously demonstrated a synergistic effect of combined Cetuximab/Trastuzumab treatment in human pancreatic carcinoma xenografts (34, 35). Although this effect might partially be due to ADCC induction by both mAbs and to the ability of Cetuximab to directly block EGF binding site, dimer disruption could also play a role in this synergistic effect. To assess this hypothesis, we first compared the ability of Cetuximab (anti-EGFR mAb), Trastuzumab (anti-HER2), Pertuzumab (anti-HER2), and combined Cetuximab + Trastuzumab to affect EGFR/HER2 dimer level in SKOV-3 cells (Fig. 3A). The irrelevant mAb Px was used as a negative control (its inability to modulate the concentration of EGFR/HER2 dimers in SKOV-3 cells was verified beforehand). SKOV-3 cells were kept in complete culture medium (i.e. neither starvation nor EGF stimulation) to be close to in vivo conditions and not to alter normal ligand level. After a 30-min treatment, cells were fixed in 10% formalin to immobilize the dimers and to avoid any modification of their status during the incubation with d2-m425 and Lumi4 Tb-FRP5. A 30-min treatment (and already after 10 min, data not shown) with the different therapeutic mAbs was sufficient to significantly reduce the concentration of EGFR/HER2 dimers (Fig. 4A). However, Pertuzumab was less efficient (24% reduction) than Trastuzumab, (44% reduction) or Cetuximab (48% reduction). The Cetuximab + Trastuzumab combination was the most effective with a 72% decrease in dimer concentration (Fig. 4B). Compared with the mAbs, treatment with increasing concentrations of the TKIs Lapatinib (EGFR and HER2 specific) or Erlotinib (EGFR specific) for 30 min had a very limited effect on the concentration of EGFR/HER2 heterodimers (less than 10% reduction) (Fig. 4C). This effect was weakly dose-dependent with Lapatinib. Conversely, in the case of Erlotinib, the EGFR/HER2 dimer quantity was slightly increased at higher doses. To verify that this minimal TKI effect was not due to a too short incubation period, Western blot analysis was performed (Fig. 4D). The dose-dependent decrease in EGFR and AKT phosphorylation upon incubation with Lapatinib and Erlotinib suggests that 30 min were enough to induce the effect of TKIs in SKOV-3 cells in the absence of EGF induction. To rule out an unspecific decrease of the TR-FRET signal due to modulation of EGFR or HER2 expression at the cell surface, we verified the stability of their expression by measuring the prompt fluorescence from bound d2-m425 (excitation at 620 nm, emission at 670 nm) and bound Lumi4 Tb-FRP5 by TRF (excitation at 337 nm, emission at 620 nm, 60 μs delay) in each experiment (supplemental Fig. S2).
FIGURE 4.
Effect of therapeutic anti-EGFR or -HER2 mAbs and TKIs on EGFR/HER2 heterodimer concentration in SKOV-3 cells. 105 cells/well were treated with increasing concentrations (from 0.1 to 100 μg/ml) of Px (irrelevant antibody), Cetuximab, Trastuzumab, Cetuximab + Trastuzumab (ratio 1:1), and Pertuzumab (A and B); or with increasing concentrations (from 0.01 to 10 μm) of the TKIs Lapatinib and Erlotinib (C). Cells were then fixed with formalin for 2 min, and EGFR/HER2 heterodimers were quantified using the antibody-based TR-FRET assay with anti-EGFR (d2-labeled m425) and anti-HER2 (Lumi4 Tb-labeled FRP5) antibodies. After extensive washes, fluorescence was measured and the TR-FRET signal was expressed as ΔF665 (%) = ΔF665/F665Terbium (see “Experimental Procedures”). ΔF665 (%) represents the concentration of EGFR/HER2 dimers normalized to HER2 expression. Data are the mean ± S.E. of three independent experiments performed in triplicate. p, ***<0.001; **<0.01, n.s. non-significant, by one- and two-way analysis of variance (B and A, respectively), with Bonferroni's multiple comparison post-tests. D, Western blot analysis of SKOV-3 cells treated with increasing concentrations of TKIs without prior induction by EGF. Antibodies against total EGFR and HER2, phosphorylated EGFR and HER2, AKT and phosphorylated AKT and GAPDH were used to detect variations in EGFR and HER2 expression/phosphorylation and AKT phosphorylation.
The Effect of Therapeutic mAbs on the Survival of Mice Xenografted with SKOV-3 Cells Correlates with Their Ability to Disturb EGFR/HER2 Dimers
Then to evaluate whether the therapeutic benefit of targeted anti-EGFR and/or HER2 therapies was correlated with their efficacy in reducing EGFR/HER2 dimers, mice xenografted with SKOV-3 cells were treated with mAbs (Trastuzumab, Pertuzumab, or Cetuximab + Trastuzumab) or the TKI Lapatinib for 4 weeks. The dosage (2 and 10 mg/kg per injection for mAbs and 100 and 300 mg/kg for the TKI) were chosen on the basis of previous experiments (34). Significant increase in the median survival was observed after treatment with Pertuzumab (p = 0.006) or Trastuzumab (p < 0.001) at 10 mg/kg in comparison to the dose at 2 mg/kg that did not have any effect (Fig. 5A). Moreover, Trastuzumab was significantly more effective than Pertuzumab (p = 0.0123) and the best inhibition of tumor growth (Fig. 5B) was obtained by combining Cetuximab and Trastuzumab at 10 mg/kg. Specifically, at 97 days post-graft, 80% of the mice treated with Cetuximab + Trastuzumab were tumor-free, a percentage much higher than with any other treatments (Fig. 5A). These results suggest that the mAb therapeutic effect (median survival and percentage of tumor-free animals) in SKOV-3 xenografted mice might be correlated with their efficiency in decreasing the concentration of EGFR/HER2 heterodimers (Fig. 4, A and C).
FIGURE 5.
Effect of therapeutic anti-EGFR and HER2 mAbs and of the TKI Lapatinib on the median survival and tumor progression in mice xenografted with SKOV-3 cells. A, median survival and tumor-free percentage of mice xenografted with 5 × 106 SKOV-3 cells and treated, when the tumor reached a minimum volume of 50 mm3, with Pertuzumab (2 or 10 mg/kg), Trastuzumab (2 or 10 mg/kg), Cetuximab + Trastuzumab (2 or 10 mg/kg of each mAb) or Lapatinib (100 or 300 mg/kg) for 4 weeks. Tumor dimensions were measured twice weekly. B, tumor progression curves in the mice treated with the highest doses of mAbs (10 mg/kg) and Lapatinib (300 mg/kg). Tumor dimensions were measured twice weekly.
Finally, mice treated with 100 mg/kg Lapatinib did not show any difference in comparison to controls. Conversely, the higher dose of Lapatinib induced an increase of median survival comparable to Pertuzumab (p = 0.689) (Fig. 5A), and Lapatinib was more efficient than Pertuzumab to slow down tumor progression (p < 0.001) as shown by the curve describing tumor volume changes at different days post-graft for the higher dose treatments (Fig. 5B).
DISCUSSION
In this study we describe a novel antibody-based TR-FRET method to detect and quantify specifically EGFR/HER2 dimers, but also EGFR/EGFR and HER2/HER2 homodimers, in intact cells (Fig. 2). In contrast to other methods, such as co-immunoprecipitation or cross-linking, our TR-FRET assay detects the dimerization process of native receptors in their biological context. Hence, this new quantitative method is more sensitive and easier to perform than those previously proposed. For instance, the proximity ligation assay allows detecting the proximity between two molecules separated by less than 40 nm. Conversely, our method allows the detection of the proximity between Lumi4 Tb and d2 molecules at less than 12 nm of distance (note that the antibody size is ∼7 nm). This difference is very important, because it would be very difficult to discriminate between a heterodimer and two co-localized receptors that do not interact by using a technology that detects proximity of 40 nm. Another quantitative method for the detection of EGFR/HER2 heterodimers is the VeraTag assay, which appears to be efficient (27). However, this assay is not currently available in any academic laboratory.
We tested our antibody-based TR-FRET method in NIH/3T3 cell lines that overexpress EGFR, HER2, or both receptors and therefore can form both homo- and heterodimers, and then validated our results in a panel of cancer cell lines that are characterized by different expression levels of EGFR and HER2. We demonstrate a good correlation between EGFR level and EGFR/HER2 dimer concentration in HER2high-expressing carcinoma cell lines (Fig. 3). Indeed, these cell lines present a high number of preformed HER2/HER2 homodimers that could be destabilized in favor of formation of heterodimers when other HER partners are available at the membrane (for instance, EGRF in our case) and upon ligand-dependent stimulation. Conversely, in carcinoma cells with more EGFR than HER2 receptors, the reciprocal correlation is less obvious. This could be due to the fact that HER2 might trigger the formation of other dimers (for instance, HER2/HER2 or HER2/HER3 in HER3+ cell lines, such as BxPC3 and A431), which affects HER2 availability for EGFR/HER2 heterodimerization.
Using our antibody-based TR-FRET assay, we then demonstrate that Trastuzumab or Cetuximab alone reduced of approximately 40% the level of EGFR/HER2 heterodimers in SKOV-3 cells, whereas Pertuzumab was less effective (24%) (Fig. 4). Pertuzumab, which targets domain II of HER2 (37), was previously shown to efficiently block ligand-mediated HER2/HER3 dimerization and this effects was correlated with the number of HER2 receptors (38). In addition, Pertuzumab can also perturb EGFR/HER2 heterodimer formation and the related signaling pathways, as demonstrated using transfected porcine aortic endothelial cells (39) and ovarian carcinoma cell lines (40). However, its murine version (2C4) (36) can also stabilize EGFR/HER2 dimers in HER2high-expressing SKBR-3 cell line and partially reduce HER2/HER3 dimerization, thus suggesting that Pertuzumab is not solely a HER dimerization blocker (41).
Trastuzumab, which targets the extracellular domain IV of HER2, has also been described to partially block HER2/HER3 dimerization in the HER2low-expressing MCF7 cell line (38, 42). Disruption of EGFR/HER2 and HER2/HER3 heterodimers by Trastuzumab has been previously described under ligand stimulation in the HER2low-expressing breast cancer MCF7, but not in the HER2high-expressing breast cancer SKBR3 cell line (43, 38). In SKBR3 cells, Trastuzumab-induced reduction of HER2/HER3 dimers is not ligand-dependent (44), as we confirm with our antibody-based TR-FRET assays that were performed with cells in complete culture medium (with growth factors provided exclusively by fetal calf serum) and without addition of exogenous ligands. We therefore suggest that the ability of Trastuzumab to reduce heterodimer concentration does not depend exclusively on HER2 expression level.
Cetuximab, which binds to EGFR extracellular domain preventing activation and dimerization, was previously reported to perturb EGFR/HER2 heterodimerization and to stabilize HER2/HER2 homodimers in gastric carcinoma and non-small cell lung cancer cell lines (45).
On the other hand, concomitant targeting of EGFR/HER2 with Trastuzumab + Cetuximab was much more effective, but nevertheless incomplete (28% of heterodimers could still be detected) in SKOV-3 cells. This remaining percentage of heterodimers could be due to residual native receptor synthesis or the presence of various conformers (quasi-dimers, tetramers, or oligomers) (46–48), which cannot be affected by antibody treatment. Moreover, the fluorescence transfer in our TR-FRET assays is abrogated only when donor and acceptor are separated by more than 12 nm. Therefore, although not associated in a dimer, EGFR and HER2 molecules could still be sufficiently in close proximity to induce a slight energy transfer between donor and acceptor.
Finally, the three mAb treatments showed a good correlation between their in vitro ability to perturb EGFR/HER2 heterodimers and their in vivo ability to reduce cell tumor growth. Specifically, Pertuzumab, which showed lower efficacy in disrupting EGFR/HER2 dimerization, was less effective than Trastuzumab in preventing tumor growth in nude mice xenografted with SKOV-3 cells. Pertuzumab was efficient in the estradiol (E2)-responsive ovarian carcinoma subgroup (49), but showed lower clinical effects in other ovarian cancers (50), particularly the E2-independent ones, such as the tumor from which the SKOV-3 cell line is derived.
In contrast, the link between in vitro dimer reduction and in vivo efficacy was not observed with TKIs, which showed a very limited effect on the dimer concentration, as previously shown for Erlotinib in pancreatic cancer cell lines (51) and for Lapatinib in breast cancer cells (43). However in other publications TKIs were reported to affect HER dimerization (52, 53). These contradictory results should be reviewed by carefully comparing the experimental procedures (for instance, culture conditions, cell lines, density of HER receptor, ligand stimulation).
In conclusion, our study demonstrates the utility of our antibody-based TR-FRET assay to analyze the effect of therapeutic antibodies on EGFR/HER2 heterodimers. This method, in addition to other commonly used investigations, such as the analysis of ADCC, phosphorylation, and proliferation, will allow us to better predict and understand the in vivo effects of targeted therapies on tumor growth.
Supplementary Material
Acknowledgments
We thank the members of our groups and J. P. Mach for insightful comments and corrections; G. Heintz and S. Bousquié for technical assistance; C. Duperray for performing the selective cell sorting; M. Brissac and I. Ait-Arsa for help with the animal experiments; L. Le Cam for kindly providing retroviral vectors, and Roche for providing pertuzumab.
This project was supported by Agence Nationale de la Recherche Grant ANR-07-RIB-003/Dim-HER.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S2.
- EGFR
- epidermal growth factor receptor
- HER
- human epidermal growth factor receptor
- TR-FRET
- time-resolved fluorescence resonance energy transfer
- RTK
- tyrosine kinase receptor
- EGF
- epidermal growth factor
- mAb
- monoclonal antibody
- TKI
- tyrosine kinase inhibitors
- Ig
- immunoglogulin
- MSCV
- murine stem cell virus
- FACS
- fluorescence-activated cell sorting
- SM
- starved medium
- ADCC
- antibody-dependent cellular cytotoxicity
- s.c.
- subcutaneously
- i.p.
- intraperitonally
- ABC
- antibody binding capacity
- QIFI
- quantitative immunofluorescence indirect assay.
REFERENCES
- 1. Lax I., Burgess W. H., Bellot F., Ullrich A., Schlessinger J., Givol D. (1988) Mol. Cell Biol. 8, 1831–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yarden Y., Sliwkowski M. X. (2001) Nat. Rev. Mol. Cell Biol. 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 3. Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Zhu H. J., Walker F., Frenkel M. J., Hoyne P. A., Jorissen R. N., Nice E. C., Burgess A. W., Ward C. W. (2002) Cell 110, 763–773 [DOI] [PubMed] [Google Scholar]
- 4. Hynes N. E., MacDonald G. (2009) Curr. Opin. Cell Biol. 21, 177–184 [DOI] [PubMed] [Google Scholar]
- 5. Burgess A. W. (2008) Growth Factors 26, 263–274 [DOI] [PubMed] [Google Scholar]
- 6. Barros F. F., Powe D. G., Ellis I. O., Green A. R. (2010) Histopathology 56, 560–572 [DOI] [PubMed] [Google Scholar]
- 7. Normanno N., Maiello M. R., De Luca A. (2003) J. Cell Physiol. 194, 13–19 [DOI] [PubMed] [Google Scholar]
- 8. Lafky J. M., Wilken J. A., Baron A. T., Maihle N. J. (2008) Biochim. Biophys. Acta 1785, 232–265 [DOI] [PubMed] [Google Scholar]
- 9. Suo Z., Risberg B., Karlsson M. G., Willman K., Tierens A., Skovlund E., Nesland J. M. (2002) J. Pathol. 196, 17–25 [DOI] [PubMed] [Google Scholar]
- 10. Wang Z., Zhang L., Yeung T. K., Chen X. (1999) Mol. Biol. Cell 10, 1621–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilkinson J. C., Staros J. V. (2002) Biochemistry 41, 8–14 [DOI] [PubMed] [Google Scholar]
- 12. Hendriks B. S., Opresko L. K., Wiley H. S., Lauffenburger D. (2003) Cancer Res. 63, 1130–1137 [PubMed] [Google Scholar]
- 13. Brandt B. H., Roetger A., Dittmar T., Nikolai G., Seeling M., Merschjann A., Nofer J. R., Dehmer-Möller G., Junker R., Assmann G., Zaenker K. S. (1999) FASEB J. 13, 1939–1949 [DOI] [PubMed] [Google Scholar]
- 14. Chen X., Yeung T. K., Wang Z. (2000) Biochem. Biophys. Res. Commun. 277, 757–763 [DOI] [PubMed] [Google Scholar]
- 15. Schmitz K. R., Ferguson K. M. (2009) Exp. Cell Res. 315, 659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lurje G., Lenz H. J. (2009) Oncology 77, 400–410 [DOI] [PubMed] [Google Scholar]
- 17. Hynes N. E., Lane H. A. (2005) Nat. Rev. Cancer 5, 341–354 [DOI] [PubMed] [Google Scholar]
- 18. Karaman M. W., Herrgard S., Treiber D. K., Gallant P., Atteridge C. E., Campbell B. T., Chan K. W., Ciceri P., Davis M. I., Edeen P. T., Faraoni R., Floyd M., Hunt J. P., Lockhart D. J., Milanov Z. V., Morrison M. J., Pallares G., Patel H. K., Pritchard S., Wodicka L. M., Zarrinkar P. P. (2008) Nat. Biotechnol. 26, 127–132 [DOI] [PubMed] [Google Scholar]
- 19. Harris M. (2004) Lancet Oncol. 5, 292–302 [DOI] [PubMed] [Google Scholar]
- 20. Scartozzi M., Bearzi I., Berardi R., Mandolesi A., Fabris G., Cascinu S. (2004) J. Clin. Oncol. 22, 4772–4778 [DOI] [PubMed] [Google Scholar]
- 21. Chung K. Y., Shia J., Kemeny N. E., Shah M., Schwartz G. K., Tse A., Hamilton A., Pan D., Schrag D., Schwartz L., Klimstra D. S., Fridman D., Kelsen D. P., Saltz L. B. (2005) J. Clin. Oncol. 23, 1803–1810 [DOI] [PubMed] [Google Scholar]
- 22. Goldstein N. S., Armin M. (2001) Cancer 92, 1331–1346 [DOI] [PubMed] [Google Scholar]
- 23. Cappuzzo F., Gregorc V., Rossi E., Cancellieri A., Magrini E., Paties C. T., Ceresoli G., Lombardo L., Bartolini S., Calandri C., de Rosa M., Villa E., Crino L. (2003) J. Clin. Oncol. 21, 2658–2663 [DOI] [PubMed] [Google Scholar]
- 24. De Pas T., Pelosi G., de Braud F., Veronesi G., Curigliano G., Leon M. E., Danesi R., Noberasco C., d'Aiuto M., Catalano G., Viale G., Spaggiari L. (2004) J. Clin. Oncol. 22, 4966–4970 [DOI] [PubMed] [Google Scholar]
- 25. Khan I. H., Zhao J., Ghosh P., Ziman M., Sweeney C., Kung H. J., Luciw P. A. (2010) Assay Drug. Dev. Technol. 8, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi Y., Huang W., Tan Y., Jin X., Dua R., Penuel E., Mukherjee A., Sperinde J., Pannu H., Chenna A., DeFazio-Eli L., Pidaparthi S., Badal Y., Wallweber G., Chen L., Williams S., Tahir H., Larson J., Goodman L., Whitcomb J., Petropoulos C., Winslow J. (2009) Diagn. Mol. Pathol. 18, 11–21 [DOI] [PubMed] [Google Scholar]
- 27. Jain A., Penuel E., Mink S., Schmidt J., Hodge A., Favero K., Tindell C., Agus D. B. (2010) Cancer Res. 70, 1989–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gajadhar A., Guha A. (2010) BioTechniques 48, 145–152 [DOI] [PubMed] [Google Scholar]
- 29. Diermeier-Daucher S., Hasmann M., Brockhoff G. (2008) Ann. N.Y. Acad. Sci. 1130, 280–286 [DOI] [PubMed] [Google Scholar]
- 30. Maurel D., Kniazeff J., Mathis G., Trinquet E., Pin J. P., Ansanay H. (2004) Anal. Biochem. 329, 253–262 [DOI] [PubMed] [Google Scholar]
- 31. Harwerth I. M., Wels W., Marte B. M., Hynes N. E. (1992) J. Biol. Chem. 267, 15160–15167 [PubMed] [Google Scholar]
- 32. Köhler G., Milstein C. (1975) Nature 256, 495–497 [DOI] [PubMed] [Google Scholar]
- 33. Zhang K., Sun J., Liu N., Wen D., Chang D., Thomason A., Yoshinaga S. K. (1996) J. Biol. Chem. 271, 3884–3890 [PubMed] [Google Scholar]
- 34. Larbouret C., Robert B., Navarro-Teulon I., Thèzenas S., Ladjemi M. Z., Morisseau S., Campigna E., Bibeau F., Mach J. P., Pèlegrin A., Azria D. (2007) Clin. Cancer Res. 13, 3356–3362 [DOI] [PubMed] [Google Scholar]
- 35. Larbouret C., Robert B., Bascoul-Mollevi C., Penault-Llorca F., Ho-Pun-Cheung A., Morisseau S., Navarro-Teulon I., Mach J. P., Pèlegrin A., Azria D. (2010) Ann. Oncol. 21, 98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adams C. W., Allison D. E., Flagella K., Presta L., Clarke J., Dybdal N., McKeever K., Sliwkowski M. X. (2006) Cancer Immunol. Immunother. 55, 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Franklin M. C., Carey K. D., Vajdos F. F., Leahy D. J., de Vos A. M., Sliwkowski M. X. (2004) Cancer Cell 5, 317–328 [DOI] [PubMed] [Google Scholar]
- 38. Agus D. B., Akita R. W., Fox W. D., Lewis G. D., Higgins B., Pisacane P. I., Lofgren J. A., Tindell C., Evans D. P., Maiese K., Scher H. I., Sliwkowski M. X. (2002) Cancer Cell 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 39. Hughes J. B., Berger C., Rødland M. S., Hasmann M., Stang E., Madshus I. H. (2009) Mol. Cancer Ther. 8, 1885–1892 [DOI] [PubMed] [Google Scholar]
- 40. Takai N., Jain A., Kawamata N., Popoviciu L. M., Said J. W., Whittaker S., Miyakawa I., Agus D. B., Koeffler H. P. (2005) Cancer 104, 2701–2708 [DOI] [PubMed] [Google Scholar]
- 41. Cai Z., Zhang G., Zhou Z., Bembas K., Drebin J. A., Greene M. I., Zhang H. (2008) Oncogene 27, 3870–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nahta R., Hung M. C., Esteva F. J. (2004) Cancer Res. 64, 2343–2346 [DOI] [PubMed] [Google Scholar]
- 43. Scaltriti M., Verma C., Guzman M., Jimenez J., Parra J. L., Pedersen K., Smith D. J., Landolfi S., Ramon y Cajal S., Arribas J., Baselga J. (2009) Oncogene 28, 803–814 [DOI] [PubMed] [Google Scholar]
- 44. Junttila T. T., Akita R. W., Parsons K., Fields C., Lewis Phillips G. D., Friedman L. S., Sampath D., Sliwkowski M. X. (2009) Cancer Cell 15, 429–440 [DOI] [PubMed] [Google Scholar]
- 45. Patel D., Bassi R., Hooper A., Prewett M., Hicklin D. J., Kang X. (2009) Int. J. Oncol. 34, 25–32 [PubMed] [Google Scholar]
- 46. Clayton A. H., Walker F., Orchard S. G., Henderson C., Fuchs D., Rothacker J., Nice E. C., Burgess A. W. (2005) J. Biol. Chem. 280, 30392–30399 [DOI] [PubMed] [Google Scholar]
- 47. Furuuchi K., Berezov A., Kumagai T., Greene M. I. (2007) J. Immunol. 178, 1021–1029 [DOI] [PubMed] [Google Scholar]
- 48. Bublil E. M., Pines G., Patel G., Fruhwirth G., Ng T., Yarden Y. (2010) FASEB J. 24, 4744–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mullen P., Cameron D. A., Hasmann M., Smyth J. F., Langdon S. P. (2007) Mol. Cancer Ther. 6, 93–100 [DOI] [PubMed] [Google Scholar]
- 50. Kristjansdottir K., Dizon D. (2010) Expert Opin. Biol. Ther. 10, 243–250 [DOI] [PubMed] [Google Scholar]
- 51. Frolov A., Schuller K., Tzeng C. W., Cannon E. E., Ku B. C., Howard J. H., Vickers S. M., Heslin M. J., Buchsbaum D. J., Arnoletti J. P. (2007) Cancer Biol. Ther. 6, 548–554 [DOI] [PubMed] [Google Scholar]
- 52. Arteaga C. L., Ramsey T. T., Shawver L. K., Guyer C. A. (1997) J. Biol. Chem. 272, 23247–23254 [DOI] [PubMed] [Google Scholar]
- 53. Gan H. K., Walker F., Burgess A. W., Rigopoulos A., Scott A. M., Johns T. G. (2007) J. Biol. Chem. 282, 2840–2850 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.