Abstract
Galectin-8 has much higher affinity for 3′-O-sulfated or 3′-O-sialylated glycoconjugates and a Lewis X-containing glycan than for oligosaccharides terminating in Galβ1→3/4GlcNAc, and this specificity is mainly attributed to the N-terminal carbohydrate recognition domain (N-domain, CRD) (Ideo, H., Seko, A., Ishizuka, I., and Yamashita, K. (2003) Glycobiology 13, 713–723). In this study, we elucidated the crystal structures of the human galectin-8-N-domain (-8N) in the absence or presence of 4 ligands. The apo molecule forms a dimer, which is different from the canonical 2-fold symmetric dimer observed for galectin-1 and -2. In a galectin-8N-lactose complex, the lactose-recognizing amino acids are highly conserved among the galectins. However, Arg45, Gln47, Arg59, and the long loop region between the S3 and S4 β-strands are unique to galectin-8N. These amino acids directly or indirectly interact with the sulfate or sialic acid moieties of 3′-sialyl- and 3′-sulfolactose complexed with galectin-8N. Furthermore, in the LNF-III-galectin-8N complex, van der Waals interactions occur between the α1–3-branched fucose and galactose and between galactose and Tyr141, and these interactions increase the affinity toward galectin-8N. Based on the findings of these x-ray crystallographic analyses, a mutagenesis study using surface plasmon resonance showed that Arg45, Gln47, and Arg59 of galectin-8N are indispensable and coordinately contribute to the strong binding of galectins-8N to sialylated and sulfated oligosaccharides. Arg59 is the most critical amino acid for binding in the S3–S4 loop region.
Keywords: Carbohydrate-binding Protein, Carbohydrate Complex, Lectin, Surface Plasmon Resonance (SPR), X-ray Crystallography, Galectin, Mutagenesis, Sialylated Glycan, Sulfated Glycan
Introduction
Galectin-8 is a member of the galectin family, which share similar carbohydrate recognition domains (CRDs)5 and affinity for β-galactosides (1, 2). Twelve galectins have been identified and characterized thus far. Each galectin has one (galectin-1, -2, -3, -5, -7, and -10) or two (galectin-4, -6, -8, -9, -11, and -12) conserved CRDs within a single molecule. According to the characteristics of their evolutionary conservation, wide tissue distribution, marked development-dependent expression, and abundance in specific tissues, galectins are presumed to function in various biological processes (3, 4).
Galectin-8 is widely expressed and made up of two CRDs joined by a short linking peptide (1, 2). Recent studies have shown that galectin-8 modulates cell adhesion, spread, growth, and apoptosis (5–8). However, the precise recognition mechanisms and physiological roles of galectin-8 have not been fully elucidated.
We determined the carbohydrate binding specificity of galectin-8 and found that it has much higher affinity for 3′-O-sulfated or 3′-O-sialylated lactose and Lewis X-containing glycans than for oligosaccharides terminating in Galβ1→3/4GlcNAc; this carbohydrate binding specificity was mainly attributed to the N-terminal CRD (N-domain) (9). The high affinity of galectin-8 for 3′-O-sialylated galactose is unique to this member of the galectin family (10); it is therefore of interest in determining its biological function via carbohydrate recognition. Because galectin-8 binds to SM3 and GM3, we hypothesize that part of galectin-8 localizes into rafts by binding to specific glycosphingolipids, thereby modulating their biological function.
X-ray crystal structures of human galectin-2, -3, -7, -9, and -10 (11–14) showed that these galectins have very similar tertiary structures. Most of these x-ray crystal structures are in complex with lactose/N-acetyllactosamine, and showed that amino acids critical for lactose binding (15) in the S4, S5, and S6 β-sheets are well conserved among galectins-1–9. Understanding the precise binding mechanisms of galectin-8 with sialylated and sulfated carbohydrates is indispensable to understanding the in vivo biological function of galectin-8. To investigate which amino acid(s) of galectin-8 interact with the Neu5Acα2→3Gal or SO3−→3Gal residues, we created a structural model of the galectin-8-N-domain (galectin-8N) using galectin-3-CRD as a template. We reported that Gln47 on the S3-β-sheet of galectin-8N is critical to its unique binding specificity (9). Because Arg45 on the S3-β-sheet of the galectin-4 N-domain is also attributed to binding specificity for sulfated glycans (16), we speculated that the amino acids on the S3 β-sheet might regulate the distinct carbohydrate binding specificity of each galectin family member. However, the mutagenesis study revealed that Gln47 of galectin-8N is necessary but not sufficient for binding enhancement of sialylated or sulfated oligosaccharides. To elucidate the precise mechanism for binding to sialylated/sulfated glycans, we performed x-ray crystallographic analysis by co-crystallization of galectin-8N with high affinity oligosaccharides.
We now present the crystal structures of galectin-8N in complexes with Galβ1→4Glc(Lac), Neu5Acα2→3Galβ1→ 4Glc (3′-SL), SO3−→3Galβ1→4Glc (3′-sulfoL), or Galβ1→ 4(Fucα1→3)GlcNAcβ1→3Galβ1→4Glc (LNF-III). Computer modeling did not anticipate the interactions of Arg59 with the sialic acid or sulfate moiety observed in x-ray crystallographic analyses. This structure reveals for the first time how galectin-8 can form a high affinity complex with sialylated/sulfated oligosaccharides, which gives them distinct and diverse carbohydrate binding specificities.
EXPERIMENTAL PROCEDURES
Materials
Carbohydrate chemicals Neu5Acα2→3Galβ1→ 4Glc (3′-SL), Galβ1→3GlcNAc (type 1), Galβ1→4GlcNAc (type 2), Galβ1→3GlcNAcβ1→3Galβ1→4Glc (LNT), Galβ1→4GlcNAcβ1→3Galβ1→4Glc (LNnT), Galβ1→ 4(Fucα1→3)GlcNAcβ1→3Galβ1→4Glc (LNF-III), Galβ1→ 3GalNAc (core 1), and Galβ1→3(GlcNAcβ1→6)GalNAcα1-O-pNP (core 2-O-pNP) were purchased from Funakoshi Co., Ltd. (Tokyo, Japan). Lactose-O-pNP was purchased from Seikagaku Co. (Tokyo, Japan). Synthesized 3′-sulfoL was purchased from Toronto Research Chemicals Inc. (North York, Canada).
Glutathione-Sepharose, MonoS HR 5/5, Superdex 200 columns, and PreScission protease were obtained from GE Healthcare (United Kingdom). Arthrobacter sialidase was from Nakarai Tesque Inc. (Kyoto, Japan). Lactose (Galβ1→4Glc) and all crystallization reagents were purchased from Hampton Research (CA) and Molecular Dimensions (Suffolk, UK). Other chemicals were obtained from Wako Pure Chemical Industries Ltd. (Japan) and Sigma.
Synthesis of Galβ1→3(Neu5Acα2→3Galβ1→4GlcNAcβ1→ 6)GalNAcα1-pNP (Siaα2→3Gal-core 2) and 3′-sulfo-lacto-N-tetraose (SO3−→3LNT)
Galβ1→3(Galβ1→4GlcNAcβ1→ 6)GalNAcα1-pNP (Gal-core 2) was prepared as described previously (17). Siaα2→3Gal-core 2 was synthesized as follows. The reaction mixture (0.7 ml), containing 50 mm sodium cacodylate (pH 6.0), 10 mm MnCl2, 2 mm Gal-core 2, and 30 milliunits of α2,3-(N)-sialyltransferase (Calbiochem), was incubated at 37 °C for 16 h. After heating at 100 °C to stop the reaction, the mixture was applied to a Sephadex G-25 gel filtration column (1.4 × 68 cm; eluted and equilibrated with EtOH/water, 5:95, v/v). The desalted oligosaccharides were applied to a Sephadex A-25 anion exchange column (0.9 × 6.3 cm; equilibrated with 3 mm Tris-HCl, pH 8.0) and eluted with a linear gradient of NaCl (0–0.1 m). The oligosaccharide-containing fractions were collected and desalted by Sephadex G-25 gel filtration. Finally, 0.64 μmol of Siaα2→3Gal-core 2 was obtained.
Sulfation of LNT was performed using recombinant Gal3ST-2. The cDNA encoding the catalytic region of human Gal3ST-2 was amplified by PCR using an expression vector as the template, as previously reported (18, 19). The oligonucleotide primers were 5′-tttgaattcGGGGGCCAGGCTGAGGG-3′ (forward primer) and 5′-tttgcggccgcAGGAGGCCTCGTC-3′ (reverse primer). The sequences in lowercase letters indicate engineered restriction sites. The amplified cDNAs were digested with EcoRI and NotI and cloned into pPIC9 (Invitrogen). The resulting plasmids were sequenced using an Applied Biosystems Prism 310 Genetic Analyzer. The pPIC9 plasmids were introduced into Pichia pastoris KM71 cells. The recombinant proteins were secreted into the culture medium and purified by nickel-nitrilotriacetic acid-agarose chromatography as described previously (17). The total activity of Gal3ST-2 from a 400-ml culture was 2.4 nmol/min.
SO3−→3LNT was prepared as follows. The reaction mixture (2 ml), containing 50 mm sodium cacodylate (pH 6.35), 10 mm MnCl2, 0.05% (v/v) Triton X-100, 0.1 m NaF, 1 mm ATP, 1 mm lacto-N-tetraose, 1.5 mm PAPS (Calbiochem), and 7.2 nmol/min of recombinant Gal3ST2, was incubated at 37 °C for 16 h. The synthesized SO3−→3LNT was purified as described above. Finally, 0.18 μmol of SO3−→3LNT was obtained.
Synthesis of SO3−→3Galβ1→4Glc (3′-SulfoL) by Gal3-O-sulfotransferase-2 (Gal3ST2)
Recombinant Gal3ST2 was prepared using a P. pastoris protein expression system (Invitrogen) (16). SO3−→3Galβ1→4Glc (3′-sulfoL) was synthesized as follows. The reaction mixture (5 ml), containing 50 mm sodium cacodylate (pH 6.35), 10 mm MnCl2, 0.1% (v/v) Triton X-100, 0.5 mm spermine, 10% (v/v) glycerol, 20 mm lactose, 0.5 mm PAPS (Calbiochem), and 12 nmol/min of recombinant Gal3ST2, was incubated at 37 °C for 16 h. After heating at 100 °C to stop the reaction, the mixture was applied to a Sephadex G-25 gel filtration column (1.4 × 68 cm; eluted and equilibrated with EtOH/water, 5:95, v/v). The desalted oligosaccharides were applied to a Sephadex A-25 anion-exchange column (0.9 × 6.3 cm; equilibrated with 3 mm Tris-HCl, pH 8.0) and eluted with a linear gradient of NaCl (0–0.1 m). The oligosaccharide-containing fractions were collected and desalted by Sephadex G-25 gel filtration. Finally, 0.44 μmol of 3′-sulfoL was obtained.
Protein Purification and Crystallization
The N-terminal CRD of human galectin-8 (galectin-8N) was expressed as described previously (9). For crystallization, DNA corresponding to galectin-8N-(1–154) was expressed as a glutathione S-transferase fusion protein in Escherichia coli strain BL21(DE3) using plasmid pGEX6p-2 (GE Healthcare). The cells were disrupted by sonication and the supernatant was applied to a glutathione S-transferase affinity column of glutathione-Sepharose 4B and washed with 50 mm Tris-HCl buffer (pH 8.0) containing 500 mm NaCl and 1 mm DTT. The fusion protein bound to the resin was eluted with 10 mm glutathione-containing buffer, glutathione S-transferase was removed from the fusion protein by cleaving with PreScission protease (GE Healthcare), and the cleaved proteins were collected for further purification by MonoS HR 5/5 and Superdex 200 columns. The purified protein was a single band on SDS-PAGE stained with Coomassie Brilliant Blue. Galectin-8N was concentrated to 10 mg/ml using Centriplus YM-10 (Milipore) in crystallization buffer containing 10 mm Hepes-NaOH, 100 mm NaCl, 1 mm DTT (pH 7.0). Crystals were grown using the hanging drop vapor diffusion method from drops containing equal volumes of the protein (10 mg/ml) in crystallization buffer and solution composed of 250 mm ammonium fluoride and 15–24% (w/v) PEG3350. The space group and cell dimensions of the respective crystals are shown in Table 1. The complex crystals with carbohydrate were prepared by 2 methods. Crystals of lactose and 3′-sulfoL complex were obtained under the same crystallization conditions, and crystals of 3′-SL complex were obtained using ammonium chloride instead of ammonium fluoride in crystallization buffer.
TABLE 1.
Data collection and refinement statics
Values in parentheses are for the highest resolution shell of the reciprocal space spherically divided to 20 bins.
| Dataset name | Iodide derivative | Apo | Lactose | 3′-SulfoL | 3′-SL | LNF-III |
|---|---|---|---|---|---|---|
| PDB ID | 3APB | 3AP5 | 3AP4 | 3AP6 | 3AP7 | 3AP9 |
| Beamline | BL38B1 (SPring-8) | NW12 (PF-AR, KEK) | BL38B1 (SPring-8) | BL6A (PF, KEK) | BL5A (PF, KEK) | NW12 (PF-AR, KEK) |
| Cryoprotectant | 30% glycerol | 30% MPDa | 30% glycerol | 30% MPDa | 30% MPDa | |
| Wavelength (Å) | 1.30000 | 1.28000 | 0.92000 | 0.97798 | 1.00000 | 1.00000 |
| Space group | P212121 | P43212 | P1 | P1 | P212121 | P212121 |
| Resolution range (Å) | 38.95–1.95 (2.01–1.95) | 34.02–1.92 (1.97–1.92) | 59.44–2.33 (2.39–2.33) | 32.93–1.58 (1.62–1.58) | 49.94–1.53 (1.57–1.53) | 34.00–1.33 (1.36–1.33) |
| Cell dimensions (Å) | a = 65.59, b = 75.87, c = 77.90 | a = b = 76.06, c = 66.51 | a = 41.53, b = 64.89, c = 71.58 | a = 41.38, b = 65.40, c = 71.69 | a = 46.36, b = 49.92, c = 70.29 | a = 46.42, b = 49.95, c = 70.21 |
| Degree | α = β = γ = 90.00 | α = β = γ = 90.00 | α = 98.41, β = 105.49, γ = 108.66 | α = 98.29, β = 105.43, γ = 108.55 | α = β = γ = 90.00 | α = β = γ = 90.00 |
| Multiplicity | 6.6 (4.3) | 13.5 (9.4) | 1.9 (1.8) | 1.9 (1.9) | 12.8 (8.5) | 7.0 (7.1) |
| Completeness (%) | 99.9 (99.6) | 99.9 (100.0) | 95.5 (76.3) | 94.4 (91.5) | 99.9 (100.0) | 98.7 (97.4) |
| Rmerge (%) | 14.9 (35.4) | 6.4 (23.9) | 4.6 (6.0) | 5.5 (12.1) | 3.4 (19.2) | 4.1 (23.1) |
| I/σ | 44.4 (3.4) | 6.7 (5.4) | 6.5 (3.1) | 14.3 (5.8) | 12.7 (3.8) | 28.8 (9.4) |
| Phasing method | Single-wavelength anomalous dispersion | MR | MR | MR | MR | MR |
| No. of heavy atom binding sites | 19 | |||||
| Overall figure of merit | 0.49 | |||||
| Refinement | ||||||
| R-factor (%) | 20.2 | 20.4 | 19.1 | 20.0 | 16.4 | 15.7 |
| Free R-factor (%) | 24.2 | 23.7 | 25.1 | 24.2 | 18.8 | 17.8 |
| Water molecules | 162 | 116 | 307 | 491 | 222 | 286 |
| Hetero molecules | I−×29 | Cl−×1 | Cl−×3, MPDa×1 | |||
| No. of protein molecules/Asym. | 2 | 1 | 4 | 4 | 1 | 1 |
| Cis peptide bonds | Ile17–Pro18 | Ile17–Pro18 | Ile17–Pro18 | Ile17–Pro18 | Ile17–Pro18 | Ile17–Pro18 |
| Lys57–Pro58 | Lys57–Pro58 | Lys57–Pro58 | Lys57–Pro58 | Lys57–Pro58 | Lys57–Pro58 | |
| R.m.s. deviation bond lengthsb (Å) | 0.012 | 0.008 | 0.009 | 0.011 | 0.009 | 0.007 |
| R.m.s. deviation bond anglesc (deg) | 1.318 | 1.155 | 1.205 | 1.423 | 1.299 | 1.242 |
| Ramachandran plot (%) | ||||||
| Most favored regions | 89.9 | 89.1 | 86.6 | 89.0 | 87.5 | 86.7 |
| Additionally allowed regions | 9.3 | 10.1 | 12.1 | 10.2 | 11.7 | 12.5 |
| Generously allowed regions | 0.4 | 0.8 | 1.2 | 0.6 | 0.8 | 0.8 |
| Disallowed regions | 0.4 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 |
a 2-Methyl-2,4-pentanediol.
b Root mean square deviation from ideal bond lengths.
c Root mean square deviation from ideal bond angles.
Data Collection, Structure Determination, and Refinement
Diffraction data were collected at beamlines at the Photon Factory, Tsukuba, Japan, and at SPring-8, Hyogo, Japan. All images were indexed and integrated using the program MOSFLM (21) and processed using SCALA (22) in the CCP4 program suite (23). Phases were obtained by the single-wavelength anomalous dispersion method using the autoSHARP program (24). Heavy atom derivatives were prepared by soaking the apo form crystal for 95 s in 1 m potassium iodide solution containing 30% (w/v) glycerol, 160 mm ammonium chloride, and 14% (w/v) PEG3350. A single crystal was flash-cooled to 100 K in a cold nitrogen-gas stream produced by a cooling device (Rigaku, Japan). The x-ray diffraction data were collected at a wavelength of 1.3000 Å with a 210-mm CCD detector (Rigaku, Japan) installed at beamline BL38B1 at SPring-8. The distance between the crystal and the detector was set to 130.0 mm, and a total of 180 images were collected from the single crystal in 1.0 degree oscillation steps. We found 19 iodide binding sites with SHELXD (25) and initial phases were obtained by SHARP (26). The overall phasing power and (centric and acentric) figures of merit calculated between 37.94 and 1.95-Å resolution were 1.22, 0.35, and 0.18, respectively. The electron density maps were subsequently improved by SOLOMON (27) and the resultant centric and acentric figures of merit were 0.36 and 0.51, respectively. A model was automatically built using ARP/wARP (28) to 293 residues, which corresponds to 90.4% of the total residues in the asymmetric unit. The remaining parts of the model were manually constructed using XtalView (29). The other structures were determined by the molecular replacement (MR) technique with the program MOLREP (30) using the single-wavelength anomalous dispersion model as a search model. Finally, crystallographic refinement was performed by REFMAC5 (31). Final statistics of the crystallographic refinement are summarized in Table 1. The figures were drawn with PyMOL (32). Coordinates for galectin-8N were deposited in the Protein Data Bank of the Research Collaboratory for Structural Bioinformatics. The Protein Data Bank accession numbers for the apo form 1, lactose complex, 3′-SL complex, 3′-sulfoL complex, and LNF-III complex structures are 3AP5, 3AP4, 3AP7, 3AP6, and 3AP9, respectively.
Comparison of Galectin-8N with Other Galectin Structures
For comparison of the overall structures, superimposition of the α carbon atoms of various galectins were processed with the program LSQKAB (33). The structure of galectin-8N, galectin-1, -2, and -7 was A chain in the asymmetric unit of lactose complex. The structure of galectin-3 was in the N-acetyllactosamine complex and the galectin-4-C-domain was the apo form structure. For comparison of the binding sites, the ring carbon atoms of the carbohydrate were selected for superimposition of the galectins. In the cases of galectin-8N and galectin-4C, we superimposed α carbon atoms (Gly79–Glu95) of galectin-4 and (Gly74–Glu90) of galectin-8N.
Site-directed Mutagenesis of Galectin-8N
Site-directed mutagenesis was performed using the QuikChangeTM Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. Mutagenic primers used were 5′-CCTAGTGACGCAGACgcATTCCAGGTGGATCTGC-3′ (sense primer for R45A mutant), 5′-GCAGATCCACCTGGAATgcGTCTGCGTCACTAGG-3′ (antisense primer for R45A mutant), 5′-GTGACGCAGACAGATTCgcGGTGGATCTGCAGAATGGC-3′ (sense primer for Q47A mutant), 5′-GCCATTCTGCAGATCCACCgcGAATCTGTCTGCGTCAC-3′ (antisense primer for Q47A mutant), 5′-GGCAGCAGCGTGAAACCTgcAGCCGATGTGGCC-3′ (sense primer for R59A mutant), and 5′-GGCCACATCGGCTgcAGGTTTCACGCTGCTGCC-3′ (antisense primer for R59A mutant). Sequences in lowercase letters indicate mismatched bases for the desired mutation. Nucleic acid sequences were analyzed on an Applied Biosystems PRISM 310 Genetic Analyzer.
Estimation of Kinetic Constants Based on SPR
The dissociation constants between galectin-8N and various carbohydrates were measured using a BIAcore 2000 instrument as described previously (34). The purified wild-type and mutant GST-galectin-8Ns were immobilized on the CM5 sensor surface at pH 5 according to the manufacturer's instructions. Various carbohydrates in HBS-EP (0.01 m Hepes-NaOH (pH 7.4), 0.15 m NaCl, 3 mm EDTA, 0.005% (v/v) polysorbate 20) buffer were introduced onto the surface at a flow rate of 20 μl/min. The interaction was monitored at 25 °C by subtracting the blank surface signal, and the dissociation constants were calculated using BIA evaluation 3.0 software.
RESULTS AND DISCUSSION
Overall Structure
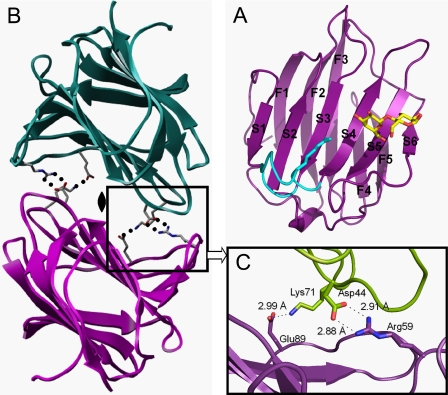
We determined the x-ray crystal structures of human galectin-8N in the absence of ligand and in the presence of 4 different carbohydrates. The x-ray crystal structure of galectin-8N shows that the protein is composed of 6-stranded (S1–S6) and 5-stranded (F1–F5) β-sheets, which together form a β-sandwich arrangement (Fig. 1A). The overall structure of galectin-8N is very similar to the structures of other galectins (11–14).
FIGURE 1.
A, ribbon model of the monomeric structure of the galectin-8N-lactose complex. The 5-stranded (F1–F5) and 6-stranded (S1–S6) β-sheets are indicated by the letter number code. The S3–S4 loop is shown in light blue. B, the dimeric structure of galectin-8N. Two monomers in an asymmetric unit in the apo crystal are shown in magenta (chain-A) and green (chain-B), respectively. C, close-up view of the dimer interface. The amino acid residues involved in dimer formation are shown in the ball-and-stick model. Oxygen and nitrogen atoms are shown in red and blue, respectively. Hydrogen bonds are depicted by dotted lines.
Apo Form Structure (Dimer Formation)
In the apo form, two molecules form a dimer, with the CRDs of each monomer facing the other (Fig. 1B). Four intermolecular hydrogen bonds between Asp44 and Arg59 and between Lys71 and Glu89 stabilize the dimer (Fig. 1C). The N and C termini of each monomer are positioned at opposite sides of the dimer interface. This suggests that the C-domain does not prevent dimer formation of the N-domain. Galectin-8N dimer formation is different from that in other galectins; galectin-1 and -2 form homodimers wherein the interfaces are formed by extended β-sheet interactions across the 2 monomers on both sides (F1 and S1) (11, 35) and mouse galectin-9-N-domain dimerizes at the S6 strand (36). Galectin-8N dimer formation has been shown in vitro (37), but it is not clear whether galectin-8 dimerizes in vivo. Because there is no space for ligand binding in the apo form structure, high affinity carbohydrates likely disrupt galectin-8N homodimers.
Galectin-8N-Lactose Complex and Comparison with Other Galectins
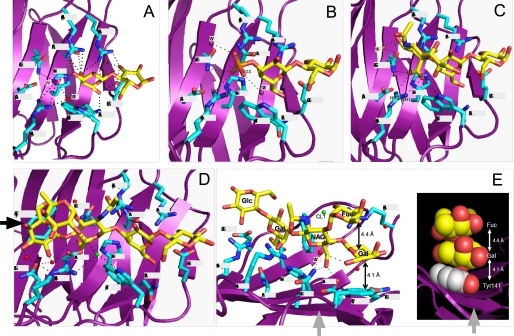
In the galectin-8N-lactose complex, 4 molecules in the asymmetric unit are almost identical. The binding networks of galectin-8N and lactose are shown in Fig. 2A. The β-galactoside moiety is most deeply buried in the binding site formed by β strands S4–S6. Arg45, His65, Asn67, Arg69, Asn79, and Glu89 directly interact with lactose via hydrogen bonding. Trp86 participates in van der Waals interactions with the galactose ring, in a manner similar to that seen in a number of other galactose- and lactose-binding lectins (37). Gln47, Trp86, and Arg59 form a water-mediated hydrogen bond with lactose.
FIGURE 2.
A–E, residues interacting with the bound lactose (A), or 3′-sulfoL (B), 3′-SL (C), and LNF-III (D and E) moiety through direct and water-mediated hydrogen bonds are shown. The bound carbohydrate moiety is shown with yellow sticks. Oxygen, nitrogen, and sulfur atoms are colored in red, blue, and orange, respectively. Water molecules are labeled W1–W3. Potential hydrogen bonds are shown as dotted lines. E, the side view (indicated by the arrow in D) of Tyr141, galactose, and fucose at the non-reducing terminal of LNF-III.
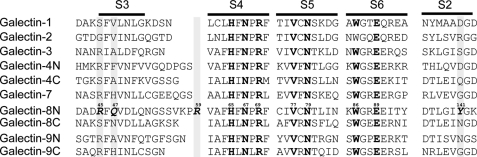
To elucidate the unique carbohydrate-binding specificity of galectin-8N, we compared the amino acid sequence and structure of the galectin-8N carbohydrate recognition site with those of other galectins. Seven amino acids directly interact with lactose, 6 of which (except Arg45) were conserved in galectins-1, -2, -3, -4, and -7 (Fig. 4). However, the amino acids located opposite the non-reducing lactose terminal are quite different from other galectins. This region of galectin-8N is more basic and Arg45 forms a hydrogen bond with galactose O4. The Arg is conserved in galectins-3 and -7, although their side chain conformations are quite different from that of galectin-8N and they interact with lactose via water-mediated hydrogen bonding (supplemental Fig. S1).
FIGURE 4.
Sequence alignment of galectins-1 to -9. Amino acid alignment of the galectin S2–S6 β-sheets. Residues that are common in all the sequences are shown in bold and amino acids that are unique to galectin-8N are in bold and italic.
However, the loop region between strands S3 and S4 are significantly different (supplemental Fig. S2). Gln47 of galectin-8N interacts with the 3-OH of the galactose moiety of lactose via water-mediated hydrogen bonding and is substituted in other galectins with Ala or Val, which do not interact with lactose. Arg59 lies within the loop region between S3 and S4 and interacts with the 3-OH of the galactose moiety of lactose via water-mediated hydrogen bonding (Fig. 2A and supplemental Fig. S3a). This S3–S4 loop has unique characteristics; it is longer than the loops of other galectins and positioned closer to the carbohydrate ligands. Other galectins do not carry a homologous Arg59 residue.
Galectin-8N Complexed with 3′-SulfoL and 3′-SL
We determined the complex structures of galectin-8N bound with 3′-sulfoL and 3′-SL to elucidate the unique recognition mechanism for sulfated and sialylated carbohydrates (Fig. 2, B and C, and supplemental Fig. S3, b and c). In the 3′-sulfoL complex, the residues involved in lactose recognition are nearly identical to those in the galectin-8N-lactose complex, and 4 residues are involved in sulfate recognition. Of the 3 oxygen molecules in sulfate, O2S and O3S hydrogen bond directly with the protein. The O3S is positioned such that it directly hydrogen bonds with Arg45, Gln47, and Arg59, whereas O2S only hydrogen bonds with Arg59. Water molecules (W2 and W3) are well conserved in the 3′-sulfoL complex as in lactose complex; however, W1 is substituted with O3S of the sulfate. Gln47 and Arg59 are important for sulfate recognition, because the distance between these elements is short enough to permit interactions. Trp86 interacts with OS2 through W2-mediated hydrogen bonding. W2 is located at almost the same position as in the lactose complex and O1S forms a W3-mediated hydrogen bond with Gln47. W3 exists in all 4 molecules of the asymmetrical unit and W1, which is conserved in all lactose complexes, is substituted with O3S. Gln47 and Arg59 are important for sulfate recognition because they are located at a short distance from the ligand and are unique to galectin-8N.
The 3′-SL complex crystallizes in space group P212121, whereas lactose and 3′-sulfoL form complex crystals in the P1 space group. In the 3′-SL complex, the residues involved in lactose recognition are almost the same as those in the lactose complex, and the terminal sialic acid-recognizing residues are very similar to those involved in sulfate recognition (Fig. 2B and supplemental Fig. S3b). Only the carboxylate in sialic acid interacts directly with galectin-8N amino acids, forming hydrogen bonds with Gln47, Arg59, and Trp86. Of these, Arg59 forms 2 independent hydrogen bonds, including the shortest hydrogen bond, suggesting that Arg59 is important for sialic acid recognition. Gln47 forms water-mediated hydrogen bonds with the oxygen atom (O11) of sialic acid; substitutions of Gln47 with Ala greatly reduced binding to 3′-SL (9). There is a large cleft on the non-reducing side of the 3′-SL; the sialic acids are not buried within this cleft, so it is free to be occupied by the extended carbohydrate. The molecular surface of the galectin-8N 3′-SL binding site shows that the galactose of the bound lactose is located within the cleft, whereas glucose is exposed to the surrounding solution. There is the space at the non-reducing carbohydrate terminal, but sialic acid is exposed to the solvent region and is not buried in this extended cleft. It is possible that the longer oligosaccharides may interact with the amino acids (Asp49, Gln51, and Tyr141) within the cleft.
LNF-III Complex
We also analyzed the high affinity galactin-8N-LNF-III complex (9). LNF-III is a pentasaccharide with lactose at the reducing terminal and an extended β-N-acetylglucosamine (β-GlcNAc) at the C3-position of galactose, with α-fucose and β-galactose attached to the extended β-GlcNAc residue. In the LNF-III complex, residues involved in lactose recognition are nearly identical to those in the lactose complex, and 4 amino acids (Gln47, Asp49, Arg59, and Tyr141) directly interact with the extended lactose moiety (Fig. 2D and supplemental Fig. S3d). Gln51 and Trp86 also interact with water-mediated and chloride-mediated hydrogen bonds, respectively. Asp49, Gln51, and Tyr141 are not involved in the recognition of lactose, 3′-SL, or 3′-sulfoL. The C6 hydroxy group of GlcNAc forms hydrogen bonds to Gln47 and Asp49. OH1 and OH2 of Gal form hydrogen bonds to Gln47 and Asp49, and OH2, OH4, and OH6 of Gal form water-mediated hydrogen bonds with Arg59, Gln51, and Gln47. The van der Waals interaction between galactose and Tyr141 is also important. The fucose attached to the C3 position of GlcNAc does not form hydrogen bonds with protein; however, Tyr141 has van der Waals interactions with galactose attached to the C4 position of GlcNAc. This galactose is situated and stabilized between Tyr141 and fucose via hydrophobic interactions. The average distance between the rings of Tyr141 and Gal and between Gal and Fuc are 4.1 and 4.4 Å, respectively (Fig. 2E). Our previous study showed that these van der Waals interactions increase the stability of the LNF-III-galectin-8N complex. The binding abilities of LNnT, which has a hydroxyl group instead of fucose and LNF-II (Galβ1→3(Fucα1→4)GlcNAcβ1→3Galβ1→4Glc), which has α1→4 fucose and β1→3 galactose instead of β1→4 galactose and α1→3 fucose, are ¼ that of LNF-III (9). Because galactose and fucose of LNF-III do not form hydrogen bonds with galectin-8N, the van der Waals interactions formed between Tyr141, galactose, and fucose are very important for galectin-8N complex formation. Proper location of the stack depends on α1→3, but not α1→4, binding of fucose.
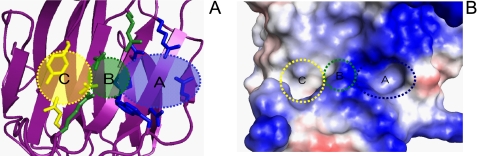
Subsites of Galectin-8N
Galectins-1, -3, and -9 have greater affinity for long oligosaccharides composed of the disaccharide repeat (3Galβ1,4GlcNAcβ1)n than for lactose or N-acetyllactosamine (10). These increases in affinity suggest that in addition to the primary binding site, which accommodates lactose/N-acetyllactosamine, there are secondary sites of interaction for more extended oligosaccharides. Such secondary sites are important determinants of lectin binding specificity, and the term subsite multivalency has been used to describe the resulting increases in affinity/specificity (38).
There are 3 subsites (referred to as A, B, and C) in the galectin-8N CRD (Fig. 3A). Subsite A is involved in lactose recognition and is conserved in the galectin family. Subsite A of galectin-8N consists of 6 amino acids (His65, Asn67, Arg69, Asn79, Trp86, and Glu89), whereas subsite A in other galectins (-1, -2, -3, -4, and -7) consist of 7–9 amino acids.
FIGURE 3.
A, subsites of galectin-8N indicated in the ribbon model. B, close-up view of subsites on the electrostatic potential of galectin-8N as seen within the carbohydrate binding site. Blue and red indicate positive and negative electrostatic potentials.
Subsite B of galectin-8N consists of 3 amino acids (Arg45, Gln47, and Arg59) and is involved in recognition of carbohydrate or acidic substitutes including sulfate and sialic acid attached to the O-3 of the non-reducing terminal galactose moiety. In the unique subsite B of galectin-8N, Arg59 is the most important amino acid because it lies within the unique long loop. The electrostatic potential of galectin-8N within the carbohydrate binding site (Fig. 3B) showed that galectin-8N have positively charged surface around subsite B.
When galectin-8N binds to longer oligosaccharides, subsite C recognizes the extended carbohydrate attached to the O-3 of the non-reducing galactose moiety. In the unique subsite C of galectin-8N, which consists of 3 amino acids (Asp49, Gln51, and Tyr141), Tyr141 is the most important amino acid, forming van der Waals interactions with the extended carbohydrate moiety. Furthermore, in the LNF-III-galectin-8N complex, van der Waals interactions are observed between the α1–3 branched fucose and galactose and between galactose and Tyr141.
The Diminishment of Siaα2→3/SO3−→3-Linked Oligosaccharide Binding by Mutagenesis of Arg45, Gln47, or Arg59
Our previous study of galectin-8 revealed that only the N-domain has affinity for sialic acid/sulfate, and substitution of Gln47 for Ala reduces this affinity (9). Amino acids in the S3 β-sheet are therefore likely to be involved in the binding of sialic acid/sulfate residues, because galactose-3-O-linked non-reducing terminal moieties interact with amino acids in the extended clefts formed by the galectin S3 β-sheet. However, substitution of Ala146 of galectin-3 (Table 2) and Phe47 of galectin-4-N-domain (corresponding to Gln47 of galectin-8N) for Gln (Table 3) does not enhance affinity for sialic acid/sulfate, as shown in supplemental Fig. S4. These results suggest that Gln47 of galectin-8N is not sufficient for binding to sialylated or sulfated oligosaccharides. X-ray crystallographic analysis of galectin-8N-lactose (Fig. 2A) showed that lactose-binding sites in β-sheets S4, S5, and S6 are well conserved in the galectin family (Fig. 4). Furthermore, x-ray crystallographic analysis of galectin-8N in complex with 3′-sulfoL and 3′-SL (Fig. 2, B and C) revealed that not only Gln47, but also Arg45 and Arg59, are involved in binding to sialylated or sulfated oligosaccharides.
TABLE 2.
Relative binding abilities of the wild-type and mutant galectin-3 for various oligosaccharides
Half-inhibition concentrations (I50) of various oligosaccharides for the wild-type and the mutant galectin-3 to the immobilized asialofetuin. The relative binding abilities are calculated by dividing the concentration (m) of lac-pNP giving 50% inhibition by the concentration (m) of the oligosaccharides giving 50% inhibition.
| Oligosaccharide | Wild-type | A146Q |
|---|---|---|
| m | ||
| Lac-O-pNP | 1.4 × 10−4 (1.0) | 1.5 × 10−4 (1.0) |
| SO3−→3Lac-O-pNP | 1.5 × 10−5 (4.7) | 3.5 × 10−5 (3.1) |
| 3′-SL | 2.4 × 10−5 (1.8) | 1.2 × 10−4 (0.91) |
| LNT | 1.7 × 10−5 (6.7) | 3.0 × 10−5 (3.7) |
| LNnT | 1.5 × 10−5 (10.0) | 2.0 × 10−5 (5.5) |
| A-tetra | 7.4 × 10−6 (35.0) | 2.9 × 10−4 (0.38) |
TABLE 3.
The KD values of galectin-4-N-domain and the mutants for various oligosaccharides
The relative binding abilities in the parenteses were calculated by dividing the KD value between Lac-O-pNP and galectin-4-N-domain at 25 °C by the KD values for the oligosaccharides.
| Oligosaccharide | Wild-type | F47A | F47Q |
|---|---|---|---|
| Lac-O-pNP | 4.9 × 10−4 (1.0) | 4.5 × 10−4 (1.1) | 5.4 × 10−4 (0.9) |
| SO3−→3Lac-O-pNP | 9.1 × 10−5 (5.4) | 7.3 × 10−5 (6.7) | 9.1 × 10−5 (5.4) |
| Lactose | 1.2 × 10−4 (4.0) | 1.1 × 10−4 (4.5) | 9.7 × 10−5 (5.0) |
| 3′-SL | 9.9 × 10−4 (0.5) | 8.6 × 10−4 (0.6) | 4.8 × 10−4 (1.0) |
| LNT | 2.1 × 10−4 (2.3) | 1.8 × 10−4 (2.7) | 4.5 × 10−4 (1.1) |
| LNnT | 2.7 × 10−4 (1.8) | 3.0 × 10−4 (1.6) | 5.4 × 10−4 (0.9) |
| A-tetra | 6.6 × 10−5 (7.4) | 1.5 × 10−5 (33) | 2.5 × 10−4 (2.0) |
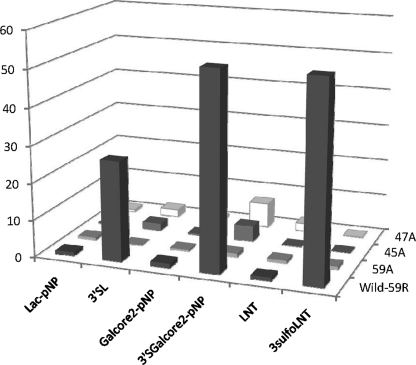
To confirm the amino acids interacting with the extended Siaα2→3 or SO3−→3 residue, we mutated Arg45, Gln47, and Arg59 of galectin-8N to Ala and analyzed carbohydrate-binding specificities (Table 4 and Fig. 5). The wild-type, Arg45 → Ala45 (R45A), Gln47 → Ala47 (Q47A), and Arg59 → Ala59 (R59A) were individually immobilized on CM5 sensor chips and their oligosaccharide-binding activities were measured by surface plasmon resonance (SPR). The wild-type protein and mutants had similarly higher KD values for Lac-O-pNP, Gal-core2-O-pNP, and LNT. Galectin-8N strongly bound to Siaα2→3Gal-core2-O-pNP, SO3−→3LNT, and Siα2→3Lac with KD values of 1.4 × 10−6, and 1.4 × 10−6, and 2.7 × 10−6 m, respectively. The KD values of the R59A, R45A, and Q47A mutants for Siα2→3Gal-core2-O-pNP were 49, 12, and 8 times higher than that of the wild-type, respectively. The KD values of the R59A, R45A, and Q47A mutants for Siα2→3Lac were also 174, 14, and 14 times higher than that of the wild-type, respectively. Similarly, the KD value of the R59A mutant for SO3−→3LNT was 50 times higher than that of the wild-type, and R45A and Q47A did not show any binding activity under the tested concentrations. These results suggest that Arg59, Arg45, and Gln47 contribute to the strong binding to sialylated and sulfated oligosaccharides; specifically, the R59A mutant lost the enhancing effect of sulfation or sialylation of neutral oligosaccharides, whereas R45A and Q47A retained their affinity enhancing effects, suggesting that Arg59 is the most critical amino acid for binding to sialylated/sulfated oligosaccharides. The binding affinities of the wild-type and mutant (R45A, Q47A, R59A) galectin-8 proteins for oligosaccharides were also measured by SPR. The mutants also lost their strong binding affinity for sialylated and sulfated oligosaccharides (data not shown), because the C-domain is not involved in binding to these oligosaccharides (9).
TABLE 4.
The KD values of galectin-8-N-domain and the mutants for various oligosaccharides
| Oligosaccharide | Wild-type | R59A | R45A | Q47A |
|---|---|---|---|---|
| m | ||||
| Lac-pNP | 7.4 × 10−5 (1.0)a | 1.0 × 10−4 (0.7) | 1.9 × 10−4 (0.4) | 1.2 × 10−4 (0.6) |
| Siaα2→3Lac (3′-SL) | 2.7 × 10−6 (27) | 4.7 × 10−4 (0.2) | 3.7 × 10−5 (2) | 3.7 × 10−5 (2) |
| Gal-core2-pNP | 5.9 × 10−5 (1.3) | 1.4 × 10−4 (0.5) | 1.7 × 10−4 (0.4) | 1.0 × 10−4 (0.7) |
| Siaα2→3Gal-core2-pNP (3′-Sgal-core2-pNP) | 1.4 × 10−6 (53) | 6.9 × 10−5 (1.1) | 1.7 × 10−5 (4.3) | 1.1 × 10−5 (6.7) |
| LNT | 7.3 × 10−5 (1.0) | 6.5 × 10−5 (0.8) | 3.2 × 10−4 (0.2) | 3.5 × 10−5 (2.1) |
| SO3−→3LNT (3-sulfoLNT) | 1.4 × 10−6 (53) | 7.0 × 10−5 (0.9) | — b | —-b |
a The relative binding abilities in the parenteses were calculated by dividing the KD value between Lac-O-pNP and galectin-8-N-domain at 25 °C by the KD values for the oligosaccharides.
b No binding up to 11 μm.
FIGURE 5.
Relative binding abilities of wild-type and mutant galectin-8N for various oligosaccharides. The relative binding abilities of wild-type and mutant galectin-8N were calculated by dividing the KD value between Lac-O-pNP and galectin-8N by the KD values for the oligosaccharides.
The S3 β-Sheet Cleft Is Important for Long Oligosaccharides
The mutagenesis study also showed the involvement of amino acids in or near the S3 β-sheet in binding to longer oligosaccharides. When Ala146 of galectin-3 and Phe47 of galectin-4N were changed to Gln, which corresponds to Gln47 of galectin-8N, affinity for sialylated and sulfated oligosaccharides was not enhanced and the affinity for longer oligosaccharides was reduced (Tables 2 and 3 and supplemental Fig. S4). On the contrary, substitution of Gln47 of galectin-8N for Ala and Phe47 of galectin-4N for Ala increased the affinity for longer oligosaccharides (Tables 3 and 4 and supplemental Fig. S4). The amino acid at this position is important for binding to longer oligosaccharides, especially for A-tetra oligosaccharides. Galectin-3, galectin-4C, galectin-8C, and galectin-9N, which have strong affinity for A-tetra, also have Ala in this position.
General Discussion
We determined the crystal structures of human galectin-8N complexed with sialylated or sulfated glycans. We used SPR to identify the amino acids that are indispensable for binding. Because Arg59 of galectin-8 is unique among the galectins, it seems to be responsible for the unique carbohydrate specificity of galectin-8.
Sulfation and sialylation of various glycoconjugates confer negatively charged characteristics to the cell surface and are important in cell-to-cell or cell-to-matrix interactions. Sialic acid moieties on the non-reducing termini of glycoproteins and glycolipids are distributed throughout the vertebrates, viruses, bacteria, plants, and other invertebrates (39) and play key roles in the various biological functions of these glycoconjugates. In fact, several influenza viruses invade via sialic acids on the surfaces of susceptible cells (40). Furthermore, there are several reports that sulfated glycosphingolipids serve as receptors for bacterial or viral toxins, such as binding of the human respiratory pathogen, Bordetella pertussis, to sulfatide (SM4) and glycosphingolipids carrying GalNAcβ1–4Gal (41). Influenza A virus binds to SM4 (42), and the neutrophil-activating protein of Helicobacter pylori also binds to SM4 (43). Because SM4 and some sulfated glycosphingolipids are expressed in the mammalian gastrointestinal tract (20), galectin-8 likely plays a role in host defense by masking the sulfated or sialylated glycosphingolipids from infection by bacterial or viral toxins.
Several potent and selective inhibitors for the influenza neuraminidase have been discovered through structure-based rational drug design. Oseltamivir (Tamiflu) has been approved for the prevention and treatment of influenza infection. The x-ray structure of the galectin-8N-sialic acid complex may yield new insight for another sialic acid-based drug design.
Supplementary Material
Acknowledgments
The synchrotron radiation experiments were performed at the Photon Factory with the approval of the Photon Factory Advisory Committee, the National Laboratory for High Energy Physics, Japan, Proposal numbers 2003S2-002, 2006S2-006, and 2007G183, and SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute, Proposal numbers 2005A0865-NL1-np-P3k and 2006A1715.
This work was supported in part by the New Energy and Industrial Technology Development Organization (NEDO) in Japan.
The atomic coordinates and structure factors (codes 3AP5, 3AP4, 3AP7, 3AP6, and 3AP9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- CRD
- carbohydrate recognition domain
- N-domain
- N-terminal carbohydrate recognition domain
- C-domain
- C-terminal carbohydrate recognition domain
- GalNAc
- N-acetylgalactosamine
- GlcNAc
- N-acetylglucosamine
- PAPS
- adenosine 3′-phosphate,5′-phosphosulfate
- pNP
- p-nitrophenyl
- SPR
- surface plasmon resonance
- type 1
- Galβ1→3GlcNAc
- type 2
- Galβ1→4GlcNAc
- core 1
- Galβ1→3GalNAc
- core 2
- Galβ1→3(GlcNAcβ1→6)GalNAc
- lactose
- Galβ1→4Glc
- 3′-SL
- Neu5Acα2→3Galβ1→4Glc
- 6′-SL
- Neu5Acα2→ 6Galβ1→4Glc
- 3′-sulfoL
- SO3−→3Galβ1→4Glc
- A-tetra
- GalNAcα1→ 3(Fucα1→2)Galβ1→4Glc
- LNT
- Galβ1→3GlcNAcβ1→3Galβ1→4Glc
- LNnT
- Galβ1→4GlcNAcβ1→3Galβ1→4Glc
- LNF-I
- Fucα1→2Galβ1→ 3GlcNAcβ1→3Galβ1→4Glc
- LNF-II
- Galβ1→3(Fucα1→4)GlcNAcβ1→ 3Galβ1→4Glc
- LNF-III
- Galβ1→4(Fucα1→3)GlcNAcβ1→3Galβ1→4Glc
- r.m.s.
- root mean square.
REFERENCES
- 1. Hadari Y. R., Paz K., Dekel R., Mestrovic T., Accili D., Zick Y. (1995) J. Biol. Chem. 270, 3447–3453 [DOI] [PubMed] [Google Scholar]
- 2. Hadari Y. R., Eisenstein M., Zakut R., Zick Y. (1997) Trends Glycosci. Glycotechnol. 9, 103–112 [Google Scholar]
- 3. Barondes S. H., Cooper D. N., Gitt M. A., Leffler H. (1994) J. Biol. Chem. 269, 20807–20810 [PubMed] [Google Scholar]
- 4. Cooper D. N., Barondes S. H. (1999) Glycobiology 9, 979–984 [DOI] [PubMed] [Google Scholar]
- 5. Levy Y., Arbel-Goren R., Hadari Y. R., Eshhar S., Ronen D., Elhanany E., Geiger B., Zick Y. (2001) J. Biol. Chem. 276, 31285–31295 [DOI] [PubMed] [Google Scholar]
- 6. Arbel-Goren R., Levy Y., Ronen D., Zick Y. (2005) J. Biol. Chem. 280, 19105–19114 [DOI] [PubMed] [Google Scholar]
- 7. Cárcamo C., Pardo E., Oyanadel C., Bravo-Zehnder M., Bull P., Cáceres M., Martínez J., Massardo L., Jacobelli S., González A., Soza A. (2006) Exp. Cell. Res. 312, 374–386 [DOI] [PubMed] [Google Scholar]
- 8. Zick Y., Eisenstein M., Goren R. A., Hadari Y. R., Levy Y., Ronen D. (2004) Glycoconj. J. 19, 517–526 [DOI] [PubMed] [Google Scholar]
- 9. Ideo H., Seko A., Ishizuka I., Yamashita K. (2003) Glycobiology 13, 713–723 [DOI] [PubMed] [Google Scholar]
- 10. Hirabayashi J., Hashidate T., Arata Y., Nishi N., Nakamura T., Hirashima M., Urashima T., Oka T., Futai M., Muller W. E., Yagi F., Kasai K. (2002) Biochim. Biophys. Acta 1572, 232–254 [DOI] [PubMed] [Google Scholar]
- 11. Lobsanov Y. D., Gitt M. A., Leffler H., Barondes S. H., Rini J. M. (1993) J. Biol. Chem. 268, 27034–27038 [DOI] [PubMed] [Google Scholar]
- 12. Seetharaman J., Kanigsberg A., Slaaby R., Leffler H., Barondes S. H., Rini J. M. (1998) J. Biol. Chem. 273, 13047–13052 [DOI] [PubMed] [Google Scholar]
- 13. Leonidas D. D., Vatzaki E. H., Vorum H., Celis J. E., Madsen P., Acharya K. R. (1998) Biochemistry 37, 13930–13940 [DOI] [PubMed] [Google Scholar]
- 14. Swaminathan G. J., Leonidas D. D., Savage M. P., Ackerman S. J., Acharya K. R. (1999) Biochemistry 38, 13837–13843 [DOI] [PubMed] [Google Scholar]
- 15. Hirabayashi J., Kasai K. (1994) Glycoconj. J. 11, 437–442 [DOI] [PubMed] [Google Scholar]
- 16. Ideo H., Seko A., Yamashita K. (2007) J. Biol. Chem. 282, 21081–21089 [DOI] [PubMed] [Google Scholar]
- 17. Seko A., Yamashita K. (2005) Glycobiology 15, 943–951 [DOI] [PubMed] [Google Scholar]
- 18. Honke K., Tsuda M., Koyota S., Wada Y., Iida-Tanaka N., Ishizuka I., Nakayama J., Taniguchi N. (2001) J. Biol. Chem. 276, 267–274 [DOI] [PubMed] [Google Scholar]
- 19. Seko A., Nagata K., Yonezawa S., Yamashita K. (2002) Jpn. J. Cancer Res. 93, 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishizuka I. (1997) Prog. Lipid Res. 36, 245–319 [DOI] [PubMed] [Google Scholar]
- 21. Powell H. R. (1999) Acta Crystallogr. D 55, 1690–1695 [DOI] [PubMed] [Google Scholar]
- 22. Evans P. R. (2006) Acta Crystallogr. D 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 23. Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 24. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2006) in Macromolecular Crystallography Protocols (Doublie S. ed) Vol. 2, pp. 215–230, Humana Press, Totowa, NJ [Google Scholar]
- 25. Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 26. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. (2003) Acta Crystallogr. D 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 27. Abrahams J. P., Leslie A. G. (1996) Acta Crystallogr. D 52, 30–42 [DOI] [PubMed] [Google Scholar]
- 28. Lamzin V. S., Wilson K. S. (1993) Acta Crystallogr. D 49, 129–147 [DOI] [PubMed] [Google Scholar]
- 29. McRee D. E. (1992) J. Mol. Graph. 10, 44–46 [Google Scholar]
- 30. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 31. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 32. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 33. Kabsch W. (1976) Acta Crystallogr. Sect. A 32, 922–923 [Google Scholar]
- 34. Ideo H., Seko A., Ohkura T., Matta K. L., Yamashita K. (2002) Glycobiology 12, 199–208 [DOI] [PubMed] [Google Scholar]
- 35. Liao D. I., Kapadia G., Ahmed H., Vasta G. R., Herzberg O. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1428–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nagae M., Nishi N., Murata T., Usui T., Nakamura T., Wakatsuki S., Kato R. (2006) J. Biol. Chem. 281, 35884–35893 [DOI] [PubMed] [Google Scholar]
- 37. Stowell S. R., Arthur C. M., Slanina K. A., Horton J. R., Smith D. F., Cummings R. D. (2008) J. Biol. Chem. 283, 20547–20559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rini J. M. (1995) Annu. Rev. Biophys. Biomol. Struct. 24, 551–577 [DOI] [PubMed] [Google Scholar]
- 39. Warren L. (1976) in Biological Roles of Sialic Acid (Rosenberg A., Schengrund C. eds) pp. 103–121, Plenum Press, New York [Google Scholar]
- 40. Suzuki Y., Ito T., Maejima Y., Masuda H., Horiike G., Suzuki T., Sato K., Hanagata G., Kiso M., Hasegawa A., Xu G., Ito T., Kawaoka Y., Webster R. G. (1996) (Brown L. E., Hampson A. W., Webster R. G. eds) pp. 443–446, Elsevier Science B. V., Amsterdam [Google Scholar]
- 41. Brennan M. J., Hannah J. H., Leininger E. (1991) J. Biol. Chem. 266, 18827–18831 [PubMed] [Google Scholar]
- 42. Suzuki T., Sometani A., Yamazaki Y., Horiike G., Mizutani Y., Masuda H., Yamada M., Tahara H., Xu G., Miyamoto D., Oku N., Okada S., Kiso M., Hasegawa A., Ito T., Kawaoka Y., Suzuki Y. (1996) Biochem. J. 318, 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Teneberg S., Miller-Podraza H., Lampert H. C., Evans D. J., Jr., Evans D. G., Danielsson D., Karlsson K. A. (1997) J. Biol. Chem. 272, 19067–19071 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.