Abstract
d-2-Hydroxyglutarate dehydrogenase (d-2HGDH) catalyzes the specific and efficient oxidation of d-2-hydroxyglutarate (d-2HG) to 2-oxoglutarate using FAD as a cofactor. In this work, we demonstrate that d-2HGDH localizes to plant mitochondria and that its expression increases gradually during developmental and dark-induced senescence in Arabidopsis thaliana, indicating an enhanced demand of respiration of alternative substrates through this enzymatic system under these conditions. Using loss-of-function mutants in d-2HGDH (d2hgdh1) and stable isotope dilution LC-MS/MS, we found that the d-isomer of 2HG accumulated in leaves of d2hgdh1 during both forms of carbon starvation. In addition to this, d2hgdh1 presented enhanced levels of most TCA cycle intermediates and free amino acids. In contrast to the deleterious effects caused by a deficiency in d-2HGDH in humans, d2hgdh1 and overexpressing lines of d-2HGDH showed normal developmental and senescence phenotypes, indicating a mild role of d-2HGDH in the tested conditions. Moreover, metabolic fingerprinting of leaves of plants grown in media supplemented with putative precursors indicated that d-2HG most probably originates during the catabolism of lysine. Finally, the l-isomer of 2HG was also detected in leaf extracts, indicating that both chiral forms of 2HG participate in plant metabolism.
Keywords: Amino Acid, Dehydrogenase, Energy Metabolism, Flavoproteins, Mitochondrial Metabolism, Amino Acid Catabolism, Carbon Starvation, d-2-Hydroxyglutarate Dehydrogenase, Plant Senescence, Tricarboxylic Acid Cycle
Introduction
2-Hydroxyglutarate (2HG3; 2-hydroxypentanedioic acid) is a five-carbon dicarboxylic acid with the hydroxy group on the α-carbon. d-2HG accumulates in humans in the inherited neurometabolic disorder 2-hydroxyglutaric aciduria (2HGA) due to a deficiency in d-2HG dehydrogenase (d-2HGDH) (1), which converts d-2HG to 2-oxoglutarate (2OG); the electron transfer flavoprotein (ETF); or the ETF-ubiquinone oxidoreductase (ETFQO) (2), both electron acceptors of d-2HGDH (1). The clinical symptoms encompass developmental retardation, neurological dysfunction, and cerebral atrophy (1). In addition to high levels of 2HG, patients with 2HGA also have high concentrations of TCA cycle intermediates. On the other hand, excess accumulation of d-2HG contributes to the formation and malignant progression of brain tumors (3). Mutations in the cytosolic enzyme IDH1 (isocitrate dehydrogenase 1) occur in ∼80% of secondary brain cancer tumors and in nearly one-tenth of acute myelogenous leukemia tumors. Normally, IDH1 catalyzes the conversion of isocitrate to 2OG. Cancer-associated mutations in IDH1 reduce the affinity of the enzyme for isocitrate and increase the affinity for NADPH and 2OG. This prevents the oxidative decarboxylation of isocitrate to 2OG and facilitates the conversion of 2OG to d-2HG. In this way, IDH1 mutations cause a gain of function, resulting in the production and accumulation of d-2HG (3).
d-2HG occurs in mammals (i) in the conversion of 2OG to d-2HG through a hydroxy acid-oxoacid transhydrogenase with the concomitant conversion of γ-hydroxybutyrate to succinic semialdehyde (4), (ii) as an intermediate in the succinate-glycine cycle between 2OG semialdehyde and 2OG (5), and (iii) in the conversion of d-2HG to 2OG by d-2HGDH (6). In bacteria, (i) a 3-phosphoglyceric acid dehydrogenase produces d-2HG from 2OG as a side reaction (7); (ii) glyoxylate condenses with propionyl-CoA, producing d-2HG (8); and (iii) 2-aminoadipate is converted to d-2HG (9). In addition to this, it has been suggested that 2HG is the end product of a sequence involving glutamate via 2OG in yeast (10).
Despite the fact that the involvement of d-2HG in metabolism in mammals and bacteria is known, the participation of this metabolite in plant metabolism was so far poorly understood. Recently, we identified the d-2HGDH from Arabidopsis thaliana and characterized it at the biochemical level (11). This enzyme catalyzes the exclusive conversion of d-2HG to 2OG, which can enter the TCA cycle, whereas electrons are transferred to the electron transport chain through the ETF-ETFQO complex (1, 11). As gene coexpression analysis indicated that d-2HGDH is in the same network as several genes involved in β-oxidation and degradation of amino acids and chlorophyll, it was proposed that d-2HGDH participates in the catabolism of d-2HG most probably during the mobilization of alternative substrates from proteolysis and/or lipid degradation (11).
Here, we demonstrate that d-2HGDH definitely localizes to plant mitochondria and that its expression increases gradually during developmental and dark-induced senescence in A. thaliana. Metabolic analysis of loss-of-function mutants (d2hgdh1) growing in media supplemented with different metabolites indicated that d-2HG would form from the free amino acid lysine. We also demonstrate that d-2HG accumulates in leaves of d2hgdh1 during natural senescence and dark-induced carbon starvation and that plants overexpressing d-2HGDH accumulate less d-2HG during the course of senescence. The metabolic and phenotypic analysis of these plants indicated a mild role of d-2HGDH in plant metabolism in the tested conditions. Moreover, the data obtained suggest fundamental differences between plants and human d-2HG metabolism both in the origin of this metabolite and how these organisms can cope with high levels of it.
EXPERIMENTAL PROCEDURES
Screening for T-DNA Insertion Lines and Genotyping
Homozygous plants of the T-DNA insertion line Salk_061383 (d2hgdh1-1) were obtained in a previous work (11). Seeds of the T-DNA insertion line GABI_127F12 (d2hgdh1-3) was obtained from the Nottingham Arabidopsis Stock Center. Plants homozygous for this insertion were isolated and confirmed by two sets of PCRs using genomic DNA as a template. The first PCR was performed using primers d-2HGDH1-3-F (ATGATGATGCAGAAATTGAGAAG) and d-2HGDH1-3-R (TAATATGCATCCTGCTTCACACAC), which are specific for the wild-type gene. The second PCR was carried out with the T-DNA left border primer GABI-KAT-LB (5′-ATAATAACGCTGCGGACATCTACATTTT-3′) in combination with d-2HGDH1-3-R. To further determine whether all native d-2HGDH transcript was absent in the insertional mutant, total RNA was isolated from 100 mg of leaves using TRIzol reagent (Invitrogen). RNA was converted into first-strand cDNA using the SuperScript II reverse transcriptase (Invitrogen). PCRs were conducted in a final volume of 10 μl using 1 μl of the transcribed product and Taq DNA polymerase (Qiagen). The pairs of primers used were d-2HGDH1-3-F and d-2HGDH1-3-R. Amplification conditions were as follows: 1.5 min of denaturation at 95 °C and 35 cycles at 95 °C for 30 s, 50 °C for 30 s and 72 °C for 2 min, followed by 5 min at 72 °C. As a control, the ACT2 gene was amplified by 28 cycles using the following primers: Actin2-F (5′-TAACTCTCCCGCTATGTATGTCGC-3′) and Actin2-R (5′-CCACTGAGCACAATGTTACCGTAC-3′).
Plant Growth Conditions and Sampling
A. thaliana wild-type (Columbia-0), T-DNA insertion lines (d2hgdh1-1 and d2hgdh1-3), and d-2HGDH overexpressor lines were grown in pots containing three parts soil (Gebrüder Patzer KG) and one part vermiculite (Basalt Feuerfest GmbH) in a growth cabinet in a 16/8-h light/dark cycle at 22 °C day/18 °C night temperatures and at a photosynthetically active photon flux density of 100 μmol quanta m−2 s−1. For dark-induced senescence, 5-week-old plants were kept in the same chamber in continuous darkness, and whole rosettes were harvested at different time points and immediately frozen in liquid nitrogen. Growth of plants in the presence of different concentrations of valine, isoleucine, lysine, propionate, and glyoxylate (0, 0.05, and 0.25 mm) was conducted on half-strength Murashige and Skoog medium with 0.8% (w/v) agar in a growth cabinet as described above. Whole rosettes were harvested in the middle of the light period from 3-week-old plants and immediately frozen in liquid nitrogen. Oxidative stress conditions were achieved by watering the plants with a solution of 300 mm NaCl and alternatively by spraying with a solution of 10 μm methyl viologen (Paraquat), and whole rosettes were harvested at different time points after treatment and immediately frozen in liquid nitrogen.
Polyacrylamide Gel Electrophoresis and Protein Determination
In-gel analysis of d-2HGDH enzymatic activity was performed using native PAGE. Buffers and gels were prepared as described previously (12), with the exception that SDS and β-mercaptoethanol were excluded from all buffers. Total leaf protein was prepared by grinding leaf tissue in liquid N2 in the presence of an equal amount (v/w) of 20 mm Tris-HCl (pH 8.0), 0.05% Triton X-100, and 1 mg/ml phenylmethylsulfonyl fluoride. The homogenate was clarified by centrifugation at 4 °C, and the protein concentration was determined according to the method of Bradford (13) using Roti-Quant solution (Roth). Twenty μg of protein was loaded in each well on 8% (w/v) polyacrylamide gels and run in the dark for 2.5 h at 100 V and 4 °C. The gels were analyzed for d-2HGDH activity with 50 mm K2PO4 (pH 8.0), 5 mm d-2HG, 0.05% nitro blue tetrazolium, and 150 μm phenazine methosulfate. Reactions were incubated at 30 °C in the dark for 30 min and stopped by washing with distilled water. To confirm equal loading of all samples, 20 μg of protein was loaded and analyzed after SDS-PAGE by staining with Coomassie Blue.
Measurement of d/l-2HG
d/l-2HG enantiomers were measured by stable isotope dilution LC-MS/MS after derivatization with diacetyl-l-tartaric anhydride (14). To 30 mg of leaf material was added 100 μl of water, and the mixture was mixed thoroughly for 15 min and centrifuged at 20,000 × g for 5 min at 4 °C. Twenty μl of the supernatant was pipetted into a glass vial, and 250 μl of methanol containing 0.004 mmol/liter [2H4]d/l-2HGA was added as an internal standard. The mixture was evaporated to dryness at 50 °C under a gentle stream of nitrogen. Diacetyltartaryl derivative was formed by treating the dry residue with 50 μl of freshly made 50 g/liter diacetyl-l-tartaric anhydride in dichloromethane/acetic acid (4:1) at 75 °C for 30 min. After the vial was cooled to room temperature, the mixture was evaporated to dryness by a nitrogen stream at room temperature. The residue was redissolved in 500 μl of distilled water, and 10 μl of the aqueous solution was injected on the LC column, which was coupled with a triple-quadrupole tandem mass spectrometer operating in the negative multiple reaction-monitoring mode.
As the concentrations of d/l-2HG in the wild-type samples were at the detection limit of the LC-MS/MS method, they were also measured with stable isotope dilution GC-MS (15). To 50 mg of leaf material was added 1 ml of water, and the mixture was mixed thoroughly for 15 min and centrifuged at 20,000 × g for 5 min at 4 °C. The clear supernatant was transferred to a tube, and 5 nmol of [2H4]d/l-2HGA was added. After acidifying the supernatant to pH <2 and saturating with NaCl, the sample was extracted with ethyl acetate/1-propanol (9:1, v/v). The organic layer was evaporated under nitrogen at 30 °C, and the dry residue was derivatized with 500 μl of (R)-(−)-2-butanol and 50 μl of HCl (37%) at 90 °C for 90 min. After cooling to room temperature, 1 ml of water was added to the sample and extracted with 4 ml of hexane. The hexane layer was evaporated under nitrogen, and the residue was derivatized with 100 μl of pyridine and 100 μl of acetic anhydride at 80 °C for 60 min. After cooling to room temperature, the mixture was evaporated under nitrogen, and the residue was redissolved in 50 μl of hexane. One μl was injected on the GC-MS system to quantify d/l-2HGA.
Metabolic Fingerprinting
For metabolite analysis during normal aging, whole rosettes and cauline leaves from 4–7-week-old plants were collected in the middle of the light period and immediately frozen in liquid nitrogen. For metabolite analysis during dark-induced carbon starvation, whole rosettes from 5-week-old plants were collected at the end of a normal night and after 24, 48, and 96 h of extension of the dark period and immediately frozen in liquid nitrogen. For metabolite analysis of plants supplemented with different metabolites, whole rosettes of 3-week-old plants were collected in the middle of the light period and immediately frozen in liquid nitrogen. Four independent biological replicates were used. The tissues were ground in a mortar, and a 50-mg fresh weight aliquot was extracted using the procedure described by Lee and Fiehn (16). Ribitol was used as an internal standard for data normalization. For GC-electron impact-TOF analysis, samples were processed and analyzed according to Lee and Fiehn (16).
Chlorophyll Fluorescence Parameters and Dark Respiratory Rate
In vivo measurements of chlorophyll a fluorescence of the upper leaf surface were performed with a PAM-2000 pulse amplitude modulation chlorophyll fluorometer (Walz GmbH) (17). Basal fluorescence (F0) was measured with modulated weak red light using leaves of plants that were dark-adapted for at least 10 min. Maximal fluorescence in the dark-adapted state (Fm) and during illumination (Fm′) was induced with a saturating white light pulse (5000 μmol m−2 s−1, 0.8-s duration). The FV/FM index was calculated as (Fm − F0)/Fm. The rate of CO2 evolution in the dark was measured according to Tomaz et al. (18) in 6-week-old plants that had been kept in the dark for 4 and 6 days. For each measurement, one fully developed leaf was enclosed in the measuring chamber of a GFS-3000 gas exchange fluorescence system (Walz GmbH) and allowed to adapt to the measurement conditions before the data points were collected. The measurement conditions were constant at 60% relative humidity, 25 °C, 380 ppm CO2, and an air flow rate of 250 μmol s−1.
Real-time RT-PCR
The relative expression levels of d-2HGDH and the senescence marker genes SAG12 and ETFQO were analyzed by real-time RT-PCR. For this purpose, leaf RNA was isolated, and first-strand cDNA synthesis was performed as described above. Subsequently, the cDNA was employed as a template using the fluorescent dye SYBR Green (Applied Biosystems) in a 7300 Real Time PCR system (Applied Biosystems). The primers used were as follows: d-2HGDH-RTfow (5′-TTTCTGATGGTGTAATCGCTC-3′) and d-2HGDH-RTrev (5′-TGCTTTCTGTAACGCCTCTG-3′), SAG12-RTfow (5′-TGCGGTAAATCAGTTTGCTG-3′) and SAG12-RTrev (5′-ACGGCGACATTTTAGTTTGG-3′), ETFQO-RTfow (5′-TGCAGATCAACGCTCAAAAC-3′) and ETFQO-RTrev (5′-ACCTTCAGGCACTGTCCACT-3′), and Actin2-RTfow (5′-ATGGAAGCTGCTGGAATCCAC-3′) and Actin2-RTrev (5′-TTGCTCATACGGTCAGCGATG-3′). The Ct, defined as the PCR cycle at which fluorescence levels significantly higher than the background fluorescence are detected, was used as a measure of the transcript level of the target genes. Relative quantification of expression levels was performed employing the comparative ΔΔCt method using A. thaliana ACT2 gene expression as a standard.
Plasmid Construction and Plant Transformation
To obtain A. thaliana d-2HGDH full-length coding sequence, leaf RNA was isolated, and first-strand cDNA synthesis was performed as described above. Using this cDNA as a template, the d-2HGDH coding sequence, without the stop codon, was amplified by PCR using primers d-2HGDH-CDS-fow (CACCATGATGATGCAGAAATTGAGAAG) and d-2HGDH-CDS-rev (GTTGGAGAAGAGAGAGTGAGGAAG). The PCR product was further cloned into the pENTR/D-Topo vector (Invitrogen) following the manufacturer's instructions and subsequently sequenced. This entry vector was further used in an LR recombination reaction (Invitrogen) to generate the vectors for overexpression (pGWB2-d-2HGDH) and a GFP translational fusion (pGWB5-d-2HGDH).
The plasmid pGWB2-d-2HGDH was introduced into A. thaliana by Agrobacterium tumefaciens (GV3101 strain)-mediated transformation using the vacuum infiltration method (19). Transformants were selected for resistance to kanamycin. DNA was extracted from leaf material collected from selected plants and used for PCR analyses. Plants containing the transgene were allowed to self-pollinate. The process was repeated to obtain non-segregating T3 transgenic lines. All further analyses were performed with homozygous T3 transgenic plants.
Analysis of d-2HGDH-GFP Translational Fusions
pGWB5-d-2HGDH and the mitochondrial control plasmid mt-rb (Invitrogen), containing a fusion of Saccharomyces cerevisiae cytochrome c oxidase IV fused to mCherry (20), were transformed into the A. tumefaciens GV3101 strain using standard protocols. Transformation of Nicotiana benthamiana was performed by injecting recombinant agrobacteria into the apoplast of whole leaves as described previously (21). After subsequently growing the plants for 3 days, protoplasts were prepared following a modified protocol from Sheen (22). Briefly, 10 leaf discs 0.75 mm in diameter were digested in enzyme solution, and protoplasts were pelleted by gravitation, resuspended, and analyzed by confocal laser-scanning microscopy using a Zeiss LSM 700 microscope.
RESULTS AND DISCUSSION
d-2HGDH Activity Increases during Carbon Starvation
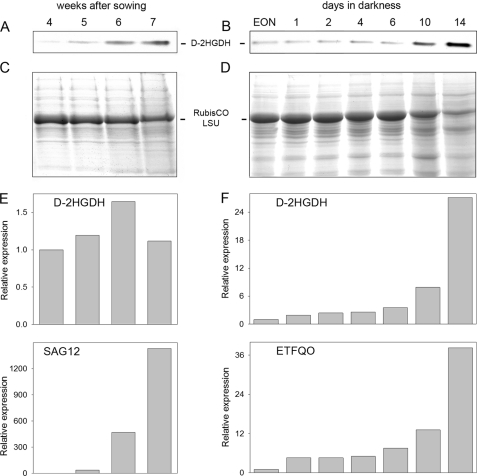
As it was previously suggested that d-2HGDH may participate in the catabolism of alternative substrates from proteolysis or lipid degradation, its activity was analyzed in A. thaliana leaves during developmental aging and during continuous darkness, where carbon starvation induces senescence (23). In-gel activity analysis of wild-type plants indicated that the activity of d-2HGDH increased gradually during natural senescence (Fig. 1A) and after transfer to continuous darkness (Fig. 1B), showing high levels by the end of both processes. The amount of ribulose-bisphosphate carboxylase/oxygenase large subunit decreases in the last weeks of development and after transfer to continuous darkness, reflecting enhanced protein degradation during the progress of senescence (Fig. 1, C and D). During development, transcript levels of d-2HGDH measured by quantitative RT-PCR showed maximal accumulation 1 week before the activity maximum (Fig. 1E), suggesting stability of the d-2HGDH protein during the last week. A constant increase in the d-2HGDH transcript level was observed during dark-induced carbon starvation (Fig. 1F), indicating that the increase in d-2HGDH activity during this process is due to enhanced transcript accumulation. The onset of carbon starvation in these experiments was further verified by the accumulation of transcripts of SAG12, whose expression is most specific for developmental senescence (Fig. 1E) (24) and of the dark-induced senescence-associated gene ETFQO (Fig. 1F) (25). Induction of the expression and higher activity of d-2HGDH in the late stages of development and during dark-induced carbon starvation suggest an increased production of the metabolite d-2HG under these conditions.
FIGURE 1.
d-2HGDH during senescence. Shown are the results from in-gel d-2HGDH activity analysis of wild-type leaf extracts during aging (A) and dark-induced carbon starvation (B). EON, end of night. The levels of ribulose-bisphosphate carboxylase/oxygenase large subunit (RubisCO LSU) analyzed after SDS-PAGE by staining with Coomassie Blue indicate the progress of senescence during aging (C) and dark-induced carbon starvation (D). Representative results of the relative expression of d-2HGDH and SAG12 during aging (E) and of d-2HGDH and ETFQO during dark-induced carbon starvation (F) were obtained by real-time RT-PCR.
In agreement with these results, the expression of the ETF-ETFQO system, which transfers the electrons donated from d-2HGDH to the electron transport chain (1, 11), is also induced during dark-induced starvation (25, 26). As this electron transport system is also induced during oxidative stress (27), we analyzed the activity of d-2HGDH in A. thaliana leaves exposed to agents causing oxidative stress such as NaCl or the redox-cycling herbicide methyl viologen. No differences in d-2HGDH activity were observed under these conditions (supplemental Fig. S1A), suggesting that the production of d-2HG may not be enhanced under these conditions.
d-2HG Accumulates to High Levels during Aging and Dark-induced Carbon Starvation in Plants Lacking d-2HGDH
The T-DNA insertion line d2hgdh1-1 was shown in our previous work to lack all d-2HGDH activity (11). In this work, the homozygous line d2hgdh1-3 was also isolated. d-2HGDH mRNA (data not shown) and activity (see Fig. 4B) were absent in these plants, demonstrating that it is a true knock-out line. Both d2hgdh1-1 and d2hgdh1-3 were used to analyze the participation of d-2HGDH in the catabolism of d-2HG during the mobilization of alternative substrates. For this, metabolic fingerprinting was conducted with wild-type and loss-of-function plants during aging and during the first 4 days of dark-induced carbon starvation.
FIGURE 4.
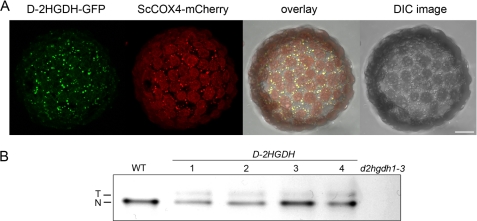
Expression of d-2HGDH. A, microscopic analysis of the subcellular localization of d-2HGDH-GFP and the mitochondrial control ScCOX4-mCherry. The overlay of GFP and mCherry signals is shown on the right. DIC, differential interference contrast. Scale bar = 10 μm. B, leaf extracts of different lines overexpressing d-2HGDH analyzed by in-gel d-2HGDH activity. As controls, leaf extracts of the wild type and d2hgdh1-3 were run in parallel. N, native d-2HGDH; T, transgenic d-2HGDH.
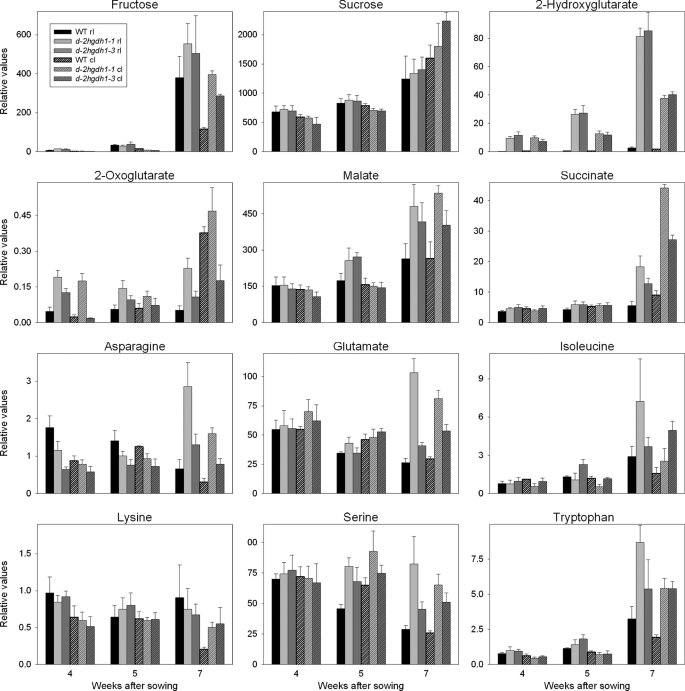
The progress of leaf aging was accompanied by a continuous increase in the levels of 2HG in wild-type and d2hgdh1-1 and d2hgdh1-3 plants (Fig. 2). Although the rate of accumulation was higher in the wild type, the knock-out mutants accumulated 28–45-fold and 18–32-fold higher steady-state levels of 2HG in rosette and cauline leaves, respectively (Fig. 2 and supplemental Tables S1 and S2). Although no differences in the levels of the TCA cycle intermediates citrate and fumarate were obvious between the genotypes, the levels of 2OG, malate, and succinate were generally higher in the last weeks of development in the knock-out mutants (Fig. 2 and supplemental Tables S1 and S2). The levels of many amino acids were significantly elevated in the knock-out mutants in the last week of development (Fig. 2 and supplemental Tables S1 and S2). In correlation with the onset of developmental senescence, the levels of asparagine, aspartate, glutamate, alanine, and serine decreased at later stages of development (24) in the wild type (Fig. 2 and supplemental Tables S1 and S2), whereas in the knock-out mutants, all these amino acids accumulated. On the other hand, the levels of amino acid markers of senescence, such as tryptophan, phenylalanine, and tyrosine, rose with development in all genotypes, but in the knock-out mutants, the markers accumulated to higher levels during the last weeks of development. Aging rosette and cauline leaves from all genotypes harvested in the middle of the light period presented similar levels of most carbohydrates with the exception of the free hexoses fructose and glucose, which accumulated at higher levels in cauline leaves of d2hgdh1 plants (Fig. 2 and supplemental Tables S1 and S2). It is worth mentioning that although all plant genotypes were grown and sampled together, some metabolites showed the same relative change with respect to the wild type but with different intensity, as in the case of 2OG in rosette leaves (Fig. 2), or responded differently in both knock-out lines, as in the case of 2OG in cauline leaves (Fig. 2). The exact mechanisms underlying these minor differences between both knock-out lines cannot be inferred from the results of this study; it cannot be excluded that they are due to positional effects of the different T-DNA insertions in the d-2HGDH gene.
FIGURE 2.
Changes of selected metabolites in rosette and cauline leaves of the wild type, d2hgdh1-1, and d2hgdh1-3 during aging assayed by GC-MS. The metabolite abundance is presented relative to the internal standard (ribitol) and was calculated as the mean ± S.E. of four replicates of eight plants each. Supplemental Tables S1 and S2 show the time points at which the samples were taken and include the results of two-tailed Student's t test (p < 0.05). rl, rosette leaves; cl, cauline leaves.
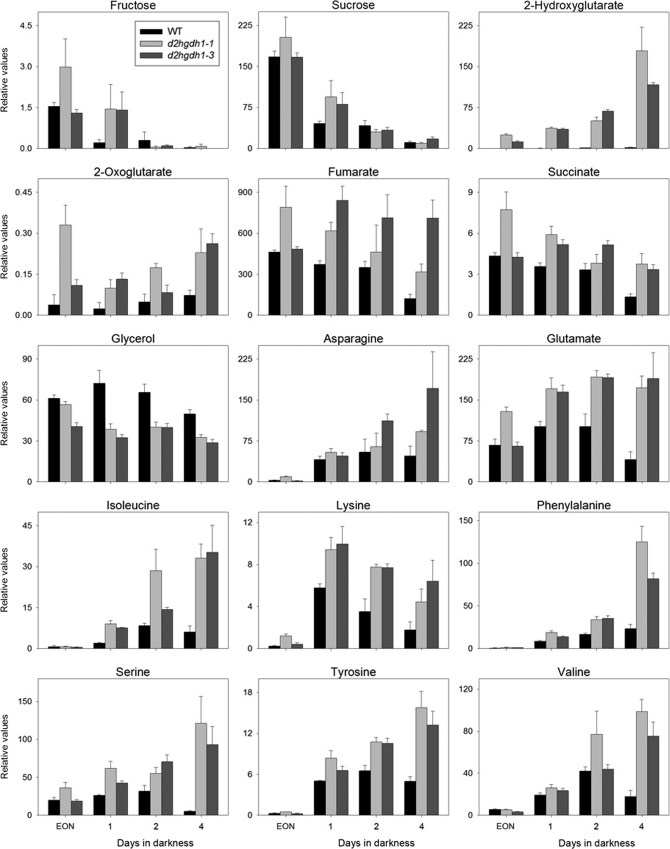
During the course of dark-induced carbon starvation, no differences in sugar metabolism were observed between d2hgdh1-1 and d2hgdh1-3 and the wild type. The levels of mannose, fructose, glucose, and sucrose declined rapidly during extended darkness in all genotypes (Fig. 3 and supplemental Table S3). The TCA cycle intermediates malate, fumarate, succinate, and 2OG were increased after 4 days in extended darkness (Fig. 3 and supplemental Table S3). In the knock-out mutants, there was a substantial accumulation of most amino acids after extension of the normal dark period (Fig. 3 and supplemental Table S3), including those known as markers of senescence (e.g. valine, isoleucine, phenylalanine, tyrosine, and tryptophan). By far the most striking difference between wild-type and knock-out plants was the high levels of 2HG in the knock-out mutants after extension of the dark period (Fig. 3). It is worth mentioning that although the rate of accumulation was higher in the wild type, the relative levels in the knock-out mutants were much higher (Fig. 3 and supplemental Table S3).
FIGURE 3.
Changes of selected metabolites in rosette leaves of the wild type, d2hgdh1-1, and d2hgdh1-3 during dark-induced carbon starvation assayed by GC-MS. The metabolite abundance is presented relative to the internal standard (ribitol) and was calculated as the mean ± S.E. of four replicates of four plants each. Supplemental Table S3 shows the time points at which the samples were taken and includes the results of two-tailed Student's t test (p < 0.05). EON, end of night.
The high accumulation of 2HG in leaves of d2hgdh1 during the course of carbon starvation was accompanied by enhanced levels of most TCA cycle intermediates and free amino acids. The question arises as to what causes this accumulation in plants lacking d-2HGDH during both developmental and dark-induced senescence. It may be due to an early onset of protein degradation in the d2hgdh1 mutants and may explain the higher levels of TCA cycle intermediates, as their withdrawal for anabolic processes under carbon starvation should be damped down. Interestingly, in humans, the deficiency of d-2HGDH is also accompanied by a moderate elevation of the levels of TCA cycle intermediates (28). In mammalian systems, d-2HG was found to cause a dose-dependent inhibition of cytochrome c oxidase activity, which may inhibit the respiratory chain in vivo and may explain the observed accumulation of TCA cycle intermediates. It is also conceivable that d-2HG may be mildly toxic to plant cells by competitively inhibiting glutamate and/or 2OG using enzymes, including transaminases (3) and/or dicarboxylic acid transporters (29). In this way, the inhibition of transaminases would explain the observed accumulation of amino acids. On the other hand, it is possible that after the onset of carbon starvation, the inability to degrade d-2HG triggers a signal that induces the accelerated use of alternative substrates for respiration. This signal could be the accumulation of d-2HG or some other intermediate of the pathway(s) producing d-2HG alone or in combination with low sugar levels.
As it cannot be excluded that not only d-2HG but also l-2HG accumulates in plant tissues, the levels of d- and l-2HG were measured by stable isotope dilution LC-MS/MS and by GC-MS after derivatization with (R)-butanol. As shown in Table 1, d-2HG accumulated during the progress of dark-induced senescence, whereas the levels of l-2HG were similar in all genotypes (0.02–0.05 nmol/mg) and invariant during the progress of carbon starvation. These results not only confirm that d-2HG is the enantiomeric form of 2HG that accumulates in wild-type and d2hgdh1 plants but also show for the first time that l-2HG is also present in plant tissues.
TABLE 1.
Levels of d-2HG in rosette leaves of wild-type and d2hgdh1 lines
The concentration of d-2HG was measured by stable isotope dilution LC-MS/MS during the course of dark-induced carbon starvation. EON, end of night.
|
d-2HG |
|||
|---|---|---|---|
| WT | d2hgdh1-1 | d2hgdh1-3 | |
| nmol/mg | |||
| EON | 0.004 ± 0.001 | 0.176 ± 0.056 | 0.242 ± 0.030 |
| 4 days in darkness | 0.025 ± 0.005 | 0.722 ± 0.100 | 0.528 ± 0.108 |
| 10 days in darkness | 0.110 ± 0.001 | 1.728 ± 0.330 | 1.714 ± 0.244 |
The metabolite quantification showed that during the progress of dark-induced senescence, A. thaliana wild-type plants accumulated d-2HG. Increments in the levels of d-2HG of 6-fold and 27-fold after 4 and 10 days in darkness in wild-type plants, together with the fact that the activity of d-2HGDH increased gradually after transfer to continuous darkness (Fig. 1A), indicate an enhanced demand of respiration of alternative substrates through this enzymatic system under these conditions. Moreover, this study indicates that wild-type plants produced d-2HG at higher rates than the knock-out mutants during the progress of dark-induced carbon starvation and that the inability of the knock-out mutants to convert d-2HG to 2OG is the cause of its high accumulation.
Plant Cells Can Cope with High Levels of d-2HG
The early onset of metabolic symptoms of carbon starvation observed in d2hgdh1-1 and d2hgdh1-3 plants is mild enough to cause no visible changes in the onset of senescence symptoms in these plants compared with the wild type under all experimental conditions tested. This correlated with (i) a similar decline in the maximal quantum efficiency of Photosystem II primary photochemistry (FV/FM ratio) of the knock-out plants with regard to the wild type during the course of senescence (supplemental Fig. S1B) and (ii) no changes in the respiratory rate of the knock-out plants with regard to the wild type late in senescence, a condition in which 2HGDH accumulates (data not shown).
The affinity of human d-2HGDH for d-2HG is high (Km < 10 μm), and the concentration of d-2HG in plasma of healthy humans is ∼1 μm. The importance of this enzyme in maintaining low d-2HG levels is illustrated by the fact that a serum concentration of 26 μm d-2HG is enough to cause severe developmental impairment in patients with d-2HGA (1). In contrast, the Km of A. thaliana d-2HGDH is ∼500 μm (11), indicating that plant tissues have higher steady-state levels of d-2HG. Taking into consideration the data from Table 1 and that cell water content is ∼90%, the levels of d-2HG in A. thaliana wild-type leaves, if homogenously distributed, would be at least 4.4 μm by the end of a normal night and 122 μm after 10 days in darkness. On the other hand, the levels of d-2HG in leaves of knock-out plants would reach values of at least 200 μm and 1.9 mm by the end of the night and after 10 days in darkness, respectively. However, most of the cellular d-2HG is likely concentrated in the mitochondria, and the in vivo concentrations of this substrate would thus be even higher. This, together with the fact that the phenotype of d2hgdh1 plants does not differ from that of the wild type, indicates that plants can cope with much higher levels of d-2HG than mammals.
d-2HGDH Localizes to Mitochondria, and Its Enhanced Expression Lowers the Accumulation of d-2HG
In silico analysis of the subcellular localization indicates that A. thaliana D-2HGDH contains an N-terminal sequence that should direct the protein to mitochondria (30). Furthermore, we have previously shown that cellular fractions enriched in mitochondria have high levels of d-2HGDH activity (11). To experimentally confirm the localization of d-2HGDH in vivo, tobacco plants were infiltrated with an expression construct carrying the full-length cDNA of A. thaliana d-2HGDH in a translational fusion to GFP. Microscopic analysis of protoplasts prepared from infiltrated leaves indicated that the green fluorescence of GFP perfectly matched the red fluorescence of the mitochondrial control targeting signal of Saccharomyces cerevisiae cytochrome c oxidase IV fused to mCherry (ScCOX4-mCherry) (20), confirming the mitochondrial localization of d-2HGDH (Fig. 4A). Here, it is worth mentioning that d-2HGDH has already been found as a mitochondrial protein in both Arabidopsis leaves and cell cultures in proteomic studies using mass spectrometry (31). In this study, At4g36400 was described as a glycolate dehydrogenase because of its close relatedness to At5g06580, which has been previously annotated as a glycolate dehydrogenase (32). However, At5g06580 was subsequently shown to be a d-lactate dehydrogenase, which participates in the methylglyoxal pathway (11).
To analyze the influence of a higher d-2HGDH activity during the progress of developmental and dark-induced senescence, non-segregating T3 transgenic lines overexpressing d-2HGDH were produced. Total leaf extracts of selected lines contained an additional protein band with d-2HGDH activity corresponding to the transgenic enzyme (Fig. 4B). The lower mobility of the overexpressed enzyme is due to the fact that this protein contains extra amino acids at the C-terminal region because, for its expression, the stop codon of the binary vector was used. Stable isotope dilution LC-MS/MS measurements indicated lower levels of d-2HG in the overexpressing plants (0.019–0.067 nmol/mg) than in the wild type (0.110 nmol/mg) after 10 days in darkness, reflecting the higher d-2HGDH activity in these plants. d-2HGDH-overexpressing lines did not show any morphological alteration during normal growth or changes in the senescence symptoms with regard to wild-type plants during the progress of developmental or dark-induced senescence (data not shown). These results indicate that the comparatively high d-2HG levels present in wild-type plants is in no way detrimental, despite being much higher than the levels detected in mammals.
d-2HGDH Is Involved in the Catabolism of d-2HG Derived from Lysine
The continuous accumulation of d-2HG in the knock-out plants during developmental senescence and after the onset of carbon starvation in continuous darkness undoubtedly indicates that d-2HGDH participates in the catabolism of d-2HG during the mobilization of substrates from proteolysis and/or lipid degradation. This is in agreement with previous results on gene coexpression analysis that indicated that the expression of d-2HGDH parallels the that of genes involved in β-oxidation and the degradation of valine, leucine, and lysine (11, 33). Moreover, the bifunctional enzyme lysine ketoglutarate reductase/saccharopine dehydrogenase, containing the first two linked enzymes of the lysine catabolic pathway, is expressed primarily in senescing tissues and under conditions of sugar starvation, in which amino acids are converted into sugars (34). In this study, to experimentally test this hypothesis, A. thaliana wild-type and d2hgdh1-1 and d2hgdh1-3 plants were grown in the presence of putative precursors of d-2HG, and metabolic fingerprinting was conducted with whole rosettes. Similar levels of d-2HG were found in all genotypes in the presence of different concentrations of propionate, glyoxylate, valine, or isoleucine (data not shown), whereas a higher accumulation was determined in the knock-out plants grown on medium supplemented with lysine (supplemental Table S4). In plants supplemented with 0.05 mm lysine, relative levels of d-2HG of 0.23, 25, and 14 were found in the wild type, d2hgdh1-1, and d2hgdh1-3, respectively, whereas plants supplemented with 0.25 mm lysine showed relative levels of d-2HG of 0.33, 105, and 102 in the wild type, d2hgdh1-1, and d2hgdh1-3, respectively. Thus, d-2HG levels in leaves of knock-out plants positively correlated with the concentration of lysine in the medium, strongly indicating that the absence of d-2HGDH represents a bottleneck in the catabolism of this amino acid. Although a higher accumulation of this lysine was not evident in the knock-out plants with regard to the wild type, it is possible that some intermediates of the lysine catabolic pathway may accumulate in these plants.
Metabolic analysis of plants growing in media supplemented with different substrates indicated that lysine is the most probable substrate generating d-2HG in vivo, as d-2HG levels in leaves of knock-out plants positively correlated with the concentration of lysine in the media. In agreement with this, a recent report on isotope tracer experiments performed with labeled lysine showed the accumulation of lysine breakdown products in isovaleryl-CoA dehydrogenase and d-2HGDH loss-of-function plants (33). The functional redundancy of these two enzymes for lysine breakdown may also explain the fact that d-2HGDH knock-out mutants did not show apparent developmental or senescence phenotypes, as isovaleryl-CoA dehydrogenase may in part complement the lack of d-2HGDH activity. In this way, d-2HGDH may play a minor role in plant metabolism, in contrast to its counterpart in humans.
Concluding Remarks
Despite the high similarity in the catabolic pathway of d-2HG in plant and mammalian mitochondria, there are fundamental differences both in the origin of this metabolite and in how these organisms can cope with high levels of it. The analysis of d-2HGDH loss-of-function and overexpressor plants performed here indicated that changes in d-2HG levels have a mild impact on other plant metabolite pool sizes. Moreover, the higher tolerance of plant tissues to d-2HG, as compared with mammals, suggests that this metabolite may not be a compound with broad toxic effects, but rather that it may be specifically toxic to certain enzymes or transporters. Further studies of these differences in tolerance and comparison of the activity of proteins that may be inhibited by d-2HG will provide important insights into the mode of toxicity of d-2HG.
Supplementary Material
Acknowledgments
We thank Frederique Breuers and Katja Wester for help with subcellular localization analysis.
This work was supported by Deutsche Forschungsgemeinschaft grants (to V. G. M. and A. P. M. W.) as part of the German Photorespiration Research Network Promics.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1–S4.
- 2HG
- 2-hydroxyglutarate
- 2HGA
- 2-hydroxyglutaric aciduria
- d-2HGDH
- d-2-hydroxyglutarate dehydrogenase
- 2OG
- 2-oxoglutarate
- ETF
- electron transfer protein
- ETFQO
- ETF-ubiquinone oxidoreductase.
REFERENCES
- 1. Struys E. A., Salomons G. S., Achouri Y., Van Schaftingen E., Grosso S., Craigen W. J., Verhoeven N. M., Jakobs C. (2005) Am. J. Hum. Genet. 76, 358–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Frerman F. E., Goodman S. I. (2001) in The Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Beaudet A. L., Valle D., Sly W. S., Childs B., Kinzler K. W., Vogelstein B. eds) 8th Ed., McGraw-Hill Medical Publishing Division, New York [Google Scholar]
- 3. Dang L., White D. W., Gross S., Bennett B. D., Bittinger M. A., Driggers E. M., Fantin V. R., Jang H. G., Jin S., Keenan M. C., Marks K. M., Prins R. M., Ward P. S., Yen K. E., Liau L. M., Rabinowitz J. D., Cantley L. C., Thompson C. B., Vander Heiden M. G., Su S. M. (2009) Nature 462, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaufman E. E., Nelson T., Miller D., Stadlan N. (1988) J. Neurochem. 51, 1079–1084 [DOI] [PubMed] [Google Scholar]
- 5. Tubbs P. K., Greville G. D. (1961) Biochem. J. 81, 104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Achouri Y., Noël G., Vertommen D., Rider M. H., Veiga-Da-Cunha M., Van Schaftingen E. (2004) Biochem. J. 381, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao G., Winkler M. E. (1996) J. Bacteriol. 178, 232–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wegener W. S., Reeves H. C., Ajl S. J. (1968) Arch. Biochem. Biophys. 123, 62–65 [DOI] [PubMed] [Google Scholar]
- 9. Kopchick J. J., Hartline R. A. (1979) J. Biol. Chem. 254, 3259–3263 [PubMed] [Google Scholar]
- 10. Albers E., Gustafsson L., Niklasson C., Lidén G. (1998) Microbiology 144, 1683–1690 [DOI] [PubMed] [Google Scholar]
- 11. Engqvist M., Drincovich M. F., Flügge U. I., Maurino V. G. (2009) J. Biol. Chem. 284, 25026–25037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 13. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 14. Struys E. A., Jansen E. E., Verhoeven N. M., Jakobs C. (2004) Clin. Chem. 50, 1391–1395 [DOI] [PubMed] [Google Scholar]
- 15. Gibson K. M., ten Brink H. J., Schor D. S., Kok R. M., Bootsma A. H., Hoffmann G. F., Jakobs C. (1993) Pediatr. Res. 34, 277–280 [DOI] [PubMed] [Google Scholar]
- 16. Lee D. Y., Fiehn O. (2008) Plant Methods 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schreiber U., Schliwa U., Bilger W. (1986) Photosynth. Res. 10, 51–62 [DOI] [PubMed] [Google Scholar]
- 18. Tomaz T., Bagard M., Pracharoenwattana I., Lindén P., Lee C. P., Carroll A. J., Ströher E., Smith S. M., Gardeström P., Millar A. H. (2010) Plant Physiol. 154, 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bechtold N., Ellis J., Pelletier G. (1993) Compt. Rendus Acad. Sci. Ser. III Sci. Vie 316, 1194–1199 [Google Scholar]
- 20. Nelson B. K., Cai X., Nebenführ A. (2007) Plant J. 51, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 21. Wydro M., Kozubek E., Lehmann P. (2006) Acta Biochim. Pol. 53, 289–298 [PubMed] [Google Scholar]
- 22. Sheen J. (2001) Plant Physiol. 127, 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- 23. Fahnenstich H., Saigo M., Niessen M., Zanor M. I., Andreo C. S., Fernie A. R., Drincovich M. F., Flügge U. I., Maurino V. G. (2007) Plant Physiol. 145, 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diaz C., Purdy S., Christ A., Morot-Gaudry J. F., Wingler A., Masclaux-Daubresse C. L. (2005) Plant Physiol. 138, 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishizaki K., Larson T. R., Schauer N., Fernie A. R., Graham I. A., Leaver C. J. (2005) Plant Cell 17, 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P. O., Nam H. G., Lin J. F., Wu S. H., Swidzinski J., Ishizaki K., Leaver C. J. (2005) Plant J. 42, 567–585 [DOI] [PubMed] [Google Scholar]
- 27. Lehmann M., Schwarzländer M., Obata T., Sirikantaramas S., Burow M., Olsen C. E., Tohge T., Fricker M. D., Møller B. L., Fernie A. R., Sweetlove L. J., Laxa M. (2009) Mol. Plant 2, 390–406 [DOI] [PubMed] [Google Scholar]
- 28. Struys E. A. (2006) J. Inherit. Metab. Dis. 29, 21–29 [DOI] [PubMed] [Google Scholar]
- 29. Struys E. A., Gibson K. M., Jakobs C. (2007) J. Inherit. Metab. Dis. 30, 690–693 [DOI] [PubMed] [Google Scholar]
- 30. Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W. B., Flügge U. I., Kunze R. (2003) Plant Physiol. 131, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee C. P., Eubel H., O'Toole N., Millar A. H. (2008) Mol. Cell. Proteomics 7, 1297–1316 [DOI] [PubMed] [Google Scholar]
- 32. Bari R., Kebeish R., Kalamajka R., Rademacher T., Peterhänsel C. (2004) J. Exp. Bot. 55, 623–630 [DOI] [PubMed] [Google Scholar]
- 33. Araújo W. L., Ishizaki K., Nunes-Nesi A., Larson T. R., Tohge T., Krahnert I., Witt S., Obata T., Schauer N., Graham I. A., Leaver C. J., Fernie A. R. (2010) Plant Cell 22, 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stepansky A., Less H., Angelovici R., Aharon R., Zhu X., Galili G. (2006) Amino Acids 30, 121–125 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.