Abstract
Although the yeast Saccharomyces cerevisiae has only one sphingolipid class with a head group based on phosphoinositol, the yeast Pichia pastoris as well as many other fungi have a second class, glucosylceramide, which has a glucose head group. These two sphingolipid classes are in addition distinguished by a characteristic structure of their ceramide backbones. Here, we investigate the mechanisms controlling substrate entry into the glucosylceramide branch of the pathway. By a combination of enzymatic in vitro studies and lipid analysis of genetically engineered yeast strains, we show that the ceramide synthase Bar1p occupies a key branching point in sphingolipid biosynthesis in P. pastoris. By preferring dihydroxy sphingoid bases and C16/C18 acyl-coenzyme A as substrates, Bar1p produces a structurally well defined group of ceramide species, which is the exclusive precursor for glucosylceramide biosynthesis. Correlating with the absence of glucosylceramide in this yeast, a gene encoding Bar1p is missing in S. cerevisiae. We could not successfully investigate the second ceramide synthase in P. pastoris that is orthologous to S. cerevisiae Lag1p/Lac1p. By analyzing the ceramide and glucosylceramide species in a collection of P. pastoris knock-out strains in which individual genes encoding enzymes involved in glucosylceramide biosynthesis were systematically deleted, we show that the ceramide species produced by Bar1p have to be modified by two additional enzymes, sphingolipid Δ4-desaturase and fatty acid α-hydroxylase, before the final addition of the glucose head group by the glucosylceramide synthase. Together, this set of four enzymes specifically defines the pathway leading to glucosylceramide biosynthesis.
Keywords: Gene Knock-out, Glycolipid Structure, Glycolipids, Lipid, Mass Spectrometry (MS), Biosynthetic Pathway, Ceramide, Ceramide Synthase, Glucosylceramide, Sphingolipid
Introduction
In eukaryotic cells, sphingolipids are essential constituents of the plasma membrane as well as of intracellular membranes (1). In addition, intermediates of their biosynthesis and breakdown are important signaling molecules (2, 3). Two classes of complex sphingolipids can be distinguished based on the nature of their polar head groups: phosphosphingolipids and glycosphingolipids. Phosphosphingolipids carry hydrophilic head groups connected to the ceramide (Cer)6 backbone via a phosphodiester bond, whereas glycosphingolipids carry sugar residues directly linked to the ceramide backbone via a glycosidic bond. The typical phosphosphingolipid in mammalian cells is sphingomyelin with a phosphocholine head group, whereas the phosphosphingolipids of plants and fungi have head groups based on phosphoinositol. The simplest compound of the latter class is inositol phosphorylceramide (IPC), but often sugar residues and additional phosphoinositol units are added to form glycosylated IPC (GIPC). Glycosylceramides, in contrast, are found in all eukaryotic kingdoms. They can be based on either glucosylceramide (GlcCer) or galactosylceramide (GalCer) with GlcCer predominating in plants and fungi (4–6). Particularly in animals, both GlcCer and GalCer may carry extended, branched, and highly variable glycan head groups (7).
Important insights into sphingolipid metabolism and functions came from studies on the yeast Saccharomyces cerevisiae (8). However, this yeast is an exception among eukaryotic organisms because it contains only one class of complex sphingolipids, namely (G)IPCs, whereas most other fungi contain both (G)IPCs and glycosylceramides (4, 9). Accordingly, the genes encoding enzymes specific to glycosylceramide biosynthesis (see Fig. 1) are missing in the S. cerevisiae genome. To investigate the biosynthesis of GlcCer, we have chosen the yeast Pichia pastoris as an alternative model organism because it produces both sphingolipid classes, and its recently sequenced genome encodes the full set of enzymes required for their biosynthesis (10, 11).
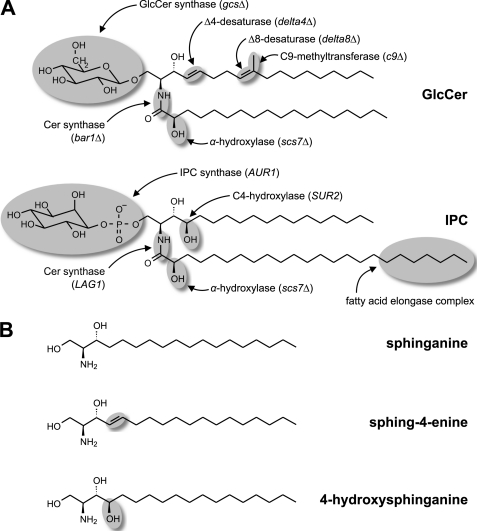
FIGURE 1.
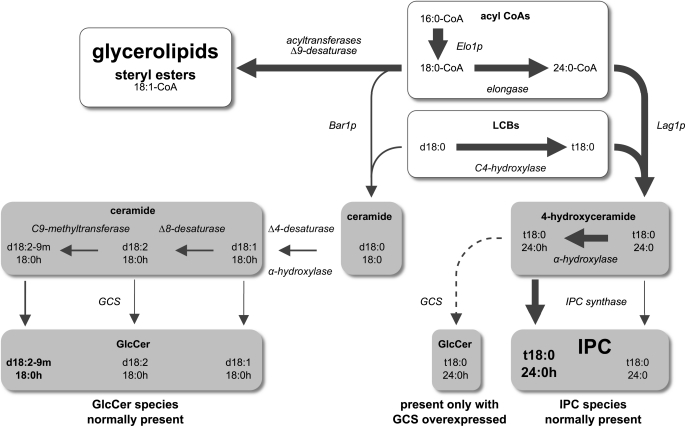
Fungi have two sphingolipid classes. A, predominant molecular species of GlcCer and IPC in P. pastoris with the structural features distinguishing the two sphingolipid classes highlighted. In addition, the enzymes introducing these structural features as well as the corresponding gene names or, where applicable, the names of the knock-out strains used in this study are given. B, commercially available LCBs used in the in vitro enzyme assays shown in Fig. 5 and supplemental Fig. S3. Lipid names are according to the recommendations of the International Lipid Classification and Nomenclature Committee (47). Sphinganine (commonly called dihydrosphingosine) is the initial LCB that serves as precursor for the biosynthesis of the other LCBs. Sphing-4-enine (sphingosine) is representative of the desaturated LCBs found in GlcCer, whereas 4-hydroxysphinganine (phytosphingosine) is the typical LCB of (G)IPCs.
As illustrated in Fig. 1, GlcCer and (G)IPCs in fungi differ not only by the nature of their polar head groups, glucose or (glycosyl)phosphoinositol, but also by the structure of their Cer backbones (4, 9). These include characteristic differences found in both the long-chain (sphingoid) base (LCB) and the amide-linked acyl group. The final structure of the LCBs found in GlcCer is defined by the enzymes introducing Δ4 and Δ8 double bonds as well as a C9-methyl group into Cer (12–14), whereas in (G)IPCs, the LCB typically carries a hydroxy group at the C4 position (15, 16). A common modification of the amide-linked acyl groups of both GlcCer and (G)IPCs is their α-hydroxylation by the fatty acid α-hydroxylase Scs7p (15). A characteristic feature distinguishing the fatty acids in GlcCer and (G)IPCs is their chain length. Although GlcCer typically has a C16 or C18 long-chain fatty acid, (G)IPCs are characterized by a very long-chain fatty acid with 24 or 26 carbons. Elongation of long-chain to very long-chain fatty acids is performed at the level of acyl-coenzyme A (CoA) by the fatty acid elongase complex.
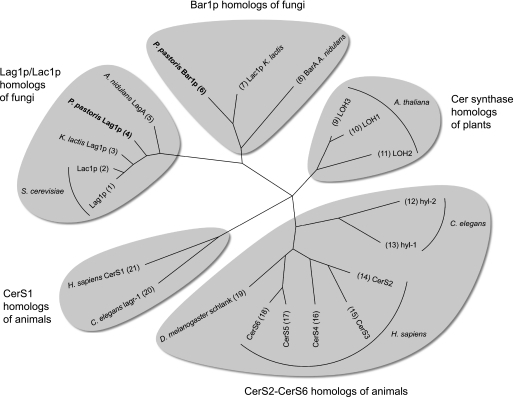
Despite the fact that all enzymes responsible for the biosynthesis of GlcCer and IPC including their specific backbone structures are known and the corresponding genes have been cloned, it is still an open question how these enzymes integrate into specific pathways leading to just two structurally well defined end products, GlcCer and IPC. In this study, we investigate the hypothesis that the separation between these two pathways is initiated by the activity of Cer synthases, which link an LCB and a CoA-activated fatty acid. The phylogenetic tree in Fig. 2 shows that many eukaryotic organisms have several Cer synthases, suggesting that individual isoforms may have evolved functional specializations. This has been investigated more closely in animal cells where Cer synthases differ regarding their preferred acyl-CoA chain length as well as in their physiological functions (17, 18).
FIGURE 2.
Most animals, plants, and fungi have multiple Cer synthases that fall into distinct phylogenetic groups. S. cerevisiae has two Cer synthases, Lag1p and Lac1p, which share a high degree of sequence identity (73%). In contrast, most fungi have two Cer synthases with a much lower degree of sequence identity (≈30%) that fall into distinct phylogenetic groups. In the text, we refer to the enzymes orthologous to S. cerevisiae Lag1p/Lac1p as Lag1p (for longevity assurance gene; Ref. 50) and to the enzymes from the phylogenetic group that is missing in S. cerevisiae as Bar1p (for biocontrol agent resistance; Ref. 51). From the latter group, A. nidulans BarA and K. lactis Lac1p (the name is misleading because this enzyme in not an ortholog of S. cerevisiae Lac1p) were investigated previously (25, 51). It is evident that, with the exception of the six mammalian Cer synthases (52), there is no consensus regarding gene nomenclature. A systematic nomenclature for Cer synthases from all organisms is urgently needed. The phylogenetic tree was constructed using Tree Puzzle (53) from an alignment of full-length protein sequences generated with T-Coffee (54). Only genes that have been investigated experimentally are depicted. UniProt identifiers are as follows: 1, P38703; 2, P28496; 3, Q6CVA7; 4, C4QWW1; 5, Q5BAG6; 6, C4R2K3; 7, Q6CP21; 8, Q5B548; 9, Q6NQI8; 10, Q9LDF2; 11, Q9LJK3; 12, Q7Z139; 13, Q9GYR9; 14, Q96G23; 15, Q8IU89; 16, Q9HA82; 17, Q8N5B7; 18, Q6ZMG9; 19, Q9W423; 20, Q9XWE9; 21, P27544. A. thaliana, Arabidopsis thaliana; H. sapiens, Homo sapiens; C. elegans, Caenorhabditis elegans; D. melanogaster; Drosophila melanogaster.
To investigate how substrate entry into the pathway for GlcCer biosynthesis is controlled, we combined in vitro investigations of the P. pastoris Cer synthase Bar1p with lipid analysis of a collection of P. pastoris strains in which the enzymes involved in GlcCer biosynthesis were systematically deleted (Fig. 1 and Table 1). It turned out that efficient GlcCer biosynthesis requires the introduction of specific functional groups into the Cer species produced by the Cer synthase Bar1p. Altogether, this defines a set of four enzymes constituting the core pathway for GlcCer biosynthesis.
TABLE 1.
Yeast strains used in this study
| Organism | Strain | Genotype | Ref. |
|---|---|---|---|
| S. cerevisiae | WBY616-LAG1 (RH6602) | ade2-1 his3-11 leu2-3–112 trp1-1 ura3-1 can1-100 lag1::HIS3 lac1::ADE2 pRS416-LAG1 | 48 |
| S. cerevisiae | WBY616-BAR1 | ade2-1 his3-11 leu2-3–112 trp1-1 ura3-1 can1-100 lag1::HIS3 lac1::ADE2 Yiplac204GPD-BAR1 | This study |
| P. pastoris | GS115 | his4 | Invitrogen |
| P. pastoris | PPY12 | his4 arg4 | 49 |
| P. pastoris | GS115-bar1Δ | his4 bar1::NAT1 | This study |
| P. pastoris | PPY12-bar1Δ | his4 arg4 bar1::NAT1 | This study |
| P. pastoris | gcsΔ | his4 gcs::Sh_ble | 24 |
| P. pastoris | scs7Δ | his4 scs7::Sh_ble | This study |
| P. pastoris | delta4Δ | his4 Δ4_desaturase::Sh_ble | 29 |
| P. pastoris | delta8Δ | his4 Δ8_desaturase::Sh_ble | This study |
| P. pastoris | c9Δ | his4 C9_methyltransferase::Sh_ble | 14 |
| P. pastoris | gcsΔGCS | his4 gcs::Sh_ble pPIC3.5-GCS | This study |
| P. pastoris | delta4ΔGCS | his4 Δ4_desaturase::Sh_ble pPIC3.5-GCS | This study |
| P. pastoris | scs7ΔGCS | his4 scs7::Sh_ble pPIC3.5-GCS | This study |
EXPERIMENTAL PROCEDURES
Yeast Strains
The construction of an S. cerevisiae strain expressing P. pastoris Bar1p under the control of the constitutive GPD1 promoter, of the P. pastoris knock-out strains shown in Fig. 1, and of the P. pastoris strains overexpressing GlcCer synthase (GCS) is described in the supplemental methods. Genotypes and references for these strains are listed in Table 1.
Yeast Culture and Lipid Extraction for Analysis of Cer and GlcCer
S. cerevisiae strains WBY616-LAG1 and WBY616-BAR1 as well as P. pastoris strains with GS115 background were grown in liquid YPD medium (1% yeast extract, 2% peptone, 2% glucose; all w/v). P. pastoris cells overexpressing GCS (strains gcsΔGCS, delta4ΔGCS, and scs7ΔGCS) were switched to MM medium (1.34% yeast nitrogen base, 0.4 μg/ml biotin, 0.5% methanol; all w/v) 24 h before harvesting to induce the AOX1 promoter. In the case of P. pastoris, cells from 100 ml of culture were harvested by centrifugation, washed with water, weighed to determine the fresh weight, resuspended, and heated in a boiling water bath for 15–20 min to inactivate lipid-metabolizing enzymes. In the case of S. cerevisiae, growth of a 100-ml culture was stopped by adding 5 g of trichloroacetic acid and incubating on ice for 15 min. The cells were harvested by centrifugation and washed twice with phosphate-buffered saline (137 mm NaCl, 2.7 mm KCl, 101 mm Na2HPO4, 17.6 mm KH2PO4) before heating in a boiling water bath for 10 min. The boiled cells were sedimented by centrifugation and resuspended in 8 (P. pastoris) or 9 ml (S. cerevisiae) of chloroform/methanol (1:2, v/v). To allow quantification of Cer and GlcCer, 15 nmol of Cer containing either a C17 fatty acid (S. cerevisiae) or a C15 fatty acid (P. pastoris) and 15 nmol of GlcCer containing a C12 fatty acid (P. pastoris only) were added as internal standards. After shaking at 4 °C for 4–5 h, the cells were sedimented, the supernatant was exchanged for chloroform/methanol (2:1, v/v), and the shaking was continued overnight. The supernatants from both extractions were combined and filtered through cotton to remove cell debris (P. pastoris only). A phase separation was induced by adding 8 (P. pastoris) or 9 ml (S. cerevisiae) of chloroform and 6 (P. pastoris) or 6.75 ml (S. cerevisiae) of 0.45% NaCl (w/v), vortexing, and centrifuging (19). The lower phase was transferred to a glass tube, the upper phase was extracted a second time with 16 (P. pastoris) or 18 ml (S. cerevisiae) of chloroform, and the solvent was evaporated under a stream of nitrogen.
Mild Alkaline Hydrolysis of Lipid Extracts
To remove glycerolipids, the dried lipid extracts were dissolved in 2 ml of 0.2 m NaOH in methanol and heated to 40 °C for 90 min. Phase separation was induced by adding 4 ml of chloroform and 1.5 ml of 0.45% NaCl (w/v), vortexing, and centrifuging (19). The lower phase was transferred to a glass tube, the upper phase was extracted a second time with 4 ml of chloroform, and the solvent was evaporated under a stream of nitrogen.
Fractionation of Lipid Extract
Before first use, a 100-mg/1-ml Strata SI-1 silica cartridge (Phenomenex, Torrance, CA) was flushed with 4 ml of chloroform, 4 ml of acetone/2-propanol (9:1, v/v), and 2 ml of methanol and then equilibrated with 1 ml of chloroform. The dried lipid extract was dissolved in 1 ml of chloroform and loaded onto the cartridge. The lipids were eluted as three separate fractions with 2 ml of chloroform, 4 ml of acetone/2-propanol (9:1, v/v), and 2 ml of methanol. The acetone/2-propanol fraction was evaporated under a stream of nitrogen, dissolved in chloroform/methanol (5:1, v/v), and stored at 4 °C until analysis by ultraperformance LC/MS (UPLC/MS). This fraction was shown by thin-layer chromatography to contain both Cer and GlcCer.
Analysis of Cer and GlcCer by UPLC/MS
The molecular species of Cer and GlcCer present in the acetone/2-propanol fraction were separated on an ACQUITY UPLCTM system coupled to an LCT PremierTM electrospray ionization time-of-flight mass spectrometer analyzer (Waters Corp., Milford, MA). Chromatography was performed on an ACQUITY UPLC BEH Shield RP18 column (1 × 100 mm; particle size, 1.7 μm; Waters Corp.) at a temperature of 50 °C and a flow rate of 0.2 ml/min. The Cer and GlcCer species were eluted under the following conditions: 80% solvent B for 0.5 min followed by a gradient from 80 to 100% solvent B in 6.5 min and finally 100% solvent B for 4 min. The column was re-equilibrated at 80% solvent B for 4 min. Solvent A was water/methanol/acetonitrile (90:5:5, v/v/v), and solvent B was acetonitrile; 0.1% formic acid was added to both solvents to facilitate ionization.
Mass spectra in the range from 500 to 1000 Da with a mass resolution of >104 were acquired by electrospray ionization-TOF-MS in positive ionization mode using “W” optics and dynamic range enhancement with a scan time of 0.5 s and an interscan delay of 0.1 s. The capillary and cone voltages were maintained at 2700 and 30 V, respectively, and the desolvation and source temperatures were maintained at 250 and 80 °C, respectively. Nitrogen was used as cone (30 liters/h) and desolvation gas (600 liters/h). For exact mass measurement of >5-ppm root mean squared, all analyses were monitored using leucine enkephalin (m/z = 556.2771; Sigma-Aldrich) and its double 13C isotopomer (m/z = 558.2828) as the lock spray reference compound at a concentration of 0.5 μg/ml in acetonitrile/water (1:1, v/v) at a flow rate of 30 μl/min (515 HPLC pump, Waters Corp.). Data were recorded in centroid format and analyzed using MassLynx software (Waters Corp.).
Identity of Cer and GlcCer Species
The retention times of Cer species produced in the in vitro Cer synthase assay with single substrates (supplemental Table S1) were used as references for the identification of Cer species in the lipid extracts from P. pastoris and S. cerevisiae. It was found that elongation of the fatty acid by two carbon atoms increases the retention time by ≈1.0 min. Concerning the functional groups on LCB and fatty acid, introduction of one double bond into the LCB decreases the retention time by ≈0.4 min, of an α-hydroxy group at the fatty acid decreases the retention time by ≈0.6 min, and of a C4-hydroxy group at the LCB decreases the retention time by ≈1.1 min. Thus, Cer species with a trihydroxy LCB and a non-hydroxylated fatty acid elute before isobaric species with a dihydroxy LCB and an α-hydroxylated fatty acid. This was supported by the lack of isomers with the longer retention times (representing the α-hydroxylated species) in the P. pastoris strain scs7Δ. This pattern was found to be consistent throughout the whole spectrum of Cer and GlcCer species. GlcCer species in P. pastoris consistently elute ≈1.5 min before the corresponding Cer species.
Yeast Culture and Lipid Extraction for Analysis of IPC, MIPC, and M(IP)2C
P. pastoris cells were grown in liquid YPD medium, harvested by centrifugation, and stored at −20 °C until lipid extraction. The cells were resuspended in 5% (w/v) ice-cold trichloroacetic acid, incubated on ice for 30 min, and washed twice with ice-cold water. 0.5–0.6 g (fresh weight) of cells was resuspended in 6 ml of ethanol, water, diethyl ether, pyridine, 2 m ammonium hydroxide (15:15:5:1:0.038, v/v/v/v/v) (20). 5 nmol of Cer containing a C15 fatty acid and 5 nmol of GlcCer containing a C12 fatty acid were added as internal standards. Glass beads were added until slightly below the meniscus, and the samples were heated at 60 °C for 30 min. During this time, each sample was vortexed twice for 3 min. The cells were sedimented by centrifugation and extracted a second time using the same procedure. The supernatants from both extractions were combined, mixed, split into two equal aliquots, and evaporated under a stream of nitrogen. To remove glycerolipids, one aliquot of each sample was dissolved in 2 ml of methanol, water, 1-butanol, 33% (w/w) methylamine in ethanol (4:3:1:5, v/v/v/v) and heated at 55 °C for 60 min. The solvent was evaporated under a stream of nitrogen.
Both hydrolyzed and non-hydrolyzed samples were dissolved in 2 ml of water-saturated 1-butanol. The samples were washed by adding 2 ml of water, vortexing for 3 min, and centrifuging for 30–60 min at 1000 × g to achieve phase separation. The upper phase was transferred to a glass tube, the lower phase was extracted a second time with 2 ml of water-saturated 1-butanol, and the solvent was evaporated under a stream of nitrogen.
Analysis of IPC, MIPC, and M(IP)2C by MS-MS
The molecular species of IPC, MIPC, and M(IP)2C present in the hydrolyzed samples were analyzed on a 320-MS triple quadrupole mass spectrometer (Varian, Santa Clara, CA) in negative ionization mode. Individual molecular species of IPC and M(IP)2C were detected by multiple reaction monitoring using a transition of 241 corresponding to phosphoinositol, whereas for MIPC species, a transition of 421 corresponding to mannosylated phosphoinositol was used. Because no standards for IPC, MIPC, and M(IP)2C were available, we used GlcCer containing a C12 fatty acid as internal standard for comparing the samples against each other. Therefore, all mol % reported in the figures have to be regarded as apparent mol %.
Quantitative Analysis of LCB Composition
LCBs were liberated from whole cells or from GlcCer purified from a lipid extract of P. pastoris GS115 by strong alkaline hydrolysis with Ba(OH)2, converted to their 2,4-dinitrophenyl derivatives, and analyzed by reverse-phase high-performance liquid chromatography (HPLC) (12, 13). See the supplemental methods for details.
Yeast Culture and Preparation of Microsomes for in Vitro Assays
Cells of the S. cerevisiae strain WBY616-BAR1 grown in 200 ml of liquid YPD medium were harvested by centrifugation, resuspended in 2 ml of Lysis Buffer (20 mm HEPES/KOH, pH 7.4, 25 mm KCl, 2 mm MgCl2, 250 mm sorbitol) and 50 μl of proteinase inhibitor mixture (Sigma-Aldrich)/g of cells, and broken by bead-bashing at 4 °C for 1.5 h. Cell debris were removed by centrifuging at 1000 × g at 4 °C for 10 min. The supernatant was loaded on a 60% (w/w) sucrose cushion and centrifuged at 100,000 × g for 1 h. The microsomes were collected from the interphase, snap frozen in liquid nitrogen, and stored at −80 °C.
In Vitro Assay of Cer Synthase Activity
The in vitro Cer synthase reaction was performed essentially as described (21, 22) with 1.5 μm defatted bovine serum albumin, 1 μm LCB, 5 μm acyl-CoA, and 6.6 μg of microsomal protein in a final volume of 120 μl made up with Lysis Buffer. In the case that several LCBs or acyl-CoAs were mixed, the concentrations were 1 μm for each LCB and 5 μm for each acyl-CoA. Preliminary experiments showed that these high total concentrations of acyl-CoAs had no negative effect. Radiolabeled substrates were diluted with the corresponding unlabeled compound to achieve the desired molarity. The reaction was preincubated by shaking at 30 °C for 5 min and started by adding either LCB or acyl-CoA depending on the conditions to be tested. After shaking at 30 °C for the times indicated under “Results” and in the legend of Fig. 5, the reaction was stopped by adding 450 μl of chloroform/methanol (1:2, v/v). A phase separation was induced by adding 150 μl of chloroform and 150 μl of water, vortexing, and centrifuging (23). The upper phase was discarded, and the lower phase was washed once with methanol/water (1:1, v/v). The lower phase was transferred to a new tube, dried under a stream of nitrogen, and dissolved in chloroform/methanol (5:1, v/v).
FIGURE 5.
Substrate preference of Bar1p matches the properties of the Cer pool with dihydroxy LCB and C16/C18 fatty acids. A, the substrate preference of Bar1p for different LCBs and acyl-CoAs was tested by a ceramide synthase assay with microsomes prepared from a lag1Δlac1Δ S. cerevisiae strain expressing FLAG-tagged P. pastoris Bar1p as its only Cer synthase (incubation time, 5 min). Non-hydroxylated acyl-CoAs with different chain lengths are compared on the left, whereas the preference for α-hydroxylated versus non-hydroxylated C18 acyl-CoA is shown in the middle. Sphing-4-enine was used as the LCB. On the right, the preference for the LCBs sphinganine, sphing-4-enine, and 4-hydroxysphinganine is shown in combination with non-hydroxylated C18 acyl-CoA. In the upper row, the substrate specificity was tested using both a single LCB and a single acyl-CoA per reaction (one reaction per combination), whereas in the lower row, substrate selectivity was tested by offering mixtures of the LCBs or acyl-CoAs to be compared against each other (one reaction per panel). Shown are averages and S.D. from three independent reactions. B, comparison of Cer species from two different S. cerevisiae strains: left, a control strain expressing c-Myc-tagged S. cerevisiae Lag1p; right, a strain expressing FLAG-tagged P. pastoris Bar1p as the only ceramide synthase. The Cer species are grouped by the number of acyl carbons, assuming a C18 (upper row of carbon numbers) or a C20 LCB (lower row; see supplemental Fig. S2), and by the hydroxylation of the LCB (dihydroxy or trihydroxy). Hydroxylation of the fatty acid is not taken into account because there were no significant differences between the strains. Shown are averages and S.D. of three independent experiments. The mol % of the sum of all Cer species was calculated separately for each strain so that the chain length distribution and the hydroxylation pattern, but not the absolute amounts, can be compared between the strains. 100% corresponds to 15 ± 2 nmol/g fresh weight for the Lag1p-expressing strain and 68 ± 8 nmol/g fresh weight for the Bar1p-expressing strain.
In the case that the reaction was performed with either 3H-labeled LCB or 14C-labeled acyl-CoA, the reaction products were separated by TLC in chloroform/methanol (100:7, v/v), detected using a Cyclone phosphorimaging system (Packard Instruments; now PerkinElmer Life Sciences) for 3H and an FLA-3000 phosphorimaging system (Fujifilm, Tokyo, Japan) for 14C, and analyzed with NIH ImageJ software. Cer was identified by co-migration with an unlabeled standard (detected by iodine staining). In the case of unlabeled substrates, the Cer species formed in the reaction were analyzed by UPLC/MS as described above.
RESULTS
Generating a Collection of P. pastoris Mutant Strains Impaired in Biosynthesis of GlcCer
To investigate the pathway leading to GlcCer biosynthesis, we determined the molecular species of Cer and GlcCer in P. pastoris wild type (WT) and in mutant strains, each impaired in the activity of one enzyme involved in GlcCer biosynthesis. For this purpose, we deleted the genes encoding the putative Cer synthase Bar1p, the fatty acid α-hydroxylase Scs7p, the sphingolipid Δ4-desaturase, the sphingolipid Δ8-desaturase, the sphingolipid-C9-methyltransferase, and the GCS by homologous recombination (Fig. 1 and Table 1). In addition, we created three different strains overexpressing recombinant GCS under the control of the strong AOX1 promoter.
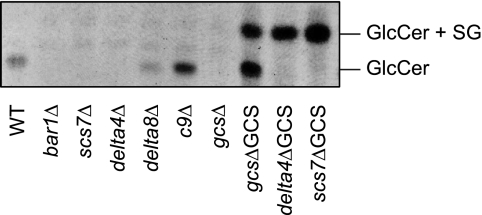
First, we checked both knock-out and overexpressing strains for the presence of GlcCer (Fig. 3). As expected, the lipid extract from WT cells showed a single band of GlcCer on the TLC plate, whereas GlcCer was undetectable in the gcsΔ strain, confirming previous results (24). Also, the deletion of the gene encoding the ceramide synthase Bar1p resulted in the loss of GlcCer. This supports our hypothesis that Bar1p is responsible for the biosynthesis of those Cer species that are used as precursors for GlcCer biosynthesis. A similar finding has been reported for a knock-out of the Bar1p ortholog in Kluyveromyces lactis (25). Surprisingly, deletion of the α-hydroxylase Scs7p or of the Δ4-desaturase also resulted in a complete loss of GlcCer. In contrast, GlcCer could still be detected in P. pastoris strains in which the sphingolipid Δ8-desaturase or the C9-methyltransferase was deleted.
FIGURE 3.
Presence or absence of GlcCer in mutants of P. pastoris with altered activities of enzymes involved in sphingolipid biosynthesis. Lipids extracted from WT and mutant lines were separated by TLC in chloroform/methanol (85:15). Glycolipids were visualized by spraying with α-naphthol/sulfuric acid and subsequent heating to 160 °C. For interpretation of the results see the text. The WT and the knock-out mutants were grown in complete medium (YPD medium), whereas the GCS-overexpressing strains (right three lanes) were grown in minimal methanol medium to induce expression of the GCS. Overexpression of the GCS resulted in the appearance of an additional glycolipid band above the normal GlcCer band that turned out to be a mixture of steryl glucoside (SG) and atypical GlcCer species (see the text and Fig. 7). Biosynthesis of steryl glucoside is typical for P. pastoris cells grown in minimal medium. One representative of several experiments is shown.
Similarly unexpected results were obtained with the P. pastoris strains overexpressing GCS. Although in the complemented strain gcsΔGCS the intensity of the “normal” GlcCer is strongly increased, this band is missing in the strains scs7ΔGCS and delta4ΔGCS lacking the α-hydroxylase or the Δ4-desaturase, respectively. Instead, a strong additional band appears above the normal band in all three strains overexpressing GCS. From the MS analysis shown below, it can be concluded that the normal band contains GlcCer species with an α-hydroxylated C16/C18 fatty acid, whereas the additional band contains untypical GlcCer species with either a non-hydroxylated C16/C18 fatty acid or an α-hydroxylated C24/C26 fatty acid.
P. pastoris Strains Lacking GlcCer Show No Obvious Phenotype
All P. pastoris knock-out strains including those that completely lack GlcCer were viable, and no obvious growth defects were observed in full (YPD) medium. Growth of the bar1Δ strain was indistinguishable from WT on YPD plates (supplemental Fig. S1A). In addition, cell wall morphology and bud formation (calcofluor white staining), organization of the actin cytoskeleton (phalloidin-rhodamine staining), and the staining pattern of fluorescent markers directed to the endoplasmic reticulum, endoplasmic reticulum exit sites, and Golgi apparatus were investigated, but no differences between the bar1Δ strain and the WT were found (supplemental Fig. S1B).
Analysis of Cer and GlcCer Species in P. pastoris Wild Type and Mutant Strains
To investigate how the enzymes highlighted in Fig. 1 cooperate during GlcCer biosynthesis, the molecular species of Cer and GlcCer were analyzed in P. pastoris WT cells and in the collection of mutant strains. For this purpose, the lipid extracts were fractionated on a silica cartridge, and the fraction containing Cer and GlcCer was analyzed by UPLC/MS. Individual Cer and GlcCer species were detected based on their retention times and exact masses (mass accuracy, <0.01 Da) and quantified relative to internal standards. The complete list of Cer and GlcCer species is shown in supplemental Table S1.
Individual selections from this data set are displayed in Figs. 4, 6, and 7 depending on the structural features to be discussed. Because the absolute amounts of Cer (and to a lesser extent GlcCer) showed a much higher sample to sample variation than the relative proportions of individual species, we decided to display the data using a relative rather than an absolute scale. In Figs. 4, 6, and 7, 100% corresponds to 70 ± 60 nmol/g fresh weight for Cer and 30 ± 2 nmol/g fresh weight for GlcCer (average and S.D. of three independent WT samples). Because of these standard deviations, only the absolute amounts of GlcCer, but not of Cer, can be reliably compared between different strains. In contrast, the relative proportions of both Cer and GlcCer species within each strain were very reproducible (Figs. 4, 6, and 7, error bars). Because the UPLC/MS method does not allow fragmentation of the Cer and GlcCer species, only the total number of carbon atoms could be determined. But because P. pastoris sphingolipids contained >95% C18 LCBs (supplemental Fig. S2), the number of acyl carbons in the Cer and GlcCer species could be easily deduced and is referred to in the following text.
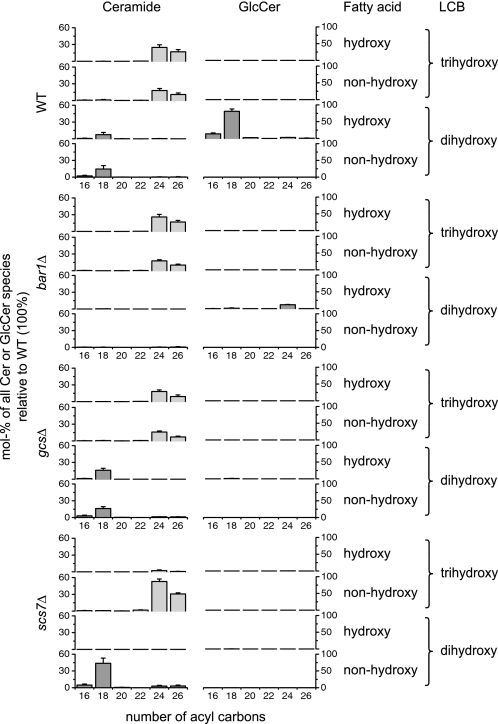
FIGURE 4.
Cer and GlcCer species can be classified into two groups that are distinguished by chain length of fatty acid and hydroxylation of LCB. Molecular species of Cer and GlcCer from WT and the knock-out strains bar1Δ, gcsΔ, and scs7Δ are grouped by the number of acyl carbons (assuming a C18 LCB) as well as by the hydroxylation of the LCB (dihydroxy or trihydroxy) and the fatty acid (hydroxy or non-hydroxy). The quantities of Cer and GlcCer in the knock-out strains are given as mol % of the sum of all species relative to the WT so that a direct comparison between the strains is possible. Shown are averages and S.D. of three independent experiments.
FIGURE 6.
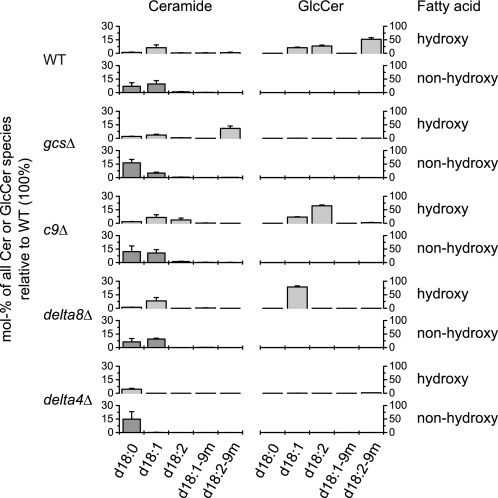
Δ4-Desaturation, but not Δ8-desaturation or C9 methylation, is essential for conversion of the Cer pool with dihydroxy LCB and C16/C18 fatty acids into GlcCer. Molecular species of Cer and GlcCer from WT and from the knock-out strains gcsΔ, c9Δ, delta8Δ, and delta4Δ are grouped by the desaturation of the LCB (d18:0, saturated; d18:1, monounsaturated; d18:2, diunsaturated; d18:1-9m, monounsaturated and C9-methylated; d18:2-9m, diunsaturated and C9-methylated) and by the hydroxylation of the fatty acid (hydroxy or non-hydroxy). Only Cer and GlcCer species with dihydroxy LCBs are shown because no species with a desaturated trihydroxy LCB could be detected. The acyl chain length of these species is almost exclusively 16 or 18 because nearly all Cer species with a C24/C26 fatty acid have a trihydroxy LCB (not included in this figure), and GlcCer species with a C24/C26 fatty acid occur only in trace amounts (Fig. 4). The quantities of Cer and GlcCer in the knock-out strains are given as mol % of the sum of all species relative to the WT so that a direct comparison between the strains is possible. Shown are averages and S.D. of three independent experiments.
FIGURE 7.
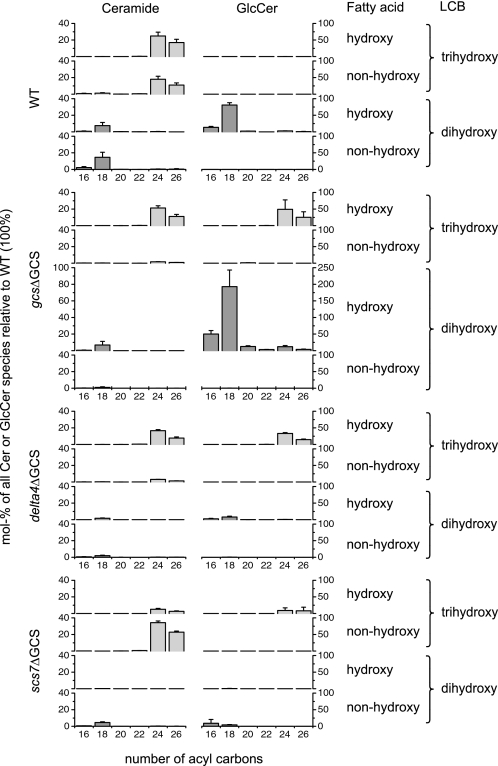
Overexpression of GCS makes the Cer pool with trihydroxy LCB and C24/C26 fatty acids accessible for GlcCer biosynthesis. Molecular species of Cer and GlcCer from WT and from the overexpressing strains gcsΔGCS, delta4ΔGCS, and scs7ΔGCS are grouped by the number of acyl carbons (assuming a C18 LCB) as well as by the hydroxylation of the LCB (dihydroxy or trihydroxy) and the fatty acid (hydroxy or non-hydroxy). The quantities of Cer and GlcCer in the overexpressing strains are given as mol % of the sum of all species relative to the WT so that a direct comparison between the strains is possible. Shown are averages and S.D. of three independent experiments.
Group of Cer Species with Dihydroxy LCB and C16/C18 Fatty Acids Is Precursor of GlcCer
Cer species in the WT strain can be divided into two groups, one with a dihydroxy LCB and C16/C18 fatty acids and the other with a trihydroxy LCB and C24/C26 fatty acids (Fig. 4). GlcCer species exclusively contained a dihydroxy LCB and C16/C18 fatty acids, indicating that Cer species from only one group can enter the pathway leading to GlcCer biosynthesis.
To test whether the Cer synthase Bar1p produces the Cer species available for GlcCer biosynthesis, we checked which Cer and GlcCer species could be detected in the bar1Δ strain (Fig. 4, bar1Δ; see supplemental Fig. S4 for minor species). Strikingly, the Cer species with a dihydroxy LCB and C16/C18 fatty acids as well as the corresponding GlcCer species disappeared, whereas the Cer species with a trihydroxy LCB and C24/C26 fatty acids were unaffected. Very likely, the latter group of Cer species is produced by the activity of the second Cer synthase encoded in the P. pastoris genome, Lag1p (Fig. 2). A tiny proportion of GlcCer with a dihydroxy LCB and a C24 fatty acid could be detected in the WT; this was increased to a small but significant proportion in the bar1Δ strain (supplemental Fig. S4). In the gcsΔ strain in which the GCS was deleted, GlcCer was missing, but both groups of Cer species were present. Surprisingly, the proportion of Cer species with a dihydroxy LCB and C16/C18 fatty acids is not significantly increased, although less Cer is used for GlcCer biosynthesis. Together, these data suggest that the Cer synthase Bar1p produces a distinct group of Cer species with a dihydroxy LCB and C16/C18 fatty acids that serve as precursors for GlcCer biosynthesis.
Bar1p Shows Cer Synthase Activity in Vitro with Preference for Dihydroxy LCBs and C16/C18 Acyl-CoAs
To confirm the specificity of Bar1p for dihydroxy LCB and C16/C18 fatty acids, in vitro Cer synthase assays were carried out. As enzyme source, an S. cerevisiae strain was engineered to express P. pastoris Bar1p as its only Cer synthase under the control of the constitutive GPD1 promoter. In the resulting strain, the genes encoding the endogenous Cer synthases Lag1p and Lac1p were deleted, but the alkaline ceramidases Ypc1p and Ydc1p were intact. Using 3H-labeled sphinganine or sphing-4-enine and unlabeled acyl-CoAs, the formation of 3H-labeled Cer could be detected with acyl chain lengths ranging from 16 to 22 carbons with a maximum at 18 (supplemental Fig. S3). Using 14C-labeled C18 acyl-CoA, the formation of 14C-labeled Cer could be detected with all three LCBs tested: sphinganine, sphing-4-enine, and 4-hydroxysphinganine (structures shown in Fig. 1B). Cer formation using sphinganine or sphing-4-enine as substrates was 3 times more efficient than with 4-hydroxysphinganine.
To more systematically investigate different combinations of LCBs and acyl-CoAs, we developed a non-radioactive version of this assay in which the Cer species produced in the reaction were analyzed by UPLC/MS. This allowed arbitrary combinations of LCBs and acyl-CoAs to be used as substrates either individually or as a mixture. With single LCBs and acyl-CoAs as substrates, Bar1p showed a clear specificity for C16 and C18 compared with longer acyl-CoAs. α-Hydroxylated species were preferred over non-hydroxylated C18 acyl-CoA, and the dihydroxy LCBs sphinganine and sphing-4-enine were preferred over the trihydroxy LCB 4-hydroxysphinganine (Fig. 5A, upper panel).
When substrate selectivity was tested using mixtures of different LCBs and acyl-CoAs, the preference for certain LCBs and acyl-CoAs became more pronounced than with individual substrates. Bar1p preferred C18 over C16 acyl-CoA and the Δ4-desaturated LCB sphing-4-enine over the saturated sphinganine, whereas 4-hydroxysphinganine was heavily discriminated against. α-Hydroxylated was strongly preferred over non-hydroxylated C18 acyl-CoA (Fig. 5A, lower panel).
S. cerevisiae Strain Expressing Bar1p Produces Cer Species with Dihydroxy LCB and C16/C18 Fatty Acids
The results above were confirmed by analysis of the Cer species produced in vivo by the same Bar1p-expressing S. cerevisiae strain that was used as enzyme source for the in vitro assays. In the control strain (expressing S. cerevisiae Lag1p), the most abundant Cer species was composed of a trihydroxy LCB and an α-hydroxy C26 fatty acid as expected (26–28). In the strain expressing P. pastoris Bar1p, however, Cer species with a dihydroxy LCB and α-hydroxy C16/C18 fatty acids were prevalent (Fig. 5B). This was despite the fact that this strain contained a WT allele encoding the sphingolipid C4-hydroxylase Sur2p, so trihydroxy LCBs should be plentiful. As in the in vitro assay, Bar1p discriminates against trihydroxy LCBs. Together with the in vitro assay, these data show that the preference of Bar1p for dihydroxy LCBs and C16/C18 acyl-CoAs is directly responsible for the characteristic structural features of the Bar1p-dependent Cer pool shown in Fig. 4.
Efficient GlcCer Biosynthesis Requires Structural Features Specific to Bar1p-dependent Cer Pool
We next addressed the question to which extent the other structural features highlighted in Fig. 1 were required for GlcCer biosynthesis. First, we consider the fatty acid hydroxylation of the Cer and GlcCer species shown in Fig. 4 (see supplemental Fig. S4 for minor species). In the WT strain, Cer species with both α-hydroxylated and non-hydroxylated fatty acids were present. In contrast, GlcCer contained exclusively α-hydroxylated fatty acids. In the scs7Δ strain in which the fatty acid α-hydroxylase is deleted, Cer species with α-hydroxylated fatty acids were missing with a concomitant increase in non-hydroxylated species. Strikingly, GlcCer was completely missing, showing that non-hydroxylated Cer cannot be converted to GlcCer. Fatty acid α-hydroxylation is thus essential for efficient GlcCer biosynthesis from the Bar1p-dependent Cer pool but not for the biosynthesis of the Bar1p-dependent Cer pool itself.
The role of the desaturation and methylation of the LCB becomes evident in Fig. 6 (see supplemental Fig. S5 for minor species). In this figure, only dihydroxy LCBs are presented because Δ4-desaturation (including the subsequent Δ8-desaturation and C9-methylation; see below) and C4-hydroxylation of the LCB are mutually exclusive. The displayed molecular species therefore belong to the Bar1p-dependent Cer pool and have C16/C18 fatty acids. In the WT strain, Cer was mainly saturated or Δ4-monounsaturated. In contrast, GlcCer was either Δ4-mono- or Δ4,8-diunsaturated, and approximately two-thirds of diunsaturated GlcCer was in addition C9-methylated. Methylated monounsaturated species were not detected in Cer or in GlcCer.
In the gcsΔ strain in which no Cer is consumed for GlcCer biosynthesis (see above), a large proportion of Δ4,8-diunsaturated Cer with a C9-methyl group could be detected. This demonstrates that both Δ8-desaturation and C9-methylation can occur at the level of Cer, which is in agreement with previous results showing that Cer, but not GlcCer, is a substrate for C9-methylation (14). Most likely, the proportion of Δ4,8-diunsaturated Cer species with a C9-methyl group was very low in the WT strain because these species were immediately consumed for GlcCer biosynthesis.
Because Δ4,8-diunsaturated Cer species with a C9-methyl branch seem to be the preferred substrates for GlcCer biosynthesis, we next asked whether GlcCer levels would be affected in the c9Δ strain in which the C9-methyltransferase is deleted. Although methylated Cer and GlcCer were missing in this strain, the proportions of the corresponding non-methylated species of both Cer and GlcCer were increased. This shows that LCB methylation is not essential for GlcCer biosynthesis in confirmation of previous results (14).
Similar results were obtained with the delta8Δ strain in which the Δ8-desaturase is deleted. Diunsaturated species of both Cer and GlcCer were replaced by equivalent proportions of monounsaturated species. Neither Cer nor GlcCer was C9-methylated in this strain because Δ8-desaturation is required for the reaction mechanism of the C9-methyltransferase (14). These results show that neither Δ8-desaturation nor C9-methylation is required for the biosynthesis of the Bar1p-dependent Cer pool or the subsequent formation of GlcCer.
In contrast, GlcCer was completely missing in the delta4Δ strain in which the Δ4-desaturase is deleted. This confirms previous results (29). Neither mono- nor diunsaturated Cer could be detected in this strain, but the level of saturated Cer was enhanced in comparison with WT. This shows that Bar1p forms a corresponding Cer pool in the delta4Δ strain but that these saturated Cer species cannot be converted to GlcCer. At the same time, this pattern shows that in P. pastoris Δ4-desaturation is a prerequisite for Δ8-desaturation. The LCB modifications in the Bar1p-dependent Cer pool thus follow the sequence Δ4-desaturation → Δ8-desaturation → C9-methylation as illustrated in Fig. 10.
FIGURE 10.
Simplified model of sphingolipid biosynthesis in P. pastoris showing qualitative and quantitative routing of substrates into Cer, GlcCer, and IPC. Intermediates of the pathway are given with the predominant molecular species indicated underneath. The width of the arrows and the letters indicates the relative quantitative proportions of the end products of lipid biosynthesis in P. pastoris (not to scale). The compounds with a gray background were subjects of our analyses. The early steps of sphingolipid biosynthesis, the late steps of GIPC biosynthesis, and degradative and recycling pathways have been omitted. For simplicity, the model only shows species containing the dominating C18 and C24 fatty acids, but it should be considered that P. pastoris also synthesizes a smaller proportion of species containing C16 and C26 fatty acids.
In summary, Δ4-desaturation of the LCB and α-hydroxylation of the fatty acid are strictly required for entry of the Bar1p-dependent Cer pool into the pathway leading to GlcCer biosynthesis. Δ8-Desaturation and C9-methylation are optional, but if present, Δ4,8-diunsaturated Cer species with a C9-methyl branch are the preferred substrates for GlcCer biosynthesis.
If Overexpressed, GCS Gains Access to Cer Pool with Trihydroxy LCB and C24/C26 Fatty Acids
Next, we wanted to see whether the loss of GlcCer in the strains gcsΔ, delta4Δ, and scs7Δ (in which the GCS, the Δ4-desaturase, and fatty acid α-hydroxylase are deleted, respectively) could be compensated for by overexpression of the GCS. For this purpose, these strains were transformed with a construct containing the P. pastoris GCS under the control of the strong, methanol-inducible AOX1 promoter. The expression of the glucosyltransferase was induced by the addition of methanol to the culture medium 24 h before harvesting of the cells.
Fig. 7 (see supplemental Fig. S6 for minor species) shows that overexpression of the GCS in the gcsΔ strain complemented the deletion because compared with WT the proportion of GlcCer was significantly higher in the resulting gcsΔGCS strain. Interestingly, the overexpressing strain contained GlcCer species with a trihydroxy LCB and C24/C26 fatty acids in addition to the regular species with a dihydroxy LCB and C16/C18 fatty acids. When GCS was overexpressed, both Cer pools could be used for GlcCer biosynthesis, but utilization of Cer species with trihydroxy LCBs was less efficient.
In the strain delta4ΔGCS in which the GCS is overexpressed and the Δ4-desaturase is deleted, the proportion of Cer and GlcCer species with dihydroxy LCB and C16/C18 fatty acids was significantly lower than that in the gcsΔGCS strain. In contrast, Cer species with trihydroxy LCB and C24/C26 fatty acids were present in similar proportions and were used at low efficiency for GlcCer biosynthesis. This shows that even when GCS is overexpressed efficient GlcCer biosynthesis from the Bar1p-dependent Cer pool requires Δ4-desaturation.
In the strain scs7ΔGCS in which the GCS is overexpressed in the absence of fatty acid α-hydroxylase activity, GlcCer was still detectable, but the levels of all GlcCer species were extremely low compared with the gcsΔGCS strain. This shows that efficient GlcCer biosynthesis from either Cer pool requires α-hydroxylated fatty acids.
Surprisingly, the levels of Cer species with a non-hydroxylated fatty acid are reduced in all three GCS-overexpressing strains compared with the WT (Fig. 7) or the scs7Δ strain (Fig. 4). This cannot be directly explained by an increased consumption because only α-hydroxylated Cer species are being used for GlcCer biosynthesis. Although an increased demand for α-hydroxylated Cer species might potentially lead to an up-regulation of the fatty acid α-hydroxylase activity, such an explanation has to be taken with care because the absolute amounts of Cer vary considerably between samples (see above). Also, an influence of the carbon source (glucose or methanol) on the extent of α-hydroxylation cannot be excluded.
In summary, both Cer pools can be used for GlcCer biosynthesis if GCS is overexpressed, but α-hydroxylation of the fatty acid and a C4 modification of the LCB (either Δ4-desaturation or C4-hydroxylation) remain important requirements. Like Δ4-desaturation, C4-hydroxylation provides an electron-dense environment at the C4 position of the LCB.
Biosynthesis of IPC, MIPC, and M(IP)2C Is Independent of GlcCer Biosynthesis
To investigate whether the biosynthesis of GlcCer and of the three phosphosphingolipids IPC, MIPC, and M(IP)2C was interconnected, their levels were investigated in the WT and in the strains bar1Δ, gcsΔ, and scs7Δ. The Cer backbone of all three phosphosphingolipids mainly consisted of 4-hydroxysphinganine as LCB and α-hydroxylated C24/C26 fatty acids except for the scs7Δ strain in which the α-hydroxylated fatty acids were replaced by the equivalent non-hydroxylated fatty acids (Fig. 8). A small but significant proportion of IPC species containing sphinganine and both α-hydroxylated and non-hydroxylated fatty acids could also be detected in all strains except for bar1Δ. This shows that the Bar1p-dependent Cer pool is incorporated into IPC to a limited extent but that these IPC species are not processed further into MIPC and M(IP)2C, suggesting an additional selection mechanism. The complete list of IPC, MIPC, and M(IP)2C species is shown in supplemental Table S2.
FIGURE 8.
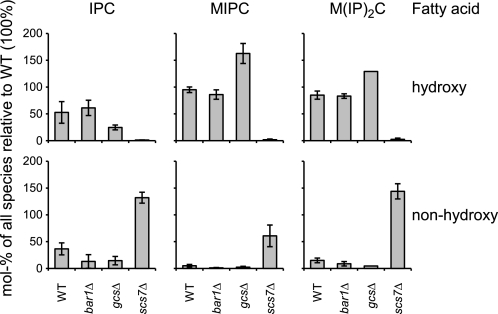
Analysis of IPC, MIPC, and M(IP)2C in P. pastoris strains lacking GlcCer. Molecular species of IPC, MIPC, and M(IP)2C from WT and from the knock-out strains bar1Δ, gcsΔ, and scs7Δ are grouped by the hydroxylation of their fatty acids. Only species containing a trihydroxy LCB are shown. The WT strain contains in addition ≈11% of IPC species with a dihydroxy LCB in confirmation of a previous study (55). MIPC and M(IP)2C species with a dihydroxy LCB were not detectable. The quantities of IPC, MIPC, and M(IP)2C are given as mol % of the sum of all species relative to the WT so that a direct comparison between the strains is possible. Shown are averages and S.D. of three independent experiments. An absolute quantification was not possible because no standards were available.
Fig. 8 shows that the levels of IPC, MIPC, and M(IP)2C were not significantly reduced in the strains bar1Δ and gcsΔ; thus, phosphosphingolipid biosynthesis proceeds in the absence of GlcCer or of the Bar1p-dependent Cer pool. In the gcsΔ strain, the levels of MIPC and M(IP)2C are even increased relative to the WT. This could potentially explain why Cer levels remain constant in this strain, although less Cer is consumed for GlcCer biosynthesis (Fig. 4). In scs7Δ, α-hydroxylated species of all three phosphosphingolipids were replaced by the equivalent non-hydroxylated species. This shows that although fatty acid α-hydroxylation is essential for GlcCer biosynthesis it is dispensable for the biosynthesis of phosphosphingolipids. In conclusion, the biosynthesis of IPC, MIPC, and M(IP)2C is independent of GlcCer biosynthesis and does not require α-hydroxylation of the fatty acid.
Abundance of the (G)IPC-specific Sphingolipid Pool Greatly Exceeds That of Bar1p-dependent Pool
A comparison of the relative proportions of GlcCer and (G)IPCs should be useful to elucidate the quantitative aspect of substrate flow into the two branches of sphingolipid biosynthesis. Unfortunately, the reported MS data are not suitable for a quantitative comparison of the GlcCer- and (G)IPC-specific sphingolipid pools. Therefore, we analyzed the total LCBs liberated from P. pastoris WT cells by HPLC and compared them with the LCBs found in purified GlcCer from the same cells (Fig. 9).
FIGURE 9.
Analysis of LCBs from WT P. pastoris. LCBs were released from sphingolipids by strong alkaline hydrolysis, converted into dinitrophenyl derivatives, and separated by HPLC. Upper panel, LCBs were released from Cer, GlcCer, and (G)IPCs of whole P. pastoris cells. The total LCBs consisted of ≈90% of the trihydroxy LCBs t18:0 and t20:0. Dihydroxy LCBs from the Bar1p-dependent Cer pool and from GlcCer made ≈10%. The left peak represents mainly t18:0 and a low proportion of d18:2, which cannot be separated by this method. Lower panel, LCBs, consisting of the three dihydroxy LCBs d18:1, d18:2, and d18:2-9m, were released from purified GlcCer from P. pastoris. Shown is one representative experiment of three.
In this analysis, the dihydroxy LCBs d18:0 (1 mol %), d18:1 (2 mol %), d18:2 (see below), and d18:2-9m (3 mol %) represent the Bar1p-dependent Cer species and GlcCer. The exact proportion of d18:2 could not be determined because it had the same retention time as t18:0. But a comparison with purified GlcCer in the lower panel shows that the proportion of d18:2 should be similar to that of d18:1, thus ≈2 mol %. Altogether, the Bar1p-dependent branch of sphingolipid metabolism made up ≈10 mol % of the total sphingoid bases. The trihydroxy LCBs t18:0 (90 mol %) and t20:0 (4 mol %) represent the branch leading to (G)IPC biosynthesis and account for ≈90 mol %.
From our enzymatic investigation of Bar1p and from qualitative and quantitative analyses of sphingolipids and their quantitative relation to other lipids, we deduced a simplified model showing the flow of precursors via ceramide pools into different sphingolipid classes in P. pastoris (Fig. 10). A complete picture should also include degradative and recycling pathways because they likely influence both the quantity and the molecular species composition of the depicted lipid pools. In the present illustration, they had to be omitted because insufficient information is available for an accurate representation.
DISCUSSION
The yeast P. pastoris contains two structurally distinct Cer pools whose main distinguishing features are the hydroxylation of the LCB (dihydroxy or trihydroxy) and the chain length of the fatty acid (C16/C18 or C24/C26). These structural differences have decisive consequences for their conversion into the two downstream products, GlcCer and (G)IPCs. Our data define a set of four enzymes that are required for GlcCer biosynthesis: Cer synthase Bar1p, sphingolipid Δ4-desaturase, fatty acid α-hydroxylase, and GCS. Deletion of any of the corresponding genes results in the complete loss of GlcCer.
Two additional enzymes, Δ8-desaturase and C9-methyltransferase, are optional in the sense that GlcCer biosynthesis can proceed in P. pastoris strains in which the corresponding genes are deleted and the corresponding functional groups are thus missing in the Cer backbone. In the following, we will discuss three aspects of GlcCer biosynthesis (formation of ceramide by the Cer synthase Bar1p, introduction of backbone modifications, and final glucosylation of Cer by the GCS) as well as the functional significance of GlcCer and GIPC biosynthesis.
Substrate Specificity of Ceramide Synthase Bar1p Provides Basis for Formation of GlcCer-specific Ceramide Backbone
The molecular species of the Cer pool with a dihydroxy LCB and C16/C18 fatty acids are specifically used for GlcCer biosynthesis (Fig. 10). The substrate specificity of Bar1p matches the structural properties of the Bar1p-dependent Cer pool, i.e. a preference for dihydroxy LCBs and C16/C18 fatty acids. The only difference is that Cer species with non-hydroxylated C16/C18 fatty acids are slightly more abundant in vivo (Fig. 4), whereas α-hydroxylated C18 acyl-CoA is clearly preferred in the in vitro assay (Fig. 5). This apparent discrepancy may be due to the fact that exclusively Cer species with α-hydroxylated fatty acids are used for GlcCer biosynthesis so that Cer species with non-hydroxylated fatty acids accumulate. Indeed, the gcsΔ strain contains approximately equal levels of Cer species with the dihydroxy LCB linked to an α-hydroxylated or non-hydroxylated fatty acid. In addition, recent results show that α-hydroxylation very likely takes place at the level of Cer instead of free acyl-CoAs.7 The preference of Bar1p for α-hydroxylated acyl-CoA thus could imply that substrates from a putative recycling pathway might be favored over non-hydroxylated acyl-CoA biosynthesized de novo.
In S. cerevisiae, both Lag1p and Lac1p produce Cer with a C24/C26 fatty acid (26–28). Unfortunately, we did not succeed in cloning the orthologous gene from P. pastoris, and the native in vitro Cer synthase activity in microsomes prepared from the P. pastoris bar1Δ strain was too low to carry out an enzymatic characterization of Lag1p. But if P. pastoris Lag1p also preferred trihydroxy over dihydroxy LCBs and C24/C26 over shorter acyl-CoAs, the structural features defining the two Cer pools, hydroxylation of the LCB and chain length of the fatty acid, could be entirely explained by the substrate specificities of the two Cer synthases, Bar1p and Lag1p.
Defined Sequence of Ceramide Backbone Modifications Precedes Final Glucosylation Step
The LCB modifications characteristic for GlcCer of P. pastoris strictly follow the sequence Δ4-desaturation → Δ8-desaturation → C9-methylation (illustrated in Fig. 10). The dependence of Δ8-desaturation on Δ4-desaturation might be caused by a specificity of the Δ8-desaturase for Δ4-desaturated LCB or Cer species. In plants, Δ4-desaturation is not a prerequisite for Δ8-desaturation (29). Instead, one of the two Δ8-desaturases is specific for C4-hydroxylated LCBs or Cer species (30), demonstrating that desaturases can indeed discriminate between different LCBs.
C9-methylation depends on Δ8-desaturation because this S-adenosylmethionine-dependent methyltransferase requires the presence of a double bond next to the methyl group to be introduced (14). This is confirmed by data showing that C9-methylated LCBs are not detectable in Candida albicans and P. pastoris strains in which the Δ8-desaturase has been deleted (Ref. 31 and this study). Although the presence of the C9-methyl group is not required for GlcCer biosynthesis, its absence has functional consequences in several fungal species (31–33). Here, it may be interesting to point out that the introduction of a methyl group into the middle of a saturated acyl group results in a substantial decrease of the phase transition temperature of the corresponding phospholipid comparable in its extent to the effect of a Z-double bond at this position (34).
GCS Uses Different Cer Substrates Depending on Its Expression Level
As discussed above, Cer species with a Δ4,8-diunsaturated, C9-methylated LCB and an α-hydroxylated fatty acid are the preferred substrates for GlcCer biosynthesis in WT cells, although only Δ4-desaturation and α-hydroxylation are strictly required. In contrast, the enzyme in addition uses Cer species with a trihydroxy LCB and C24/C26 fatty acids in the P. pastoris strains overexpressing GCS. This Cer pool, which normally serves as substrate for IPC biosynthesis, is now to a limited extent used for GlcCer biosynthesis. This implies that after the specific generation of the two Cer pools by Bar1p and (probably) Lag1p, a second mechanism must exist to ensure the separation of the two branches shown in Fig. 10. This mechanism (but not the requirement for a C4 modification of the LCB and α-hydroxylation of the fatty acid) can be circumvented by overexpression of the GCS.
Only Small Proportions of C18 Acyl-CoA and Sphinganine Pools Are Recruited for GlcCer Biosynthesis
In addition to the qualitative aspects discussed so far, the quantitative control of substrate entry into the two branches of the sphingolipid biosynthetic pathway is of similar importance (Fig. 10). Under steady state conditions, the resulting sphingolipid classes are present in very different quantities: in P. pastoris WT cells, the GlcCer branch represents only ≈10 mol % of the amount of (G)IPCs (determined by the ratio of GlcCer-specific LCBs to 4-hydroxysphinganine in Fig. 9). As illustrated in Fig. 10, this qualitatively and quantitatively very unequal flux may originate at one major branching point. At this step, Bar1p competes directly with several other enzymes for its two preferred substrates, sphinganine (d18:0) and 18:0-CoA, respectively. Bar1p, the fatty acid elongase, the Δ9-fatty acid desaturase, and several acyltransferases involved in glycerolipid and sterol ester biosynthesis compete for 18:0-CoA. According to the low proportion of GlcCer compared with (G)IPCs and the glycerolipids, one would infer a significantly higher consumption of 18:0-CoA by the elongases, the Δ9-desaturase, and the acyltransferases than by Bar1p. In addition, Bar1p has to compete with the C4-hydroxylase Sur2p for its second substrate, sphinganine. Also here, Bar1p can only recruit a minor fraction for the biosynthesis of the Cer species d18:0/18:0.
Our data on the specificity and selectivity of Bar1p can fully explain the qualitative separation of the two Cer pools into the two branches of sphingolipid biosynthesis. In contrast, the quantities of the end products might be controlled by the activity ratios of the competing enzymes just mentioned. So far, no kinetic data are available for these enzymes. Our data therefore represent steady state levels resulting from the integration of all individual enzyme activities involved in the pathway. In addition, both the quantity and the molecular species composition of (G)IPCs and GlcCer as well as of the two Cer pools might be influenced by degradative and recycling pathways (not included in Fig. 10). As an example, the apparent discrepancy between the extent of α-hydroxylation of the Cer species produced by Bar1p in vivo (Fig. 4) and in vitro (Fig. 5) might be explained by preferential recycling of α-hydroxylated fatty acids (see above). In the future, metabolic labeling experiments as well as knock-out strains in which degradative or recycling enzymes are inactivated could help resolve these issues.
Functional Significance of GlcCer and (G)IPC Biosynthesis
A number of studies in fungi indicate that GlcCer and GIPCs fulfill different functions. These experiments have been performed with inhibitor-treated cells or with knock-out mutants impaired in the activity of the enzymes involved in GlcCer or IPC biosynthesis, respectively.
In our study, all P. pastoris mutant strains impaired in GlcCer biosynthesis were viable and did not show any growth defects. This is consistent with previous reports showing that a bar1Δ strain of K. lactis (BAR1 is called LAC1 in that study) and a delta4Δ strain of Schizosaccharomyces pombe are viable (25, 35). These observations are in contrast to studies with dimorphic or filamentous fungi: deletion of different genes involved in GlcCer biosynthesis in such fungi, e.g. C. albicans, Cryptococcus neoformans, and Fusarium graminearum, resulted in viable mutants with widely varying phenotypes regarding growth, morphology, and host colonization (24, 31–33, 36–38). Importantly, the proportions of GlcCer relative to (G)IPCs in P. pastoris are similar to those in filamentous fungi (39). These phenotypes therefore suggest that although GlcCer may be dispensable under optimal laboratory conditions it may become important in more complex growth modes, under non-optimal environmental conditions, or to carry out specialized functions.
On the other hand, deletion or pharmacological inhibition of the IPC synthase of S. cerevisiae is either lethal or results in severe growth defects, although the severity of the phenotype is partially attributable to the accumulation of Cer as the precursor of IPC biosynthesis (27, 40–45). We did not systematically investigate the enzymes involved in the biosynthesis of (G)IPCs in P. pastoris. Despite repeated efforts, it was not possible to delete the genes encoding the Cer synthase Lag1p and the C4-hydroxylase Sur2p. This may suggest that (G)IPCs are essential for viability in P. pastoris and that the C4-hydroxylation of the LCB is required for the Lag1p-dependent Cer pool to be used for (G)IPC biosynthesis. Interestingly, sur2Δ strains of S. cerevisiae are viable, and C4-hydroxylation of the LCB is not required for (G)IPC biosynthesis (15, 16), whereas an equivalent knock-out in Aspergillus nidulans shows a severe growth defect (46). This suggests that the requirements for C4-hydroxylation of the LCB differ between fungal species.
Conclusions
Cer species finally found as backbones in GlcCer have to pass two filters imposing both qualitative and quantitative controls. At the first point, the Cer synthase Bar1p selectively uses sphinganine and 18:0-CoA in competition with other enzymes, which use the bulk of both substrates. The second control point in GlcCer synthesis is exerted by the GCS. This enzyme most likely does not accept the Cer species d18:0/18:0, the primary product of Bar1p, but requires Δ4-desaturation of the LCB and α-hydroxylation of the fatty acid.
Supplementary Material
Acknowledgments
We thank Sharon Epstein, University of Geneva, Geneva, Switzerland for help with the Cer synthase assay and for discussions; Dr. Cornelia Göbel and Sabine Freitag, Georg August University, Göttingen, Germany for assisting with the analytical procedures; and Wiebke Hellmeyer, University of Hamburg, Hamburg, Germany, for technical assistance. The plasmid pSLNat was kindly provided by Christoph Basse, Max Planck Institute for Terrestrial Microbiology, Marburg, Germany. The marker constructs for the secretory pathway in P. pastoris were kindly provided by Benjamin Glick, University of Chicago, Chicago, IL.
This work was supported in part by Deutsche Forschungsgemeinschaft Grants TE 491/3-1 (to P. T.) and SFB470 (to S. A. and T. W.) and by the Swiss National Science Foundation and SystemsX.ch (evaluated by the Swiss National Science Foundation) (to H. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental methods, Figs. S1–S6, and Tables S1 and S2.
H. Riezman, unpublished observations.
- Cer
- ceramide
- GalCer
- galactosylceramide
- GCS
- GlcCer synthase
- GIPC
- glycosylated IPC
- GlcCer
- glucosylceramide
- IPC
- inositol phosphorylceramide
- LCB
- long-chain (sphingoid) base
- UPLC
- ultraperformance LC
- MIPC
- mannose inositol phosphorylceramide
- M(IP)2C
- mannose (inositol phosphoryl)2-ceramide.
REFERENCES
- 1. van Meer G., Voelker D. R., Feigenson G. W. (2008) Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 3. Chalfant C., Del Poeta M. (eds) (2010) Sphingolipids as Signaling and Regulatory Molecules, Landes Bioscience, Austin, TX [Google Scholar]
- 4. Warnecke D., Heinz E. (2003) Cell. Mol. Life Sci. 60, 919–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pata M. O., Hannun Y. A., Ng C. K. (2010) New Phytol. 185, 611–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zäuner S., Ternes P., Warnecke D. (2010) Adv. Exp. Med. Biol. 688, 249–263 [DOI] [PubMed] [Google Scholar]
- 7. Wennekes T., van den Berg R. J., Boot R. G., van der Marel G. A., Overkleeft H. S., Aerts J. M. (2009) Angew. Chem. Int. Ed. Engl. 48, 8848–8869 [DOI] [PubMed] [Google Scholar]
- 8. Dickson R. C. (2008) J. Lipid Res. 49, 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dickson R. C., Lester R. L. (1999) Biochim. Biophys. Acta 1426, 347–357 [DOI] [PubMed] [Google Scholar]
- 10. Mattanovich D., Graf A., Stadlmann J., Dragosits M., Redl A., Maurer M., Kleinheinz M., Sauer M., Altmann F., Gasser B. (2009) Microb. Cell Fact. 8, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Schutter K., Lin Y. C., Tiels P., Van Hecke A., Glinka S., Weber-Lehmann J., Rouzé P., Van de Peer Y., Callewaert N. (2009) Nat. Biotechnol. 27, 561–566 [DOI] [PubMed] [Google Scholar]
- 12. Sperling P., Zähringer U., Heinz E. (1998) J. Biol. Chem. 273, 28590–28596 [DOI] [PubMed] [Google Scholar]
- 13. Ternes P., Franke S., Zähringer U., Sperling P., Heinz E. (2002) J. Biol. Chem. 277, 25512–25518 [DOI] [PubMed] [Google Scholar]
- 14. Ternes P., Sperling P., Albrecht S., Franke S., Cregg J. M., Warnecke D., Heinz E. (2006) J. Biol. Chem. 281, 5582–5592 [DOI] [PubMed] [Google Scholar]
- 15. Haak D., Gable K., Beeler T., Dunn T. (1997) J. Biol. Chem. 272, 29704–29710 [DOI] [PubMed] [Google Scholar]
- 16. Grilley M. M., Stock S. D., Dickson R. C., Lester R. L., Takemoto J. Y. (1998) J. Biol. Chem. 273, 11062–11068 [DOI] [PubMed] [Google Scholar]
- 17. Menuz V., Howell K. S., Gentina S., Epstein S., Riezman I., Fornallaz-Mulhauser M., Hengartner M. O., Gomez M., Riezman H., Martinou J. C. (2009) Science 324, 381–384 [DOI] [PubMed] [Google Scholar]
- 18. Levy M., Futerman A. H. (2010) IUBMB Life 62, 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Folch J., Lees M., Sloane Stanley G. H. (1957) J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 20. Guan X. L., Wenk M. R. (2006) Yeast 23, 465–477 [DOI] [PubMed] [Google Scholar]
- 21. Hirschberg K., Rodger J., Futerman A. H. (1993) Biochem. J. 290, 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lahiri S., Lee H., Mesicek J., Fuks Z., Haimovitz-Friedman A., Kolesnick R. N., Futerman A. H. (2007) FEBS Lett. 581, 5289–5294 [DOI] [PubMed] [Google Scholar]
- 23. Blight E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 24. Leipelt M., Warnecke D., Zähringer U., Ott C., Müller F., Hube B., Heinz E. (2001) J. Biol. Chem. 276, 33621–33629 [DOI] [PubMed] [Google Scholar]
- 25. Takakuwa N., Ohnishi M., Oda Y. (2008) FEMS Yeast Res. 8, 839–845 [DOI] [PubMed] [Google Scholar]
- 26. Guillas I., Kirchman P. A., Chuard R., Pfefferli M., Jiang J. C., Jazwinski S. M., Conzelmann A. (2001) EMBO J. 20, 2655–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schorling S., Vallée B., Barz W. P., Riezman H., Oesterhelt D. (2001) Mol. Biol. Cell 12, 3417–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cerantola V., Vionnet C., Aebischer O. F., Jenny T., Knudsen J., Conzelmann A. (2007) Biochem. J. 401, 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Michaelson L. V., Zäuner S., Markham J. E., Haslam R. P., Desikan R., Mugford S., Albrecht S., Warnecke D., Sperling P., Heinz E., Napier J. A. (2009) Plant Physiol. 149, 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sperling P., Blume A., Zähringer U., Heinz E. (2000) Biochem. Soc. Trans. 28, 638–641 [PubMed] [Google Scholar]
- 31. Oura T., Kajiwara S. (2010) Microbiology 156, 1234–1243 [DOI] [PubMed] [Google Scholar]
- 32. Ramamoorthy V., Cahoon E. B., Li J., Thokala M., Minto R. E., Shah D. M. (2007) Mol. Microbiol. 66, 771–786 [DOI] [PubMed] [Google Scholar]
- 33. Noble S. M., French S., Kohn L. A., Chen V., Johnson A. D. (2010) Nat. Genet. 42, 590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marsh D. (2010) Biochim. Biophys. Acta 1798, 40–51 [DOI] [PubMed] [Google Scholar]
- 35. Garton S., Michaelson L. V., Beaudoin F., Beale M. H., Napier J. A. (2003) FEBS Lett. 538, 192–196 [DOI] [PubMed] [Google Scholar]
- 36. Rittershaus P. C., Kechichian T. B., Allegood J. C., Merrill A. H., Jr., Hennig M., Luberto C., Del Poeta M. (2006) J. Clin. Investig. 116, 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oura T., Kajiwara S. (2008) Microbiology 154, 3795–3803 [DOI] [PubMed] [Google Scholar]
- 38. Ramamoorthy V., Cahoon E. B., Thokala M., Kaur J., Li J., Shah D. M. (2009) Eukaryot. Cell 8, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sakaki T., Zähringer U., Warnecke D. C., Fahl A., Knogge W., Heinz E. (2001) Yeast 18, 679–695 [DOI] [PubMed] [Google Scholar]
- 40. Heidler S. A., Radding J. A. (1995) Antimicrob. Agents Chemother. 39, 2765–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nagiec M. M., Nagiec E. E., Baltisberger J. A., Wells G. B., Lester R. L., Dickson R. C. (1997) J. Biol. Chem. 272, 9809–9817 [DOI] [PubMed] [Google Scholar]
- 42. Luberto C., Toffaletti D. L., Wills E. A., Tucker S. C., Casadevall A., Perfect J. R., Hannun Y. A., Del Poeta M. (2001) Genes Dev. 15, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cheng J., Park T. S., Fischl A. S., Ye X. S. (2001) Mol. Cell. Biol. 21, 6198–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cerantola V., Guillas I., Roubaty C., Vionnet C., Uldry D., Knudsen J., Conzelmann A. (2009) Mol. Microbiol. 71, 1523–1537 [DOI] [PubMed] [Google Scholar]
- 45. Tani M., Kuge O. (2010) J. Biochem. 148, 565–571 [DOI] [PubMed] [Google Scholar]
- 46. Li S., Bao D., Yuen G., Harris S. D., Calvo A. M. (2007) Genetics 176, 243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fahy E., Subramaniam S., Brown H. A., Glass C. K., Merrill A. H., Jr., Murphy R. C., Raetz C. R., Russell D. W., Seyama Y., Shaw W., Shimizu T., Spener F., van Meer G., VanNieuwenhze M. S., White S. H., Witztum J. L., Dennis E. A. (2005) J. Lipid Res. 46, 839–861 [DOI] [PubMed] [Google Scholar]
- 48. Kageyama-Yahara N., Riezman H. (2006) Biochem. J. 398, 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gould S. J., McCollum D., Spong A. P., Heyman J. A., Subramani S. (1992) Yeast 8, 613–628 [DOI] [PubMed] [Google Scholar]
- 50. D'mello N. P., Childress A. M., Franklin D. S., Kale S. P., Pinswasdi C., Jazwinski S. M. (1994) J. Biol. Chem. 269, 15451–15459 [PubMed] [Google Scholar]
- 51. Li S., Du L., Yuen G., Harris S. D. (2006) Mol. Biol. Cell 17, 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pewzner-Jung Y., Ben-Dor S., Futerman A. H. (2006) J. Biol. Chem. 281, 25001–25005 [DOI] [PubMed] [Google Scholar]
- 53. Schmidt H. A., Strimmer K., Vingron M., von Haeseler A. (2002) Bioinformatics 18, 502–504 [DOI] [PubMed] [Google Scholar]
- 54. Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 55. Ejsing C. S., Moehring T., Bahr U., Duchoslav E., Karas M., Simons K., Shevchenko A. (2006) J. Mass Spectrom. 41, 372–389 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.