Abstract
Notch signaling is active in many sites, and its diverse activities must require tissue-specific intermediaries, which are largely unknown. In the intestinal epithelium, Notch promotes crypt cell proliferation and inhibits goblet cell differentiation. Pharmacologic studies suggest that the latter effect occurs through the transcription factor Math1/Atoh1, which specifies all intestinal secretory cells. We tested this hypothesis using mouse mutants. Genetic loss of the Notch effector RBP-Jκ alone increases all intestinal secretory lineages, with variation between proximal and distal gut segments. This secretory cell excess observed with RBP-Jκ loss was blocked in the absence of Math1 in RBP-JκFl/Fl;Math1Fl/Fl;Villin-Cre(ER-T2) mice. Loss of both factors also restored progenitor replication, proving that Math1 is epistatic to Notch signaling in both secretory cell differentiation and crypt cell proliferation. Investigating mechanisms downstream of Math1, we found that expression of the known Notch effector protein Hes1 was predictably lost in RBP-Jκ−/− mice but surprisingly recovered in RBP-Jκ;Math1 compound conditional mutants. Furthermore, the cell cycle inhibitors p27Kip1 and p57Kip2 were selectively overexpressed in duodenal and ileal crypts, respectively, in RBP-Jκ-deficient mice. Regional activation of these products was completely abrogated in the absence of Math1. Thus, all intestinal Notch effects channel through the tissue-restricted factor Math1, which promotes secretory differentiation and cell cycle exit by regionally distinct mechanisms. Our data further suggest that, besides transmitting Notch signals, the transcription factor Hes1 acts downstream of Math1 to regulate expression of cell cycle inhibitors and intestinal crypt cell replication.
Keywords: Cell Cycle, Cell Differentiation, Colon Cancer, Development, Helix-Loop-Helix Transcription Factors, Mouse Genetics, Notch Pathway, Stem Cell
Introduction
Stem and progenitor cells proliferate briskly in the crypts of the self-renewing gut epithelium and differentiate into two major lineages, absorptive and secretory cells, which populate villi in the small intestine (1). Absorptive enterocytes of a single type digest and absorb nutrients, whereas three distinct types of secretory cells, goblet, Paneth, and enteroendocrine (EE),2 produce mucus, antimicrobial peptides, and various hormones, respectively. Against a background of rapid cell turnover, replication and differentiation of progenitors must be coordinated precisely. The Wnt signaling pathway plays a seminal role in these events and in intestinal cancers (2). Recent studies further implicate Notch signaling in intestinal epithelial homeostasis, again with possible implications for cancer. Zebrafish mutants deficient in Notch signaling and mouse embryos lacking the Notch target and effector product Hes1 increase secretory cell numbers at the expense of enterocytes (3, 4), suggesting that the Notch pathway directs cell fate choices. Gain and loss of Notch function in mice provide further evidence of its role in repressing secretory differentiation (5–7), although it is unclear if Notch is absolutely necessary for enterocyte specification or maturation. Additionally, inhibition of Notch activity by drugs or gene targeting potently arrests proliferation of crypt progenitors and induces impressive goblet cell metaplasia. Pharmacologic inhibition of Notch converts adenomas in ApcMin mice into postmitotic goblet cells (5). Thus, the sum effect of Notch signaling in the mammalian intestine is to promote crypt cell proliferation and inhibit secretory lineage differentiation.
The cell fate decisions and progenitor properties that Notch controls in diverse tissues (8, 9) are probably mediated by distinct tissue-specific factors, most of them unknown. The intestine- and nervous system-specific basic helix-loop-helix transcription factor Math1 is a good candidate mediator of cell lineage function because it is required to specify intestinal secretory cells. Math1−/− secretory progenitors differentiate into enterocytes (10, 11). Moreover, an inverse relationship between Math1 expression and Notch activity is well documented. Loss of Notch function induces Math1 expression in intestinal crypts (5), and gain of Notch activity represses Math1 expression (7). Two groups recently reported that Math1−/− intestines resist the goblet cell metaplasia effects of γ-secretase inhibitors (12, 13). Although the latter studies support the idea that Math1 functions downstream of Notch signaling in allocating intestinal epithelial lineages, they lacked the design necessary to establish a definitive epistatic relationship. Furthermore, because the full spectrum of γ-secretase inhibitor activity is unknown, it remains uncertain if the distinct Notch functions in cell replication and differentiation channel through the same downstream effectors. Genetic analysis should help overcome these limitations.
We used a genetic approach to determine how Notch might accomplish its diverse intestinal functions, studying conditional compound mutant mice that lack intestinal expression of both RBP-Jκ, the core transcriptional effector of Notch signaling (14), and Math1. Combined genetic loss of Math1 and RBP-Jκ completely rescued both the secretory cell metaplasia and cell cycle arrest that occur with isolated loss of RBP-Jκ. Surprisingly, predictable loss of expression of the Notch target and effector product Hes1 in RBP-Jκ-null intestines was also restored in the absence of Math1, indicating complex pathway interactions. Finally, we uncovered regional differences in the effects and mechanisms of Notch activity in the proximal and distal intestine. On a wild-type Math1 background, although loss of RBP-Jκ/Notch activity rapidly induced Math1 in crypts throughout the intestine, the cell cycle regulator p27Kip1 appeared only in the duodenum and not in the ileum, whereas p57Kip2 appeared in the ileum and not in the duodenum. Abnormal activation of these presumptive direct Hes1 target genes was abrogated in intestines doubly deficient for RBP-Jκ and Math1.
Our results establish a bona fide genetic hierarchy wherein Notch signaling inhibits secretory cell differentiation and stimulates crypt cell proliferation through obligate repression of Math1. Downstream of this unifying molecular mechanism, Notch activity represses distinct cell cycle inhibitors to regulate crypt cell replication in the proximal and distal intestine.
EXPERIMENTAL PROCEDURES
Experimental Animals and Drug Administration
Math1Fl and ROSA26Lox-Stop-Lox-EYFP mice were purchased from The Jackson Laboratories (Bar Harbor, ME). RBP-JκFl mice (15) were generously provided by S. Artavanis-Tsakonas, with permission from T. Honjo. Villin-Cre(ER-T2) transgenic mice (16) were kindly provided by S. Robine. Cre recombinase was induced by 5 consecutive days of intraperitoneal injection of tamoxifen (Invitrogen), 1 mg dissolved in 10% ethanol and sunflower oil; mice were euthanized 4 days after the last injection. BrdU (BD Biosciences, 50 mg/kg) was injected into the peritoneum of adult mice 1 h before euthanasia. The γ-secretase inhibitor dibenzazepine was synthesized at Syncom (Groningen, Holland), suspended in 0.5% (w/v) hydroxypropylmethylcellulose (Methocel E4M, Dow Chemicals, Michigan) and 0.1% (w/v) Tween 80 in water, and injected into the peritoneum of adult mice at the indicated doses.
Histology and Immunohistochemistry
Paraffin-embedded 5-μm sections of tissues previously fixed in 4% paraformaldehyde overnight at 4 °C were rehydrated, stained in Alcian blue solution (pH 2.5), and counterstained with nuclear fast red. For immunohistochemistry, antigens were retrieved in 10 mm Na citrate buffer (pH 6.0), and endogenous peroxidase activity was blocked in methanol and 3% H2O2. Sections were treated with FBS before incubation with one of the following antibodies: ChromograninA (1:500, Abcam, Cambridge, MA), Ki67 (clone MM1, 1:1000, Vector Laboratories, Burlingame, CA), Math1 (1:100, gift of Dr. J. Johnson), Hes1 (1:500, gift of Dr. N. Brown), phospho-Ezrin/Radixin/Moesin (1:100, Cell Signaling Technology, Danvers MA), BrdU (1:300, AbD Serotec, Raleigh, NC), Crs4c1 (1:1000, gift of Dr. Andre Ouellette), p27kip1 (1:100, BD Biosciences), p57kip2 (1:50, Abcam), or green fluorescent protein (1:200, Cell Signaling Technology). After washing, samples were incubated with biotin-conjugated anti-mouse or anti-rabbit IgG (1:300, Vector Laboratories) and color reactions developed with the Vectastain avidin-biotin complex ABC kit (Vector Laboratories) and diaminobenzidine substrate (Sigma). Slides were examined with an Olympus BX41 upright microscope and images captured with a charge-coupled device camera (QCapture, Surrey, BC, Canada) using Adobe Photoshop 7.0. ChromograninA+ endocrine cells, and Crs4c (cryptdin)-expressing Paneth cells were counted in 50 representative crypts and villi in wild-type and RBP-JκFl/Fl;Villin-Cre(ER-T2) mice (n = 3 each). BrdU+-replicating cells were counted in 50 representative crypts of wild-type, RBP-JκFl/Fl;Villin-Cre(ER-T2), RBP-JκFl/Fl;Math1Fl/Fl; Villin-Cre(ER-T2), and Math1Fl/Fl;Villin-Cre(ER-T2) mice (n = 3 each). Mean cell counts and S.D. were calculated using Microsoft Excel.
RESULTS
Notch Signaling Inhibits Differentiation of All Secretory Cell Types, with Regional Differences along the Intestinal Length
Genetic inhibition of Notch signaling arrests crypt cell proliferation and causes profound goblet cell metaplasia but is reported to affect other secretory lineages minimally, if at all (5). Because accurate assessment of Notch function requires distinction between unilineage and multilineage effects, we critically examined all secretory cell types in a genetic model for the loss of Notch signaling. We used a conditional, Cre recombinase-dependent allele of the core Notch transcriptional effector RBP-Jκ (15) and Villin-Cre(ER-T2) transgenic mice (16), which respond to tamoxifen with efficient activation of Cre throughout the small bowel epithelium. In addition to a massive increase in goblet cells (see Alcian blue (AB) staining in Fig. 2A), similar to the scale reported previously (5), tamoxifen-treated RBP-JκFl/Fl;Villin-Cre(ER-T2) mice also showed increased fractions of EE and Paneth cells (Fig. 1), confirming that Notch signaling normally represses all intestinal secretory lineages. Furthermore, loss of Notch activity had quantitatively different effects in the proximal and distal gut. The numbers of EE cells marked by ChromograninA expression were increased 4-fold in the duodenum, compared with 1.5-fold in the distal ileum. In contrast, cryptdin-expressing Paneth cells were increased about 2.5-fold in the ileum but less than 2-fold in the duodenum (Fig. 1). This variation mirrors basal differences in cell proportions in the small intestine, where EE cells are more plentiful in the duodenum and Paneth cells more so in the ileum (17, 18). Exposure of wild-type mice to the Notch/γ-secretase inhibitor dibenzazepine (19) also increased the numbers of all intestinal secretory cells (supplemental Fig. 1), as recently reported in mice treated with a different inhibitor, GSI-20 (12). Thus, Notch inhibition affects the full secretory lineage, with discernible differences between the proximal and distal intestine.
FIGURE 2.
Requirement for Math1 in secretory cell metaplasia induced by loss of Notch activity. A, Alcian blue-positive goblet cells increased substantially after tamoxifen treatment of RBP-JκFl/Fl;Villin-Cre(ER-T2) mice but not of Math1Fl/Fl;Villin-Cre(ER-T2) (top) or RBP-JκFl/Fl;Math1Fl/Fl;Villin-Cre(ER-T2) (bottom) mice. Similarly, Math1Fl/Fl;Villin-Cre(ER-T2), and RBP-JκFl/Fl;Math1Fl/Fl;Villin-Cre(ER-T2) small intestines lacked all Crs4c1-expressing Paneth and ChromograninA (ChgA)-positive EE cells, indicating loss of all secretory lineages with lone loss of Math1 or combined absence of Math1 and RBP-Jκ. Failure of secretory cell metaplasia was equally evident in the duodenum (top rows) and ileum (bottom rows). B, expression of intestinal alkaline phosphatase (AP) and the microvillus brush border marker phospho-Ezrin/Radixin/Moesin (p-ERM) was notably reduced in RBP-JκFl/Fl;Villin-Cre(ER-T2) mice after tamoxifen treatment, reflecting significant enterocyte loss, but restored in RBP-JκFl/Fl;Math1Fl/Fl;Villin-Cre(ER-T2) mice and preserved in Math1Fl/Fl;Villin-Cre(ER-T2) mice. The results indicate that Notch signaling is dispensable for enterocyte maturation, at least when Math1 is absent. Scale bars = 50 μm.
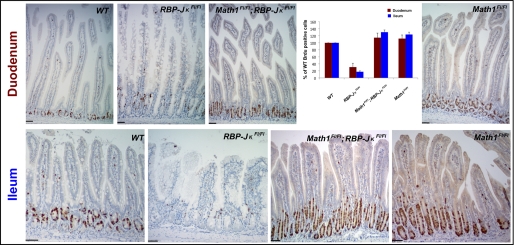
FIGURE 1.
Role for Notch signaling in all three intestinal secretory cell lineages. Tamoxifen treatment increased the numbers of ChromograninA-positive EE cells (top row) as well as Paneth cells expressing the cryptdin Crs4c1 (bottom row) in RBP-JκFl/Fl;Villin-Cre(ER-T2) mice, compared with wild-type littermates. Insets in the duodenal panels show high-magnification images of the boxed areas, and quantitation of immunohistochemical data is shown (right). The duodenum showed a larger increase in EE cells, whereas Paneth cells were more significantly increased in the ileum. Scale bars = 50 μm.
Epistatic Test of the Role of Math1 in Notch Signaling and in the Choice between Secretory or Absorptive Cell Differentiation
The basic helix-loop-helix transcription factor Math1 is required for differentiation of all intestinal secretory cells, possibly at the level of specifying a common secretory progenitor (10, 11). Moreover, intestinal Math1 expression levels drop upon Notch pathway activation and increase when Notch signaling is blocked (5, 7). Our analysis of Math1 expression in mice treated with dibenzazepine confirmed the latter result (supplemental Fig. 2). We therefore postulated that the increase in numbers of all secretory cell types after Notch inhibition (Fig. 1) might represent a direct consequence of increased Math1 expression. To test the requirement for Math1 in Notch-dependent cell differentiation, we crossed mice carrying conditional, Cre-dependent null alleles for RBP-Jκ and Math1 to generate compound RBP-JκFl/Fl;Math1Fl/Fl;Villin-Cre(ER-T2) mutants. In contrast to a Cre-expressing strain previously used to assess Math1 function, which was limited by mosaicism (11), Villin-Cre(ER-T2);ROSA26Lox-Stop-Lox-EYFP mice verified efficient Cre activation throughout the small bowel epithelium (supplemental Fig. 3).
In both the duodenum and ileum, the goblet cell metaplasia that always followed tamoxifen treatment of RBP-JκFl/Fl;Villin-Cre(ER-T2) mice was completely absent when the Math1 gene was also disrupted (Fig. 2A). Indeed, as the phenotype of single-mutant Math1−/− mice (11) might have predicted, RBP-JκFl/Fl;Math1Fl/Fl;Villin-Cre(ER-T2) compound mutants lacked all intestinal secretory cells (Fig. 2A). This phenocopy of the absence of Math1 reveals a crucial requirement for Math1 in the full effect of Notch inhibition on secretory cell differentiation and establishes that Math1 is epistatic to Notch signaling in secretory cell specification.
Although high Notch activity is necessary for enterocyte differentiation in Drosophila (20), Notch signaling requirements for the absorptive lineage in the mammalian gut are unclear. In principle, enterocyte differentiation may represent the default outcome when Notch signaling represses the secretory lineage. The combination of conditional-null RBP-Jκ and Math1 alleles in the same mice allowed us to assess Notch pathway requirements when absence of Math1 favors enterocyte differentiation at the expense of secretory cells. Enterocyte morphology and the expression and distribution of intestinal alkaline phosphatase or the brush border marker phospho-Ezrin/Radixin/Moesin (21) were unaffected in tamoxifen-treated RBP-JκFl/Fl;Math1Fl/Fl;Villin-Cre(ER-T2) intestines (Fig. 2B). Thus, in the absence of Math1, Notch signaling is dispensable for proper maturation or maintenance of absorptive intestinal cells.
Notch Activity in Intestinal Progenitor Cell Replication Also Operates through Math1
Blockade of Notch signaling in the mouse intestine potently arrests crypt cell replication (5), implicating Notch activity in progenitor cell proliferation. Recent evidence further suggests that Notch and Wnt signals cooperate in intestinal tumorigenesis (22, 23). Notch pathway inhibition could block cell proliferation either by virtue of an intrinsic role in progenitor cells, as is commonly assumed (9), or as a secondary consequence of excessive, Math1-dependent secretory cell differentiation. Although secretory cells are totally absent in Math1−/− mice, cell proliferation is affected only subtly (10, 11), making Math1 repression an improbable mediator of the cell proliferation effects of Notch signaling. We monitored cell proliferation using the S-phase reagent bromodeoxyuridine (BrdU), whose transit-amplifying progenitors lying near the base of small intestine crypts incorporate rapidly (Fig. 3). In the absence of RBP-Jκ, BrdU uptake was significantly reduced in all crypts and absent from many. The few remaining BrdU+ cells were positioned higher in the crypts because of an expanded Paneth cell compartment at the base (Fig. 3). To our surprise, the cell cycle effects of isolated RBP-Jκ loss were, like the secretory cell metaplasia, also fully rescued in RBP-JκFl/Fl;Math1Fl/Fl;Villin-Cre(ER-T2) intestines (Fig. 3). In fact, compared with the wild-type distal ileum, pulse BrdU labeling revealed an approximately 30% increase in the fraction of cycling RBP-Jκ and Math1-double deficient cells, a value very similar to the increased fraction of cycling cells in Math1-deficient intestines (Fig. 3). These results establish that Math1 is also epistatic to Notch signaling for intestinal crypt cell proliferation.
FIGURE 3.
Math1 acts downstream of Notch signaling as an obligate regulator of intestinal crypt cell replication. Assessment of crypt cell replication by incorporation of the S-phase marker BrdU. Tamoxifen-induced arrest of cell proliferation in RBP-JκFl/Fl;Villin-Cre(ER-T2) mice was fully rescued in the absence of Math1, in both duodenum (top row) and ileum (bottom row). Absence of Math1 alone modestly increased cell proliferation in both regions, and this degree of increase persisted in RBP-JκFl/Fl;Math1Fl/Fl;Villin-Cre(ER-T2) mice. Immunohistochemical data are quantified (top right). Scale bars = 50 μm.
Math1 haplodeficiency was inadequate to rescue the intestinal epithelial defects associated with absence of Notch signaling. Tamoxifen-treated Math1+/Fl;RBP-JκFl/Fl;Villin-Cre(ER-T2) mice showed a persistent and considerable excess of all intestinal secretory cell types, although goblet cell numbers were slightly reduced compared with their Math1+/+ counterparts, and minimal BrdU uptake (supplemental Fig. 4).
Unexpected Hes1 Expression with Combined Absence of Notch Signaling and Math1
To investigate how Math1 might implement a cell cycle arrest in the absence of Notch activity, we first examined expression of Hes1, which has been characterized extensively as a target and effector of Notch signaling in many tissues (24, 25). However, mice with forebrain-specific knockout of RBP-Jκ show loss of Hes5 but retain normal Hes1 expression (26), indicating that in certain contexts Hes1 can be expressed independent of Notch signaling and RBP-Jκ. Furthermore, Math1 depletion in cultured colon cancer cells results in Hes1 overexpression (12). We therefore considered the possibility that Math1 might normally inhibit Hes1 independent of RBP-Jκ activity. As expected for a Notch target gene, and compared with wild-type controls, Hes1 expression was virtually absent in RBP-Jκ−/−mice in both the proximal and distal intestine (Fig. 4A). To our surprise, Hes1 expression recovered significantly in RBP-Jκ−/−;Math1−/− mice, indicating derepression in the absence of Math1. These results reveal an unexpectedly complex interplay among transcription factors acting downstream of Notch signaling (Fig. 4B). In particular, whereas Hes1 is widely considered an obligate effector and target of Notch activity, and our genetic analysis reveals Math1 as a second obligate effector in the intestine (Figs. 2–3), these data suggest a separate, repressive effect of Math1 over Hes1 expression in intestinal crypts.
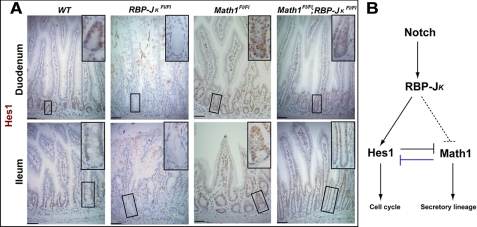
FIGURE 4.
Recovery of Hes1 expression in the combined absence of RBP-Jκ and Math1. A, Hes1 immunohistochemistry reveals that its expression is abolished after tamoxifen treatment of RBP-JκFl/Fl;Villin-Cre(ER-T2) intestines, preserved or slightly increased in Math1Fl/Fl;Villin-Cre(ER-T2) intestines, and restored in RBP-JκFl/Fl;Math1Fl/Fl;Villin-Cre(ER-T2) duodenum and ileum. Insets in each panel show high-magnification images of the boxed crypts. Scale bars = 50 μm. B, schematic diagram illustrating that, in addition to its known role as a direct Notch target and effector protein, Hes1 is also regulated by Math1. Previously known relationships are indicated with solid black lines and the relationship uncovered in the present study with a solid blue line. The dashed lines reflect uncertainty whether RBP-Jκ represses Math1 directly or through Hes1.
Regional Differences in Expression of Cell Cycle Regulators Downstream of Notch and Math1
Hes1 behaves as a transcriptional repressor and its gene targets include the cell cycle-dependent kinase inhibitors p27Kip1 and p57Kip2. Expression of these cell cycle regulators often increases when Notch signaling is blocked (27–29), suggesting a Hes1-dependent molecular pathway in replication of intestinal epithelial progenitors. To appreciate the basis of cell proliferation effects in RBP-Jκ and Math1 mutant intestines, we examined p27Kip1 and p57Kip2 expression. Isolated, tamoxifen-induced loss of RBP-Jκ resulted in a robust increase in crypt cell expression of p27Kip1 in the duodenum but not in the ileum. Conversely, p57Kip2 expression was markedly increased in ileal but not in duodenal crypts (Fig. 5). This aberrant activation of cell cycle inhibitors and known Hes1 targets was fully reversed in the intestines of tamoxifen-treated RBP-JκFl/Fl;Math1Fl/Fl;Villin-Cre(ER-T2) mice (Fig. 5). These results reveal regionally distinct mechanisms acting downstream of Notch and implicate Math1 in regulating p27Kip1 and p57Kip2 expression.
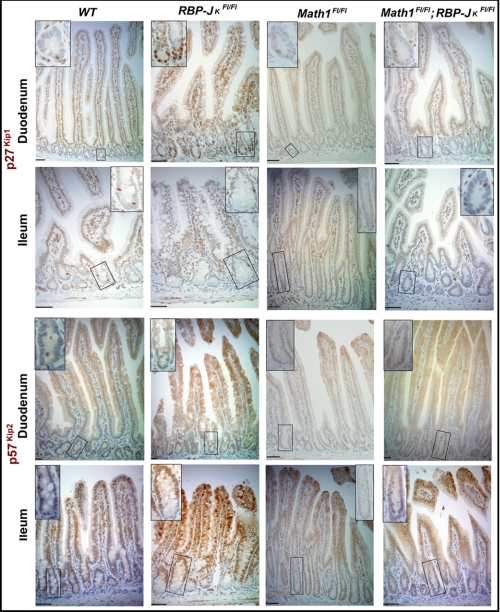
FIGURE 5.
Differential Notch-induced repression of cell cycle inhibitors in the proximal and distal small intestine. Expression of the endogenous cell cycle inhibitor p27Kip1, normally confined to a few crypt cells in tamoxifen-treated wild-type and Math1Fl/Fl;Villin-Cre(ER-T2) mice, was significantly increased in duodenal (top row) but not ileal (bottom row) crypts in RBP-JκFl/Fl;Villin-Cre(ER-T2) mice. This abnormal increase in p27kip1 expression was not observed in the absence of Math1. Expression of the alternative cell cycle inhibitor p57Kip2, also largely absent from tamoxifen-treated wild-type and Math1Fl/Fl;Villin-Cre(ER-T2) crypts, increased significantly in ileal (bottom row) but not duodenal (top row) crypts in RBP-JκFl/Fl;Villin-Cre(ER-T2) mice. Ileal p57Kip2 expression did not increase in the absence of Math1. Insets in each panel show high-magnification images of the boxed crypts. Scale bars = 50 μm.
DISCUSSION
Although early insights into Notch pathway functions in the intestine drew on genetic mouse models, recent conclusions regarding the role of the transcription factor Math1 rely mainly on pharmacologic inhibition of Notch signaling (12, 13). Our activation of Cre recombinase in the intestinal epithelium of compound mutant RBP-JκFl/Fl;Math1Fl/Fl mice clarifies Notch functions in this tissue and provides firm genetic evidence for repression of Math1 as a core molecular mechanism controlling both proliferation and fate selection in progenitor cells. Furthermore, we show that Notch-mediated repression of Math1 inhibits differentiation of all secretory cell types but is not required for de novo specification or maturation of enterocytes, implying that absorptive cells may represent a default, Notch-independent lineage. Whereas absence of secretory cell differentiation in the absence of both RBP-Jκ and Math1 may have been predictable from the known phenotype of Math1−/− mice (10), there was little precedent to expect that absence of Math1 would also fully rescue the dramatic crypt cell proliferation arrest that occurs upon interference with Notch activity. Our results with genetic crosses and epistasis hence place Math1 at a crucial juncture downstream of Notch signaling in the intestinal epithelium. Its obligate repression serves as the predominant mode of action both in specifying the secretory lineage and in forcing crypt progenitors to exit the cell cycle.
In principle, it is formally possible that Math1 acts independent of Notch signaling and that loss of Math1 function somehow masks both the cell cycle arrest and secretory metaplasia induced by Notch inhibition. However, this is highly unlikely in light of the known inverse relationship between Math1 expression and Notch activity (5, 7) and the resistance of Math1-null mice to the intestinal effects of γ-secretase inhibitors (12, 13), findings that support a role for Math1 directly downstream of Notch. Moreover, VanDussen and Samuelson (30) recently showed that overexpression of Math1 in the intestines of transgenic mice induces secretory metaplasia, nearly complete enterocyte loss, and dramatically reduced epithelial proliferation. These defects exactly match those observed upon loss of Notch activity and provide additional in vivo evidence that intestinal Math1 functions are linked to Notch signaling. Taken together, the weight of evidence hence strongly favors a direct role for Math1 in mediating intestinal Notch functions.
In the combined absence of RBP-Jκ and Math1, we also observed that Hes1 expression, normally lost in the absence of Notch activity, was restored, revealing a surprising regulation of Hes1 expression by Math1 in intestinal crypt cells. One unifying possibility is that all intestinal Notch effects, including Hes1 expression, channel through Math1. However, because Hes1 appears to be a nearly universal Notch target, even in tissues lacking Math1, we favor the alternative model depicted in Fig. 4B. In this model, Hes1 is a direct Notch target, whereas Math1, another downstream effector of Notch signaling, may be regulated directly or indirectly, perhaps even through Hes1. Moreover, Math1 mRNA levels are increased in Hes1−/− mice (4), suggesting the possibility of mutual repression. The surprising result from our study is that Math1 suppresses Hes1 expression independent from Notch activity and that absence of Math1 allows Hes1 expression in RBP-Jκ-deficient intestines. This result provides a satisfying explanation for the otherwise puzzling observation that Math1 loss rescues both the secretory differentiation and cell proliferation effects of the absence of Notch signaling.
The potent effect of Math1 loss on secretory cell differentiation in RBP-Jκ-null intestine is readily explained by its known requirement in specification of secretory progenitors (10, 11). However, at least when the Notch pathway is active, Math1 must exert only a subtle influence over crypt cell proliferation. Indeed, a measurable shortfall is elicited in Math1-null mice only when they are challenged with surgical bowel resection (11). How then does the absence of Math1 restore normal crypt cell proliferation (Fig. 3)? Notch-independent regulation of Hes1 and its downstream targets p27Kip1 and p57Kip2 by Math1 seems to provide at least a partial, and possibly complete, answer (Fig. 4B). Expression of Hes1 and its target cell cycle-dependent kinase inhibitors seems to be critical, as their expression patterns mirror in vivo cell proliferation defects. We also observed differences along the rostrocaudal axis of the intestine, with selective expression of p27Kip1 in duodenal crypts and of p57Kip2 in ileal crypts in the absence of Notch signaling (Fig. 5). Together with quantitative differences in Notch pathway effects on Paneth and EE cells in the duodenum and ileum (Fig. 1), these results highlight the varied functions and mechanisms of an important signaling pathway in different segments of the digestive tract.
Notch and Wnt signaling are believed to cooperate in intestinal epithelial tumorigenesis (7, 22, 23). Independently, silencing of the ATOH1 locus is reported in some human colorectal cancers, and Math1 loss enhances intestinal tumorigenesis in mice, implicating it as a tumor suppressor (31). Recent evidence that Math1 is necessary to observe the growth and differentiation effects of γ-secretase inhibitors in APCMin mice reinforces this idea (12). Our results on Notch-dependent cell proliferation in a genetic mouse model further suggest that cooperation between Notch and Wnt signaling in intestinal tumorigenesis may occur through Math1 and reflect complex interactions with Hes1.
Supplementary Material
Acknowledgments
We thank T. Honjo, S. Robine, and S. Artavanis-Tsakonas for providing mice; and J. Johnson, N. Brown, and A. Ouellette for gifts of antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK081113 (to R. A. S.). This work was also supported by a gift from the Caring for Carcinoid Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- EE
- enteroendocrine.
REFERENCES
- 1. van der Flier L. G., Clevers H. (2009) Annu. Rev. Physiol. 71, 241–260 [DOI] [PubMed] [Google Scholar]
- 2. Clevers H. (2006) Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 3. Crosnier C., Vargesson N., Gschmeissner S., Ariza-McNaughton L., Morrison A., Lewis J. (2005) Development 132, 1093–1104 [DOI] [PubMed] [Google Scholar]
- 4. Jensen J., Pedersen E. E., Galante P., Hald J., Heller R. S., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O. D. (2000) Nat. Genet. 24, 36–44 [DOI] [PubMed] [Google Scholar]
- 5. van Es J. H., van Gijn M. E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D. J., Radtke F., Clevers H. (2005) Nature 435, 959–963 [DOI] [PubMed] [Google Scholar]
- 6. Stanger B. Z., Datar R., Murtaugh L. C., Melton D. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12443–12448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fre S., Huyghe M., Mourikis P., Robine S., Louvard D., Artavanis-Tsakonas S. (2005) Nature 435, 964–968 [DOI] [PubMed] [Google Scholar]
- 8. Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 9. Wilson A., Radtke F. (2006) FEBS Lett. 580, 2860–2868 [DOI] [PubMed] [Google Scholar]
- 10. Yang Q., Bermingham N. A., Finegold M. J., Zoghbi H. Y. (2001) Science 294, 2155–2158 [DOI] [PubMed] [Google Scholar]
- 11. Shroyer N. F., Helmrath M. A., Wang V. Y., Antalffy B., Henning S. J., Zoghbi H. Y. (2007) Gastroenterology 132, 2478–2488 [DOI] [PubMed] [Google Scholar]
- 12. Kazanjian A., Noah T., Brown D., Burkart J., Shroyer N. F. (2010) Gastroenterology 139, 918–928, 928 e1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Es J. H., de Geest N., van de Born M., Clevers H., Hassan B. A. (2010) Nat. Commun. 1, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. (1995) Nature 377, 355–358 [DOI] [PubMed] [Google Scholar]
- 15. Han H., Tanigaki K., Yamamoto N., Kuroda K., Yoshimoto M., Nakahata T., Ikuta K., Honjo T. (2002) Int. Immunol. 14, 637–645 [DOI] [PubMed] [Google Scholar]
- 16. el Marjou F., Janssen K. P., Chang B. H., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., Robine S. (2004) Genesis 39, 186–193 [DOI] [PubMed] [Google Scholar]
- 17. Rubin D. C., Swietlicki E., Roth K. A., Gordon J. I. (1992) J. Biol. Chem. 267, 15122–15133 [PubMed] [Google Scholar]
- 18. Bry L., Falk P., Huttner K., Ouellette A., Midtvedt T., Gordon J. I. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10335–10339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Milano J., McKay J., Dagenais C., Foster-Brown L., Pognan F., Gadient R., Jacobs R. T., Zacco A., Greenberg B., Ciaccio P. J. (2004) Toxicol. Sci. 82, 341–358 [DOI] [PubMed] [Google Scholar]
- 20. Ohlstein B., Spradling A. (2007) Science 315, 988–992 [DOI] [PubMed] [Google Scholar]
- 21. Saotome I., Curto M., McClatchey A. I. (2004) Dev. Cell 6, 855–864 [DOI] [PubMed] [Google Scholar]
- 22. Fre S., Pallavi S. K., Huyghe M., Laé M., Janssen K. P., Robine S., Artavanis-Tsakonas S., Louvard D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6309–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodilla V., Villanueva A., Obrador-Hevia A., Robert-Moreno A., Fernández-Majada V., Grilli A., López-Bigas N., Bellora N., Albà M. M., Torres F., Duñach M., Sanjuan X., Gonzalez S., Gridley T., Capella G., Bigas A., Espinosa L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6315–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ohtsuka T., Ishibashi M., Gradwohl G., Nakanishi S., Guillemot F., Kageyama R. (1999) EMBO J. 18, 2196–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ross D. A., Rao P. K., Kadesch T. (2004) Mol. Cell. Biol. 24, 3505–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Imayoshi I., Shimogori T., Ohtsuka T., Kageyama R. (2008) Development 135, 2531–2541 [DOI] [PubMed] [Google Scholar]
- 27. Riccio O., van Gijn M. E., Bezdek A. C., Pellegrinet L., van Es J. H., Zimber-Strobl U., Strobl L. J., Honjo T., Clevers H., Radtke F. (2008) EMBO. Rep. 9, 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Georgia S., Soliz R., Li M., Zhang P., Bhushan A. (2006) Dev. Biol. 298, 22–31 [DOI] [PubMed] [Google Scholar]
- 29. Monahan P., Rybak S., Raetzman L. T. (2009) Endocrinology 150, 4386–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. VanDussen K. L., Samuelson L. C. (2010) Dev. Biol. 346, 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bossuyt W., Kazanjian A., De Geest N., Van Kelst S., De Hertogh G., Geboes K., Boivin G. P., Luciani J., Fuks F., Chuah M., VandenDriessche T., Marynen P., Cools J., Shroyer N. F., Hassan B. A. (2009) PloS. Biol. 7, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.