Abstract
The endoplasmic reticulum (ER) harbors elaborate quality control mechanisms to ensure proper folding and post-translational modifications of polypeptides targeted to this organelle. Once an aberrant protein is detected, it is dislocated from the ER and routed to the proteasome for destruction. Autophagy has been recently implicated in the elevation of the ER stress response; however, the involvement of this pathway in selective removal of ER-associated degradation (ERAD) substrates has not been demonstrated. In the present study, we show that an ER membrane lesion, associated with the accumulation of the yeast ERAD-M substrate 6Myc-Hmg2p elicits the recruitment of Atg8 and elements of the cytosol to vacuole targeting (CVT) to the membrane, leading to attenuation in the degradation process. Deletion of peptide:N-glycanase (PNG1) stabilizes this association, a process accompanied by slowdown of 6Myc-Hmg2p degradation. Truncation of the unstructured C-terminal 23 amino acids of 6Myc-Hmg2p rendered its degradation PNG1-independent and allowed its partial delivery to the vacuole in an autophagy-dependent manner. These findings demonstrate a new conduit for the selective vacuolar/lysosomal removal of ERAD misfolded proteins by an autophagy-related machinery acting concomitantly with the proteasome.
Keywords: Autophagy, ER Stress, Proteasome, Protein Degradation, Ubiquitination, CVT, ERAD
Introduction
The endoplasmic reticulum (ER)3 is the port of entry into the secretory pathway. A fraction of newly synthesized polypeptides remains in the ER to serve in various metabolic and structural functions. Almost all proteins processed in the ER acquire co- or post-translational modifications, of which the most common and diverse is glycosylation (1). Correctly folded proteins that pass the ER quality control checkpoints continue to their final destination, whereas misfolded proteins are retained in the ER and subsequently eliminated. Their removal entails dislocation across the ER membrane, ubiquitination, extraction into the cytosol, and proteasomal degradation (2). Protein breakdown by ER-associated degradation (ERAD) is not a uniform process. ERAD substrates are divided into three classes, depending on the location of the lesion within the aberrant protein; ERAD-L substrates are either soluble or membrane proteins with a luminal lesion, whereas ERAD-M and ERAD-C substrates are transmembrane proteins with a misfolded transmembrane or cytosolic domain, respectively (3, 4).
Degradation of all ERAD substrates begins with recognition of the misfolded polypeptides within the ER by chaperones, which detect exposed hydrophobic patches of misfolded proteins, aberrant disulfide bonds, or unassembled protein complexes (2). N-Linked glycans have a key role in quality control and in the recognition of improperly folded proteins. Within the ER, glycans are recognized by lectins, such as the yeast Htm1/Mnl1 and Yos9 and their mammalian orthologs EDEM and EDEM2 (5–11). Glycans may also be essential for retrotranslocation of ER-misfolded glycoproteins to the cytosol. For example, removal of the terminal glycan from the ERAD substrate CPY* (mutated yeast vacuolar carboxyl peptidase Y), which is decorated with four N-linked glycans, results in inhibition of its degradation and its consequent accumulation in the ER (12, 13). Moreover, certain E3 ubiquitin ligases (Fbs1 and Fbs2) bind glycans of misfolded glycoproteins in the cytosol and promote their ubiquitination and proteasomal degradation (14, 15).
ER stress ensues when the amount of client proteins that emerges into the ER exceeds its overall folding capacity or upon overload of the ER with damaged or misfolded polypeptides. This challenge activates an intricate cytoprotective ER-to-nucleus signaling cascade, collectively termed the unfolded protein response, which in turn up-regulates the ER capacity to handle the load of misfolded proteins (16–18). Another potential cellular route for clearing ER-misfolded polypeptides that exceed the capacity of the ERAD machinery may involve routing of these misfolded proteins to the vacuole/lysosome. In fact, previous studies have demonstrated that upon overexpression of CPY*, a portion of this ERAD-L substrate is transported to the vacuole for degradation (19).
Autophagy may also serve to deliver some of the aberrant proteins for vacuolar/lysosomal removal. This pathway, engaged in the removal of superfluous or damaged organelles and cytosolic proteins, was found important for development, cellular maintenance, and remodeling (20, 21). The yeast ubiquitin-like protein Atg8 and its mammalian orthologs LC-3, GATE-16, and GABARAP serve as key components of autophagy (22–24). In the early stages of autophagosome formation, Atg8 becomes lipidated and accumulates in preautophagosomal structures, which are the nucleation sites for autophagosomes (25). Atg8 remains attached to the autophagosome and is delivered to the vacuole (22, 26–28). Autophagy is normally elicited under stress conditions, such as nutrient or growth factor deprivation, and fulfills a protective role in human diseases, including cancer, some types of neurodegenerative diseases, and muscular disorders (29, 30). A typical scenario for such conditions may involve aggregation of misfolded proteins (i.e. polyglutamin, mutants of ataxin3, Huntingtin, mutant α-synuclein, and different forms of Tau) that escape proteasomal degradation. In such circumstances, macroautophagy may replace the proteasome and become the major clearance pathway (30–36). Shuttle proteins, such as P62/SQSTM1 and HDAC6, potentially serve in recruiting polyubiquitinated aggregated proteins into autophagic vesicles (34, 37–42). Autophagy has also been implicated in ER quality control, providing an alternative mechanism for the clearance of misfolded proteins that accumulate in the ER lumen. For instance, the Z variant of human α-1 antitrypsin (ATZ), a protease inhibitor produced in the liver, may misfold and accumulate in the ER, causing liver disease. Recent studies have indicated that degradation of the misfolded ATZ involves both the general ER quality control and the autophagic systems (43–46). Moreover, ER stress in yeast and mammals leads to induction of autophagy (47, 48).
Because the majority of ERAD substrates are decorated by N-linked glycans, we postulated that attenuation in the degradation of certain ERAD substrates observed in the absence of PNG1 may lead to the recruitment of autophagic components to the ER lesion. To unravel a potential function of autophagy in the removal of glycosylated ER-misfolded proteins, we searched for a known ERAD substrate whose proteasomal clearance is attenuated in the Δpng1 strain in an autophagy-dependent manner. We found that in the absence of PNG1, the degradation of 6Myc-Hmg2p, an ERAD-M substrate, is attenuated. This inhibition is inferred from the formation of a stable complex between the ERAD substrate and components of the autophagic machinery. Finally, we demonstrate that upon deletion of the unstructured 23 C-terminal residues of 6Myc-Hmg2p, a portion of this substrate travels to the vacuole in an autophagy-dependent manner. Taken together, these results provide evidence for a new role for the autophagic machinery in ER quality control of transmembrane proteins.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
Wild type Saccharomyces cerevisiae and single open reading frame (ORF) deletion mutants were obtained from Euroscarf (BY4741; Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; ORF(X)::kanMX4). Double deletion mutants of PNG1 and ATG/CVT genes were prepared by replacing the PNG1 gene with a hygromycin B resistance cassette (amplified from the pAG32 plasmid; Euroscarf) in single ATG/CVT deletion mutants. 6Myc-Hmg2p was expressed from plasmid pRH244 (49). Mutants 6Myc-Hmg2p-RFP, 6Myc-Hmg2p-1–1022, 6Myc-Hmg2p-1–1022-RFP, 6Myc-Hmg2p-FLAG, and 6Myc-Hmg2p-FLAG-RFP were expressed from the same vector. The mutant 6Myc-HMG2-RFP was generated by insertion of SwaI and NotI restriction sites instead of the original stop codon of 6Myc-HMG2 into which the RFP coding sequence with the same restriction sites was ligated in frame. 6Myc-HMG2–1-1022 and 6Myc-HMG2–1-1022-RFP were generated by deletion. A FLAG tag was introduced by insertion to generate 6MYC-HMG2-FLAG and 6MYC-HMG2-FLAG-RFP using 6MYC-HMG2 and 6MYC-HMG2-RFP as templates, respectively. HA-PNG1–6His, GFP-ATG8, FLAG-Ubiquitin, APE1-HA, and AMS1-GFP were cloned into pAD54 (Leu2, 2μ, ADH promoter), kindly provided by J. Gerst. The plasmid pSM1082 encoding Ste6p-166-HA was kindly provided by S. Michaelis (50). A plasmid encoding GFP-Atg8 from the ATG8 promoter was generated by subcloning a SpeI-EcoRI fragment from pRS316-GFP-Atg8 (25) into pRS313. Yeast transformants were obtained by standard plasmid transformation techniques (51).
Cycloheximide Chase Analysis
Yeast were grown in 3 ml of selective synthetic medium containing 2% glucose (SD) until the culture reached an A600 of 0.6. The SD medium was then replaced by YPD medium, and the culture was grown for additional 2 h. Cycloheximide (100 μg/ml) was added, and equal samples of yeast were removed at the indicated time points, pelleted by centrifugation, and snap-frozen in liquid nitrogen. Lysates were then prepared by NaOH/trichloroacetic acid (TCA) precipitation and separated by SDS-PAGE followed by Western blot analysis.
Cell Lysis by NaOH/TCA Precipitation
Frozen samples were resuspended in 300 μl of 1.85 m NaOH and 7% β-mercaptoethanol and vortexed for 1 min. TCA was then added to each sample to a final concentration of 33%, followed by a 5-min incubation on ice. The samples were then centrifuged for 10 min (4 °C, 14,000 rpm). The protein pellets were washed with 1 m Tris, resuspended in 2.5% SDS, 0.5 mm EDTA, 1 mm PMSF and frozen at −20 °C. The samples were then incubated for 5 min at 65 °C, followed by centrifugation (5 min, room temperature, 14,000 rpm). The supernatant was supplemented with SDS-PAGE sample buffer, incubated at 65 °C, and separated on SDS-PAGE followed by Western blot analysis. The immunoblots were developed either by exposure to x-ray films or by the Bio-Rad Chemidoc XRS system, followed by quantification using the Quantity One program (Bio-Rad).
Cross-linking and Immunoprecipitation
Fifty ml of A600 = 0.4 cultures were shifted from SD medium to YPD and grown for 4 h. The cells were then washed and resuspended in 8 ml of 0.1 m KH2PO4/K2HPO4, pH 7.5, 1.2 m sorbitol, 5 mm EDTA supplemented with 1 mg of zymolase. Following a 1-h incubation with gentle agitation at 30 °C, the cells were harvested by centrifugation and resuspended in 20 mm HEPES, pH 7.5, 100 mm NaCl, and 1 mm 3′-dithiobis(sulfosuccinimidylpropionate) (Pierce). The cells were lysed using a Dounce homogenizer, and the lysates were incubated on ice for 30 min. Access cross-linker was quenched by the addition of Tris, pH 7.5, to a final concentration of 50 mm and incubation for an additional 15 min. SDS was added to the lysates to a final concentration of 2.5%. The lysates were then vigorously vortexed and frozen at −20 °C. Next, the lysates were heated to 65 °C for 10 min and centrifuged (14,000 rpm), and the supernatant fraction was used for immunoprecipitation (IP). For IP, agarose beads conjugated with anti-Myc antibodies were washed with IP buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100) and incubated with IP buffer containing 1% BSA for 30 min at 4 °C. The beads were washed, and lysates containing 5 mg of protein diluted with IP buffer in a ratio of 1:10 were added and incubated for 2 h at 4 °C with gentle agitation. The immunoprecipitates were washed and supplemented with SDS-PAGE sample buffer. Following 5 min at 65 °C, the samples were separated by SDS-PAGE and analyzed by Western blot.
Cell Fractionation
Fifty ml of yeast were grown in SD medium until the A600 of the culture reached 0.4. The SD medium was then replaced by YPD medium, and the cultures were grown for an additional 4 h. The yeast were centrifuged, washed with distilled water, resuspended with lysis buffer (20 mm HEPES, pH 7.5, 250 mm sorbitol, 150 mm NaCl, 5 mm MgAc, 1 mm PMSF), and lysed by vortexing with glass beads for 5 min. The lysates were centrifuged for 5 min at 3,000 rpm, and the supernatant was separated and centrifuged for 30 min at 4 °C at 80,000 rpm. The supernatant was separated from the pellet fraction, and the pellet was either subjected to NaOH/TCA precipitation or resuspended with the indicated solution. Where indicated, immunoprecipitation was performed from the supernatant and pellet fractions using anti-Myc antibodies.
Fluorescent Microscopy
For analysis of GFP-Atg8 localization, fluorescent microscopy was performed using a confocal microscope (Olympus FV500+IX70) or a DeltaVision microscope (Applied Precision). Cells were grown in SD medium to midexponential phase and shifted to SD-N starvation medium. For visualization of 6Myc-Hmg2p-RFP and Ams1-GFP, a DeltaVision microscope was used. DeltaVision images were deconvolved using the Soft-Worx software (Applied Precision). For Hoechst staining, yeasts were treated with zymolase (2.5 mg/1 g of yeast) for 15 min. Following treatment with zymolase, the spheroplasts were incubated in SD medium containing 1.2 m sorbitol for 2 h. Hoechst reagent for DNA staining was added at the last 3 min of incubation.
Transmission Electron Microscopy
A culture of early log phase was concentrated by centrifugation, after which a drop of pellet was placed in an aluminum disc with a 100-μm deep cavity and covered with a flat disc. The sample was then high pressure-frozen in a HPM010 high pressure freezing machine (Bal-Tec, Liechtenstein). Cells were subsequently freeze-substituted in a Leica AFS2 device in anhydrous acetone containing 1% osmium tetroxide and 0.2% uranyl acetate for 3 days at −90 °C and then warmed up to −30 °C over 24 h and washed three times in anhydrous acetone at room temperature. Samples were then infiltrated in a series of increasing concentrations of Epon (SPI Supplies). After polymerization at 60 °C, 60–80-nm sections were stained with uranyl acetate and lead citrate and examined in a Tecnai T12 electron microscope (FEI, Eindhoven, The Netherlands) operating at 120 kV. Images were recorded with an Eagle 2048 × 2048-pixel CCD camera (FEI).
35S Pulse-Chase Experiments
Two A600 yeast cultures expressing 6Myc-Hmg2p were starved for methionine and cysteine for 30 min and pulse-labeled with 400 μCi of [35S]methionine/cysteine mixture for 30 min. The cells were then washed and resuspended in fresh medium containing 5 μg/ml cold methionine and cysteine. Samples of 0.5 A600 were removed at various time points and lysed by NaOH/TCA precipitation. The lysates were then diluted in a 1:10 ratio with IP buffer and used for immunoprecipitation with anti-Myc antibodies as described above. The immunoprecipitates were separated on SDS-PAGE and transferred to a nitrocellulose membrane that was exposed to an x-ray film for 24 h. The radiograms were quantified using the Quantity One program (Bio-Rad).
Real-time PCR Analysis
Total RNA from a 2-ml culture of mid-log phase yeast was extracted (total RNA isolation kit; Epicenter) and treated with RNase-free DNase to remove residual DNA contaminations. cDNA was then prepared from 2 μg of RNA template using a cDNA synthesis kit containing a mixture of random hexamer and oligo(dT) primers (Beit Haemek). For RT-PCR, triplicates of the cDNA and a standard curve prepared from serial dilutions of pooled cDNA samples were used. Transcripts were detected using SYBR Green 1 according to the manufacturer's instructions (Finzymes) and normalized to the levels of ACT1, as an internal control. Quantification of cDNA targets was performed using an Applied Biosystems 7300 real-time PCR system and analyzed utilizing the Applied Biosystems software.
Total Protein Degradation under Starvation
The procedures for the protein degradation assay were performed as described previously (52).
RESULTS
Png1 Is Essential for Rapid Extraction and Degradation of 6Myc-Hmg2p
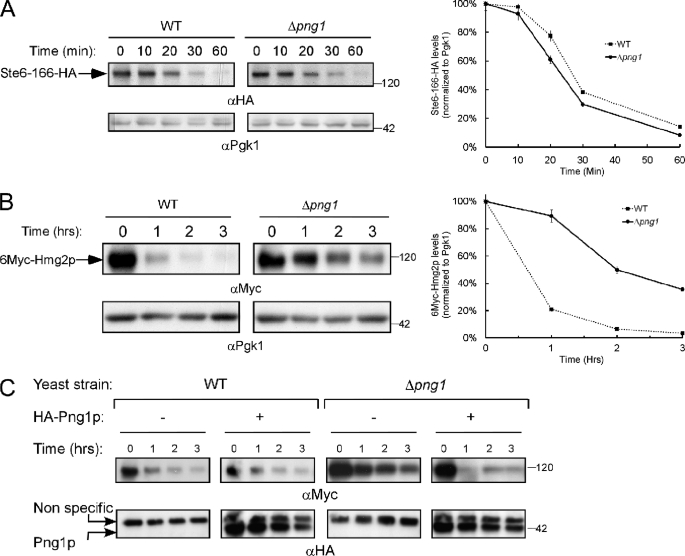
The significance of glycan removal from ERAD substrates en route to proteasomal degradation remains unclear. In yeast, the degradation of the ERAD-L substrate RTA (a ricin A-chain non-toxic mutant) is impaired in the absence of PNG1 (53, 54). However, PNG1 deficiency had only a minor effect on the degradation of the ERAD-L substrate CPY* (55). Similar results were obtained when we analyzed the degradation of the ERAD-C substrate Ste6p-166 (Fig. 1A), a transmembrane ERAD substrate containing a point mutation within its cytoplasmic domain (56). However, the half-life of the ERAD-M target 6Myc-Hmg2p significantly increased when expressed in the Δpng1 strain (Fig. 1B). Hmg2p was converted into a rapidly degraded ERAD substrate through the insertion of a 6Myc tag between its two N-terminal transmembrane domains (denoted as 6Myc-Hmg2p) (49). As shown in Fig. 1C, add back of wild type Png1 rescued the inhibitory phenotype PNG1 deletion conferred on the breakdown of 6Myc-Hmg2p. Taken together, these experiments indicate that the presence of Png1 is important for the rapid removal of 6Myc-Hmg2p.
FIGURE 1.
Degradation of 6Myc-Hmg2p is attenuated in Δpng1 yeast. A and B, evaluation of the half-life of Ste6p-166 (A) and 6Myc-Hmg2p (B) by cycloheximide chase. The magnitude of the HA (A) or Myc (B) signal was quantified relative to the PGK signal. C, Δpng1 yeast expressing 6Myc-Hmg2p were transformed with an HA-tagged PNG1-encoding plasmid and subjected to cycloheximide chase. The amounts of the heterogeneous Png1p were estimated by Western blot with anti-HA antibodies (bottom). Error bars, S.D.
Ubiquitinated 6Myc-Hmg2p Accumulates on the Peripheral ER in the Absence of PNG1
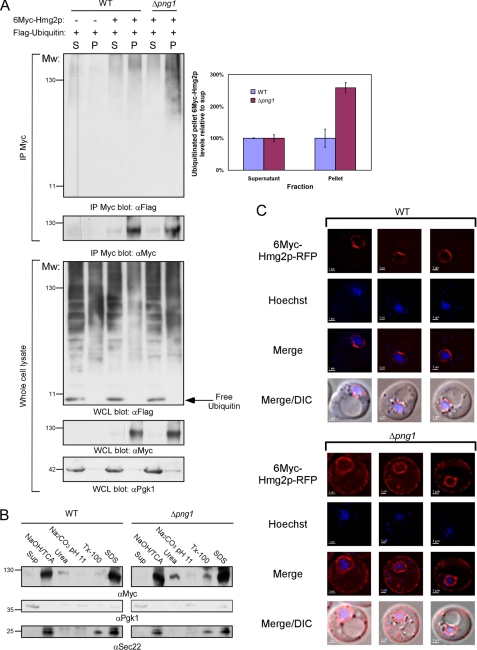
ER dislocation of target substrates en route to proteasomal degradation requires their ubiquitination. The Hrd1 E3 ubiquitin ligase complex was found to be responsible for the ubiquitination of 6Myc-Hmg2p (49, 57, 58). Once ubiquitinated, 6Myc-Hmg2p is extracted from the membrane in a CDC48-dependent manner and delivered to the proteasome (59). Subcellular fractionations followed by immunoprecipitations with anti-Myc antibodies revealed that in the absence of PNG1, 6Myc-Hmg2p accumulates in its ubiquitinated form (Fig. 2A). We conclude that the ubiquitinated 6Myc-Hmg2p aggregates, accumulated in the PNG1-deficient strain, remain integrated in the membrane because these were only solubilized by 1% SDS and not by 2 m urea, 100 mm Na2CO3, pH 11, or 1% Triton X-100 (Fig. 2B, top).
FIGURE 2.
Ubiquitinated 6Myc-Hmg2p accumulates in the peripheral ER membrane in Δpng1 cells. A, cytosolic fraction (S) and membrane fraction (P) were prepared from wild-type and Δpng1 yeast expressing either FLAG-ubiquitin or a combination of FLAG-ubiquitin and 6Myc-Hmg2p. Proteins were extracted from the pellet fraction by NaOH/TCA precipitation. Pellet and supernatant fractions were subjected to immunoprecipitation with anti-Myc antibodies, followed by Western blot with the indicated antibodies. Quantifications of the ubiquitinated 6Myc-Hmg2p fractions relative to the total immunoprecipitated 6Myc-Hmg2p levels are presented in the top right panel. B, proteins from the pellet fractions were either extracted by NaOH/TCA precipitation or resuspension with 2 m urea, 100 mm Na2CO3, pH 11, 1% Triton X-100, or 1% SDS. The amounts of 6Myc-Hmg2p in the original supernatant and those that were extracted by the specific treatments from the pellet were estimated by Western blot analysis with anti-Myc antibodies. The fractionation quality was estimated by reblotting with anti-Sec22 (membrane) and anti-Pgk1 (soluble) antibodies. C, localization of 6Myc-Hmg2p-RFP in wild-type and Δpng1 cells. The nuclei were stained using Hoechst reagent prior to visualization by a fluorescent microscope. DIC, differential interference contrast. Error bars, S.D.
To follow the localization of 6Myc-Hmg2p microscopically, we fused RFP to the C terminus of 6Myc-Hmg2p. Initially, we verified that 6Myc-Hmg2p-RFP degradation proceeded similarly to that of 6Myc-Hmg2p in wild-type yeast and is attenuated in the Δpng1 strain (supplemental Fig. S1). As shown in Fig. 2C, in wild-type yeast, the fluorescent signal accumulated on the nuclear ER, whereas in the Δpng1 strain, 6Myc-Hmg2p-RFP was detected both in the nuclear ER and in the cortical ER (Fig. 2C, bottom). Electron microscopy analysis of wild-type and Δpng1 yeast revealed membrane proliferations in both strains only upon expression of 6Myc-Hmg2p; however, a marked increase in membrane expansion was evident upon expression of the ERAD-M substrate in Δpng1 yeast (supplemental Fig. S2). This result correlates with the elevated levels of 6Myc-Hmg2p detected biochemically and fluorescently in this strain (Figs. 1B and 2C). Notably, such membrane proliferations were observed previously in wild-type yeast expressing exceptionally high levels of wild-type Hmg2p (60, 61) as well as in mammalian cells overexpressing its ortholog HMGR (62–64). In summary, we conclude that the slowdown in 6Myc-Hmg2p degradation observed in the absence of PNG1 leads to its accumulation in the cortical ER membrane.
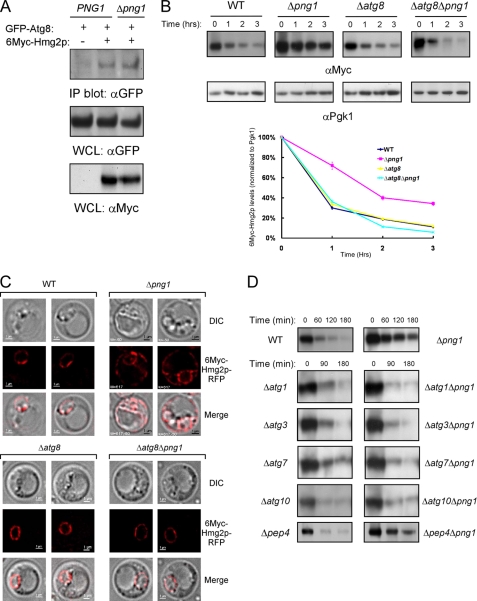
6Myc-Hmg2p Stabilization in Δpng1 Yeast Ensues from Atg8 Recruitment to the Aberrant Protein
We next tested the hypothesis that the accumulation of ubiquitinated transmembrane proteins on the ER serves as a signal for recruiting the autophagic machinery to these lesions. We therefore examined whether Atg8, a key autophagic factor, interacts with 6Myc-Hmg2p. As shown in Fig. 3A, GFP-Atg8 co-immunoprecipitated with 6Myc-Hmg2p in both wild-type and Δpng1 strains. Similar results were obtained with the endogenous, unmodified Atg8 (supplemental Fig. S3, top). These findings point to the involvement of the autophagic machinery in 6Myc-Hmg2p degradation attenuation, observed in Δpng1 yeast, constituting a checkpoint that regulates the extraction rate of 6Myc-Hmg2p. Next, we examined the degradation of 6Myc-Hmg2p in both Δatg8 and Δatg8Δpng1 strains. As depicted in Fig. 3B, deletion of ATG8 by itself did not affect 6Myc-Hmg2p degradation, whereas deletion of ATG8 in the background of the Δpng1 strain (Δatg8Δpng1) restored rapid 6Myc-Hmg2p turnover. These rather unexpected findings are in line with the notion that recruitment of an autophagy-related component, and not the lack of deglycosylation activity per se, is responsible for the attenuated 6Myc-Hmg2p degradation observed in Δpng1 yeast. Consistently, 6Myc-Hmg2p-RFP expressed in Δatg8Δpng1 strain was only detected in the nuclear ER (Fig. 3C). To examine whether other autophagic factors participate in the inhibition of 6Myc-Hmg2p degradation upon PNG1 deletion, this protein was expressed in a series of representative ATG deletion mutants on the background of Δpng1. As shown in Fig. 3D, deletion of ATG1, -3, -7, and -10 had a similar effect observed in the absence of ATG8. However, deletion of PEP4, a vacuolar aspartyl protease responsible for the activation of numerous vacuolar proteases, did not restore rapid 6Myc-Hmg2p degradation. Similar results were obtained when the turnover rate of 6Myc-Hmg2p was monitored in a 35S pulse-chase experiment, thus excluding the possibility that some of our findings are related to the cycloheximide itself (supplemental Fig. S4). In aggregate, these findings demonstrate the involvement of the autophagic machinery in the attenuation of 6Myc-Hmg2p degradation.
FIGURE 3.
Inhibition of the autophagic machinery restores the fast elimination of 6Myc-Hmg2p in Δpng1 cells. A, lysates from either Δatg8 (PNG1) or Δatg8Δpng1 (Δpng1) cells expressing GFP-Atg8 and 6Myc-Hmg2p were prepared in the presence of 1 mm 3′-dithiobis(sulfosuccinimidylpropionate). The proteins cross-linked to 6Myc-Hmg2p were co-immunoprecipitated with anti-Myc antibodies and analyzed by Western blot with the indicated antibodies. B, evaluation of the half-life of 6Myc-Hmg2p by cycloheximide chase in the indicated yeast strains by immunoblotting with anti-Myc antibodies. Quantification of the decay of the magnitude of the Myc signal relative to the Pgk1 signal over time is presented. C, visualization of the cellular distribution of 6Myc-Hmg2p-RFP in the indicated strains was ascertained by fluorescence microscopy. D, impairment of the autophagic pathway by deletion of essential components overturns the degradation attenuation of 6Myc-Hmg2p observed in Δpng1 cells. Determination of 6Myc-Hmg2p amounts was performed by Western blot upon cycloheximide chase in the indicated single and double deletion yeast strains. DIC, differential interference contrast; WCL, whole cell lysate. Error bars, S.D.
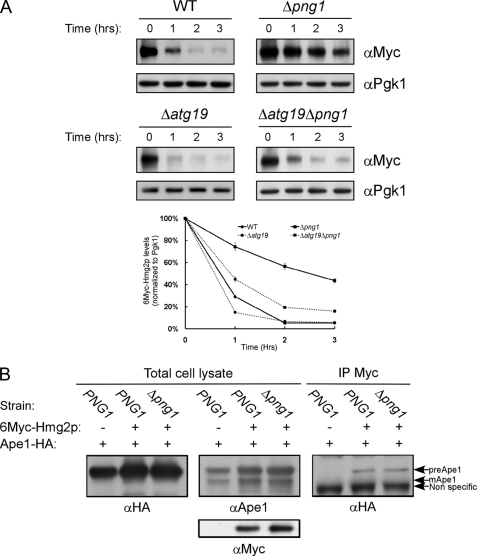
6Myc-Hmg2p Interacts with the CVT Complex
In yeast, autophagy is activated under stress conditions, such as nutrient deprivation, in a process that entails up-regulation of ATG8 and ATG1 transcripts (two hallmarks of autophagy activation) (22). As shown in supplemental Fig. S5, mRNA levels of ATG8 and ATG1 increased more than 8-fold when the yeasts were starved for nitrogen. However, expression of 6Myc-Hmg2p in either the wild-type or Δpng1 strains resulted only in moderate elevations in ATG8 and ATG1 transcription levels (2.6- and 1.4-fold, respectively, in wild type), suggesting that autophagy per se was not elicited. Yeasts also possess a housekeeping/constitutively active pathway known as CVT (cytoplasm to vacuole targeting), responsible for the delivery of at least two hydrolases, Ape1 (amino peptidase 1) and Ams1 (α-mannosidase 1) from the cytoplasm to the vacuole (65). To determine whether the CVT pathway is involved in the attenuated proteasomal removal of 6Myc-Hmg2p, we tested if deletion of ATG19 (a CVT-specific factor) rescues the phenotype observed in the absence of PNG1. As shown in Fig. 4A, deletion of ATG19 (Δatg19 and Δatg19Δpng1 strains) restored rapid degradation of 6Myc-Hmg2p. Thus, as with Atg8, the presence of the CVT cargo receptor, Atg19, is necessary for 6Myc-Hmg2p accumulation observed in the Δpng1 strain.
FIGURE 4.
The CVT machinery is required for the attenuation of 6Myc-Hmg2p degradation in Δpng1 yeast. A, evaluation of the half-life of 6Myc-Hmg2p by cycloheximide chase in the indicated yeast strains by immunoblotting with anti-Myc antibodies. Quantification of the decay of the magnitude of the Myc signal relative to the Pgk1 signal over time is presented. B, Δape1 (PNG1) or Δape1Δpng1 (Δpng1) yeast expressing Ape1-HA and 6Myc-Hmg2p were lysed in the presence of 1 mm 3′-dithiobis(sulfosuccinimidylpropionate). The lysates were subjected to immunoprecipitation with anti-Myc antibodies and analyzed by Western blot with the indicated antibodies. Error bars, S.D.
We have shown above that 6Myc-Hmg2p interacts with Atg8 (Fig. 3A). To test whether other CVT-related factors also associate with this ERAD-M substrate, a co-immunoprecipitation approach was taken. As shown in Fig. 4B, 6Myc-Hmg2p interacts with the cytosolic unprocessed form of Ape1, as detected by the anti-HA antibody and further confirmed by anti-Ape1 antibodies. Similar interaction between 6Myc-Hmg2p and the endogenous unprocessed Ape1 was also observed (supplemental Fig. S3). Based on these findings, we hypothesize that the CVT machinery is specifically involved in the attenuation of 6Myc-Hmg2p degradation observed in Δpng1 yeast.
The Atg8-6Myc-Hmg2p Complex Is Stabilized in the Absence of PNG1
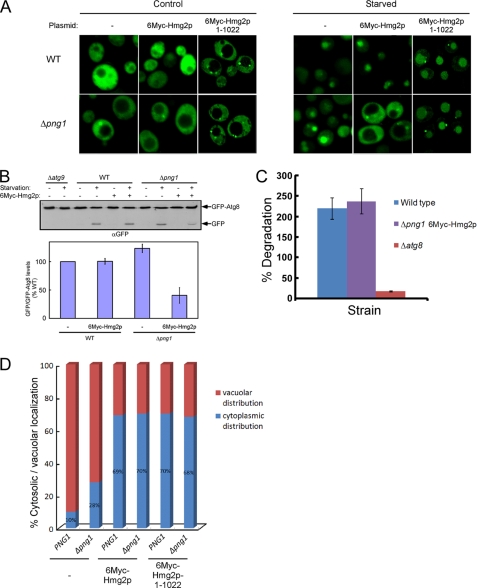
Thus far, we have shown that Png1 activity promotes fast proteasomal elimination of 6Myc-Hmg2p, which is regulated by Atg8 that forms a complex with this substrate. However, as shown in Fig. 3A, this complex was only detected in a PNG1-independent manner. These results may indicate that in the absence of PNG1, the transient complex between Atg8 and 6Myc-Hmg2p is stabilized. To test this hypothesis, we determined autophagic activity in different strains ectopically expressing GFP-Atg8. Initially, we studied the effect of 6Myc-Hmg2p expression on the localization of GFP-Atg8 in wild-type and Δpng1 yeast strains. In the absence of 6Myc-Hmg2p, GFP-Atg8 was localized to the cytosol, and upon nitrogen starvation, it accumulated within the vacuole in both yeast strains (Fig. 5A), indicating that PNG1 is not essential for starvation-induced autophagy. However, expression of 6Myc-Hmg2p blocked vacuolar delivery of GFP-Atg8 under starvation conditions in the Δpng1 strain (Fig. 5A). In a complementary experiment, we monitored the delivery of Atg8 into the vacuole by assessing GFP-Atg8 cleavage (29). Following delivery of GFP-Atg8 to the vacuole, the Atg8 portion is degraded, and the GFP moiety is spared. A marked reduction in starvation-induced cleavage of GFP-Atg8 was observed in Δpng1 yeast expressing 6Myc-Hmg2p, whereas expression of this ERAD substrate did not alter GFP-Atg8 cleavage in wild-type cells (Fig. 5B). This suggests that in the absence of PNG1, 6Myc-Hmg2p forms a stable long lasting complex with Atg8. This hypothesis implies that newly synthesized endogenous untagged Atg8 overly expressed under starvation would not chase the already bound GFP-Atg8, as illustrated in Fig. 5A. Moreover, autophagic activity measured by long lived protein degradation under starvation remains unaffected in Δpng1 cells expressing 6Myc-Hmg2p (Fig. 5C). Thus, Atg8 expressed under basal conditions is stably recruited to the 6Myc-Hmg2p lesions on the ER formed in the absence of PNG1.
FIGURE 5.
6Myc-Hmg2p expression impairs starvation-induced autophagy and CVT. A, the indicated yeast strains expressing GFP-Atg8 from an ADH1 promoter and either expressing or not 6Myc-Hmg2p were transferred to SD-N starvation medium or left in rich medium. After 4 h, the distribution of GFP in the cells was visualized by fluorescence microscopy. A control experiment in a Δatg9 strain is presented in supplemental Fig. S6. B, estimation of the extent of GFP-Atg8 processing (expressed from an ADH1 promoter) by monitoring the formation of GFP upon 6Myc-Hmg2p expression and 4 h of nitrogen starvation in the indicated yeast strains by immunoblotting with anti-GFP antibodies. A quantification of the cleaved GFP levels relative to the total amount of GFP-Atg8 is presented in the lower panel. The quantified levels represent the average of four repeated experiments. C, cells were pulse-labeled with [35S]methionine/cysteine and chased on non-radioactive nitrogen starvation medium. Aliquots were taken after 3 h, and the amounts of acid-soluble small peptides generated by proteolysis were determined. D, the distribution of the cellular Ams1 was determined in the indicated strains by visualizing a fluorescent derivative of Ams1 (Ams1-GFP). A statistical summary of the localization of the GFP signal of 100 cells randomly selected from each strain is presented. A typical fluorescent image illustrating cytoplasmic and vacuolar GFP signal is presented in supplemental Fig. S8. Error bars, S.D.
Furthermore, these findings predict that the CVT pathway, which relies on the basal levels of Atg8, is affected by the expression of 6Myc-Hmg2p in Δpng1. This was ascertained by monitoring the delivery of Ams1-GFP to the vacuole by fluorescent microscopy. In the majority of cells of both PNG1 and Δpng1 strains, Ams1-GFP reached the vacuole. However, upon expression of 6Myc-Hmg2p in these strains, Ams1-GFP remained in the cytosol with intense staining of the preautophagosomal structure (supplemental Fig. S8). Statistical analysis of these experiments is presented in Fig. 5D.
The Effect of Png1 on the Turnover of 6Myc-Hmg2p Is Mediated by 6Myc-Hmg2p C Terminus
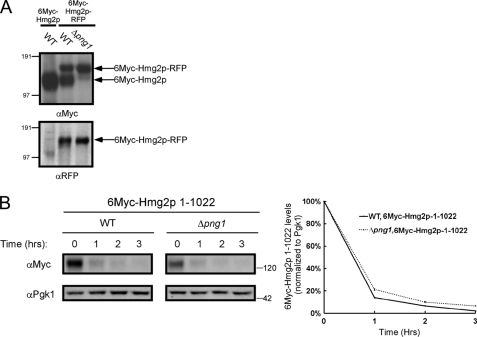
When lysates obtained from wild-type yeast expressing 6Myc-Hmg2p-RFP were separated by SDS-PAGE, two forms of the protein were revealed: a high molecular weight form corresponding to the full-length RFP-fused protein (that reacted with both anti-Myc and anti-RFP antibodies) (Fig. 6A) and a lower molecular weight form that reacted only with the anti-Myc antibodies, suggesting a loss of the RFP domain (Fig. 6A). When expressed in Δpng1 yeast, the 6Myc-Hmg2p-RFP fusion protein appeared resistant to this cleavage (Fig. 6A). A cleavage close to the C terminus would be undetectable unless a large moiety such as RFP is fused to the carboxyl terminus of the protein as in our experiments (Fig. 6A). Although the three-dimensional structural information of the yeast Hmg2p is not available, the crystal structure of the C terminus of its mammalian counterpart (HMGR) was solved (66). Sequence alignment of the C terminus of the yeast and human proteins indicates that the last 23 residues of the yeast Hmg2p might be unstructured and therefore were not detected in the crystal structure (supplemental Fig. S9).
FIGURE 6.
The C terminus of 6Myc-Hmg2p is essential for the Png1-mediated degradation attenuation. A, Western blot analysis of 6Myc-Hmg2p and 6Myc-Hmg2p-RFP in wild-type and Δpng1 cells with the indicated antibodies. B, evaluation of the half-life of 6Myc-Hmg2p-1–1022 by cycloheximide chase in the indicated yeast strains by immunoblotting with anti-Myc antibodies. The magnitude of the Myc signal was quantified relative to the PGK signal.
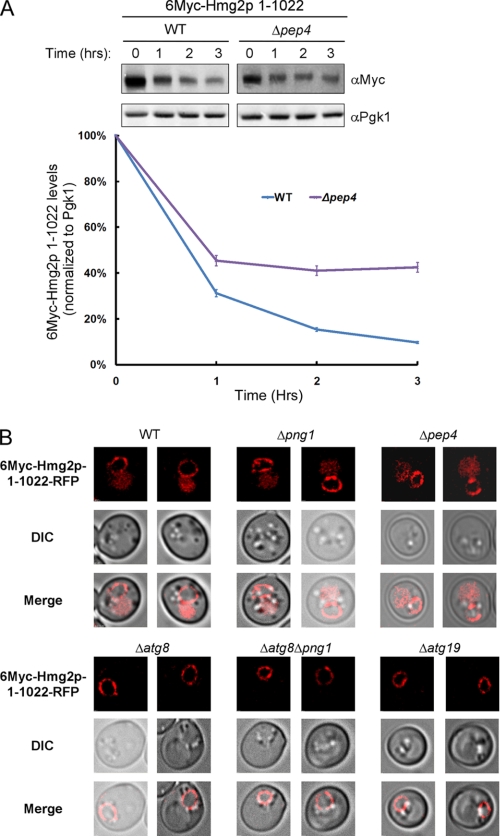
These findings lend support to the hypothesis that 6Myc-Hmg2p C terminus controls the rate of degradation linked to Png1 activity. The rationale guiding this hypothesis relies on an earlier report indicating that a loose terminus is critical for extraction from the ER membrane during ERAD (67). To test this, 6Myc-Hmg2p lacking the last 23 residues (denoted 6Myc-Hmg2p-1–1022) was expressed in the different yeast strains. As shown in Fig. 6B, deletion of the last 23 residues renders the elimination of this derivative independent of PNG1. The 6Myc-Hmg2p-1–1022 mutant kept its ability to interact with Atg8 (supplemental Fig. S10); however, its expression did not compromise vacuolar delivery of GFP-Atg8 under starvation conditions in both wild type and Δpng1 yeast (Fig. 5A). Furthermore, about 40% of the 6Myc-Hmg2p-1–1022 itself was delivered to the vacuole for degradation, as evident by its impaired degradation in a Δpep4 strain (Fig. 7A). This vacuolar delivery may account for the rapid decay of 6Myc-Hmg2p-1–1022 observed in both wild-type and Δpng1 yeast (Fig. 6B). To test the involvement of the autophagic/CVT machinery in vacuolar delivery of 6Myc-Hmg2p-1–1022-RFP, we examined its localization in Δatg8, Δatg8/Δpng1, and Δatg19 yeast. As shown in Fig. 7B, 6Myc-Hmg2p-1–1022-RFP was detected in the vacuole only when expressed in wild-type or Δpng1 strains but not in strains lacking ATG8 or ATG19. These results indicate that a significant portion of this mutant form of Hmg2p is delivered for vacuolar degradation in a CVT-dependent manner. Notably, Wolf and colleagues (68) have recently demonstrated that insertion of a 3HA tag at the C terminus of the ER-associated protein, Dfm1, transforms this stable protein into a rapidly degraded substrate.
FIGURE 7.
The autophagic/CVT machineries mediate vacuolar transport of 6Myc-Hmg2p-1–1022. A, evaluation of the half-life of 6Myc-Hmg2p-1–1022 by cycloheximide chase in the indicated strains. Quantification of the Western blots is presented in the lower panel. B, cellular distribution of 6Myc-Hmg2p-1–1022-RFP was determined in the indicated strains by visualizing RFP fluorescence. DIC, differential interference contrast. Error bars, S.D.
To test whether the mere presence or rather the integrity of the C terminus of 6Myc-Hmg2p is responsible for the effect of Png1 on 6Myc-Hmg2p turnover, we performed a similar experiment in which we inserted a FLAG tag at the C terminus of the full-length 6Myc-Hmg2p. Again, similar to the results obtained with 6Myc-Hmg2p-1–1022, the degradation of 6Myc-Hmg2p-FLAG was rapid in WT yeast as well as in the Δpng1 strain (supplemental Fig. S11). In addition, 6Myc-Hmg2p-Flag-RFP was also detected in the vacuole in both WT and Δpng1 cells (supplemental Fig. S11). These findings emphasize the notion that the 6Myc-Hmg2p C terminus is essential for the role of Png1 in this process. Thus, removal or modification of the 6Myc-Hmg2p C terminus allows the delivery of this substrate for vacuolar degradation in a Png1-independent manner.
DISCUSSION
Although the role of the proteasome in the degradation of ER-misfolded proteins has been established, the involvement of the lysosomes/vacuole, the other major degradative cellular pathway, has remained unclear. Here we present evidence for an alternative route for clearance of an ERAD substrate by the vacuole. By investigating the removal of 6Myc-Hmg2p, one of the most studied ERAD-M substrates, we identified an interaction between the misfolded ubiquitinated Hmg2p and elements of the CVT machinery on the ER membrane (Figs. 3A and 4B). This interaction is destabilized by the action of Png1, the cytosolic N-glycanase, previously implicated in ER-associated degradation (53–55). In the absence of PNG1, a stable complex is formed, which in turn leads to a slowdown in proteasomal degradation of 6Myc-Hmg2p (Fig. 1B). Concomitant removal of PNG1 and one of several key autophagic factors resumes fast proteasomal degradation of 6Myc-Hmg2p (Figs. 3, B and D, and 4A). We also demonstrate that the regulatory role of Png1 is mediated by the C terminus of Hmg2p. Truncation of the last 23 residues of the 6Myc-Hmg2p as well as the addition of a FLAG tag at its C terminus decoupled Png1 activity and proteasomal degradation of 6Myc-Hmg2p (Figs. 6B and 8A). This allows delivery of a portion of the truncated ERAD-M substrate 6Myc-Hmg2p-1–1022 to vacuolar degradation in an ATG8- and ATG19-dependent manner (Fig. 7). Based on these findings, we postulate the existence of a new route responsible for the delivery of ERAD substrates to the vacuole acting in parallel to the proteasome.
FIGURE 8.
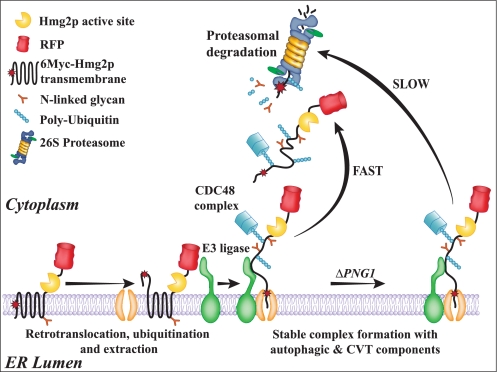
A model for the involvement of Png1 and the autophagic machinery in regulating the degradation of 6Myc-Hmg2p. In the presence of an active Png1, 6Myc-Hmg2p is removed from the ER membrane to the cytosol, where it is rapidly degraded by the proteasome. This process entails retrotranslocation, ubiquitination, and extraction from the ER membrane followed by proteasomal degradation. In the absence of PNG1, autophagy- and CVT-specific components attenuate the degradation process, leading to the accumulation of ubiquitinated 6Myc-Hmg2p on the ER membrane.
In the present study, we identified for the first time a physical interaction between an ERAD-M target and Atg8, a key autophagic factor (27). Atg8 has been implicated previously in selective recruitment of Ape1 and mitochondria into autophagosomes by interacting with Atg19 and Atg32, respectively (69–71). Notably, we also found that 6Myc-Hmg2p co-immunoprecipitated with the precursor form of Ape1 (Fig. 4B), suggesting that these proteins together with Atg8 and potentially other factors form a complex on the ER membrane. Functional evidence for this interaction is also provided. Accordingly, the removal of either ATG8 or ATG19 alleviated the attenuated proteasomal degradation in Δpng1 (Figs. 3A and 4A). Likewise, Atg3 and Atg7, factors mediating Atg8 conjugation to phosphatidylethanolamine (27), are also involved in this process, indicating that lipidation of Atg8 may also be required. Moreover, delivery of 6Myc-Hmg2p lacking the last 23 amino acids to the vacuole was found to be fully dependent on ATG8 and ATG19 (Fig. 7B). These findings are consistent with a recent report implicating the CVT pathway in degradation of a mutated misfolded form of Pma1 (72).
One of the consequences of elevated 6Myc-Hmg2p levels in the ER membrane was reflected by prevention of vacuolar transport of GFP-Atg8 and CVT cargo (Fig. 5, A, B, and D). Most likely, under normal growth conditions, crucial components of these pathways present in limiting amounts, such as Atg8 and Atg19, are recruited to the ER membrane lesion, rendering them unavailable for the constitutive CVT. Consistently, expression of 6Myc-Hmg2p in wild-type cells led to a 2.6-fold increase in the transcription level of ATG8 (supplemental Fig. S5), perhaps to partially compensate for the sequestration of Atg8. Blockage of starvation-induced transport of GFP-Atg8 supports the notion that in the absence of PNG1, a stable complex between 6Myc-Hmg2p and Atg8 is formed. Our findings indicate that both expression of 6Myc-Hmg2p and the absence of PNG1 are necessary for the attenuation of vacuolar delivery of GFP-Atg8 upon starvation (Fig. 5, A and B). This was unexpected because nitrogen depletion induces strong up-regulation of the ATG8 transcript (22) (supplemental Fig. S5) that should compensate for the fraction of GFP-Atg8 recruited to 6Myc-Hmg2p. Together, these findings suggest that the GFP-Atg8 sequestrated by the ERAD-M substrate under normal growth conditions is tightly bound. Thus, upon starvation, the elevated endogenous Atg8 expression did not chase off GFP-Atg8 from the ER membrane, thereby not reaching the vacuole. In support, when GFP-Atg8 was expressed under the endogenous ATG8 promoter, its levels were elevated upon starvation, and GFP was detected in the vacuole in Δpng1 yeast expressing 6Myc-Hmg2p (supplemental Fig. S7). Based on these findings, we conclude that in the absence of PNG1, the complex between Atg8 and 6Myc-Hmg2p is stabilized.
Here we provide evidence for a new role of the CVT machinery in surveying the ER membrane for potential aberration. Our data imply that this system acts differently depending on the character of the misfolded protein found on the ER membrane. In one scenario, where 6Myc-Hmg2p is expressed in cells lacking PNG1, the CVT machinery may fulfill a protective function, sequestering the lesion from the bulk cytosol. Indeed, Bernales et al. (47) reported that an elevation in cellular autophagy in response to ER stress might also lead to the appearance of membrane expansions and autophagosome-like structures. Under these conditions, the vesicles are thought to play a protective role in response to ER stress induced by the addition of DTT or tunicamycin rather than delivering the content for vacuolar degradation. Truncation of the 6Myc-Hmg2p C-terminal 23-amino acid stretch constituted a different scenario whereby a significant portion of this ERAD substrate is delivered to vacuolar degradation in a CVT-dependent manner. Either way, our data imply that recruitment and commitment of the autophagic system to a membrane lesion in the ER is relatively slow compared with the proteasomal degradation associated with ER dislocation. Upon slowdown in the extraction of the defective protein, the association with Atg8 and the CVT complex is stabilized, thus providing an autophagy-dependent checkpoint for the extraction and degradation process of 6Myc-Hmg2p (Fig. 8).
ER chaperones were previously demonstrated to maintain ERAD substrates in a retrotranslocation-competent state (73, 74). Moreover, these studies concluded that the ERAD machinery is unable to process aggregated targets. It appears that a selective set of chaperones is required for retrotranslocation of different ER aberrant proteins (75). Interestingly, mammalian PNGase was shown to associate with the ERAD machinery promoting retrotranslocation (76, 77). Accordingly, Png1 promotes efficient extraction of 6Myc-Hmg2p, thus acting as chaperone in this system. In fact, we show here that upon deletion of PNG1, the ERAD-M substrate accumulates on the membrane decorated with polyubiquitin, in Triton X-100-resistant aggregates (Fig. 2B). Deletion of the 6Myc-Hmg2p C terminus (6Myc-Hmg2p-1–1022-RFP) renders the protein insensitive to the presence of Png1, leading to its partial delivery to the vacuole (Fig. 7). We argue that the fraction of 6Myc-Hmg2p-1–1022-RFP that was delivered to the vacuole may form aggregates on the ER inaccessible to the ERAD machinery.
In the present study, we provide evidence of a new role for an autophagy-related mechanism in the selective removal of an ERAD-M substrate. Support for the selectivity of the process emerges from the fact that Atg19, a protein implicated in the selective delivery of Ape1 from cytosol to vacuole, was found to be essential for the delivery of 6Myc-Hmg2p-1–1022-RFP to the vacuole. Moreover, we show that Ape1, in its premature form, associates with 6Myc-Hmg2p on the ER membrane to form a complex that also contains Atg8. Based on these findings and the functional analysis of the effect of the different ATG null mutants on 6Myc-Hmg2p degradation, we hypothesize that factors of the CVT machinery constantly survey the ER membrane for possible lesions caused by the accumulation of transmembrane proteins. Once such a lesion is recognized, possibly by interaction of Atg8 and Ape1 with ubiquitinated ERAD-M substrates, a complex is formed. Under normal conditions, Png1, acting as a chaperone, is responsible for the dissociation of this complex, thus allowing the retrotranslocation of the ERAD-M substrate and its delivery for proteasomal degradation. In the absence of Png1, the complex between 6Myc-Hmg2p and factors of the CVT machinery is stabilized, leading to a slowdown in the degradation of the ERAD-M substrate, possibly playing a protective role of the ER lesions.
Autophagy was originally thought to act non-selectively; however, a growing body of evidence provides examples of molecular mechanisms for cargo specificity. In yeast, numerous selective autophagy pathways were recently described. For instance, Atg19 acts as a specific receptor for the delivery of Ape1 and Ams1 from the cytosol to the vacuole via the CVT pathway (71, 78). Peroxisome recruitment into autophagosomes depends on Pex3 and Pex14, two peroxisomal proteins essential for targeting peroxisomes for autophagy (79). The expansion of the ER triggered by the unfolded protein response is remodeled to its homeostatic volume by selective autophagy (47). Selective degradation of the 60 S ribosomal subunit by the autophagic pathway is regulated by the ubiquitin protease Ubp3 and its activator Bre5 (80). Finally, Atg32, a mitochondrial peripheral membrane protein, was recently reported to target autophagosomes to mitochondria by interacting with Atg8 and Atg11 (69, 70). Based on our findings, we propose that the autophagic machinery selectively recognizes the ERAD M substrate 6Myc-Hmg2p, thus implicating selective autophagy during ERAD. Future experiments are required to determine whether this system represents part of a more general function of the CVT machinery in quality control of the ER membrane.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S11.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- IP
- immunoprecipitation.
REFERENCES
- 1. Helenius A., Aebi M. (2001) Science 291, 2364–2369 [DOI] [PubMed] [Google Scholar]
- 2. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carvalho P., Goder V., Rapoport T. A. (2006) Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 4. Vashist S., Ng D. T. (2004) J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mast S. W., Diekman K., Karaveg K., Davis A., Sifers R. N., Moremen K. W. (2005) Glycobiology 15, 421–436 [DOI] [PubMed] [Google Scholar]
- 6. Hosokawa N., Wada I., Hasegawa K., Yorihuzi T., Tremblay L. O., Herscovics A., Nagata K. (2001) EMBO Rep. 2, 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olivari S., Galli C., Alanen H., Ruddock L., Molinari M. (2005) J. Biol. Chem. 280, 2424–2428 [DOI] [PubMed] [Google Scholar]
- 8. Nakatsukasa K., Nishikawa S., Hosokawa N., Nagata K., Endo T. (2001) J. Biol. Chem. 276, 8635–8638 [DOI] [PubMed] [Google Scholar]
- 9. Bhamidipati A., Denic V., Quan E. M., Weissman J. S. (2005) Mol. Cell 19, 741–751 [DOI] [PubMed] [Google Scholar]
- 10. Jakob C. A., Bodmer D., Spirig U., Battig P., Marcil A., Dignard D., Bergeron J. J., Thomas D. Y., Aebi M. (2001) EMBO Rep. 2, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oda Y., Hosokawa N., Wada I., Nagata K. (2003) Science 299, 1394–1397 [DOI] [PubMed] [Google Scholar]
- 12. Spear E. D., Ng D. T. (2005) J. Cell Biol. 169, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kostova Z., Wolf D. H. (2005) J. Cell Sci. 118, 1485–1492 [DOI] [PubMed] [Google Scholar]
- 14. Yoshida Y., Tokunaga F., Chiba T., Iwai K., Tanaka K., Tai T. (2003) J. Biol. Chem. 278, 43877–43884 [DOI] [PubMed] [Google Scholar]
- 15. Yoshida Y., Chiba T., Tokunaga F., Kawasaki H., Iwai K., Suzuki T., Ito Y., Matsuoka K., Yoshida M., Tanaka K., Tai T. (2002) Nature 418, 438–442 [DOI] [PubMed] [Google Scholar]
- 16. Cox J. S., Walter P. (1996) Cell 87, 391–404 [DOI] [PubMed] [Google Scholar]
- 17. Mori K. (2000) Cell 101, 451–454 [DOI] [PubMed] [Google Scholar]
- 18. Sidrauski C., Cox J. S., Walter P. (1996) Cell 87, 405–413 [DOI] [PubMed] [Google Scholar]
- 19. Spear E. D., Ng D. T. (2003) Mol. Biol. Cell 14, 2756–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levine B., Klionsky D. J. (2004) Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 21. Meijer A. J., Codogno P. (2006) Mol. Aspects Med. 27, 411–425 [DOI] [PubMed] [Google Scholar]
- 22. Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999) J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. (2004) J. Cell Sci. 117, 2805–2812 [DOI] [PubMed] [Google Scholar]
- 24. Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. (2010) EMBO J. 29, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang W. P., Scott S. V., Kim J., Klionsky D. J. (2000) J. Biol. Chem. 275, 5845–5851 [DOI] [PubMed] [Google Scholar]
- 27. Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000) Nature 408, 488–492 [DOI] [PubMed] [Google Scholar]
- 28. Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. (2000) J. Cell Biol. 151, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shintani T., Klionsky D. J. (2004) J. Biol. Chem. 279, 29889–29894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berger Z., Ravikumar B., Menzies F. M., Oroz L. G., Underwood B. R., Pangalos M. N., Schmitt I., Wullner U., Evert B. O., O'Kane C. J., Rubinsztein D. C. (2006) Hum. Mol. Genet 15, 433–442 [DOI] [PubMed] [Google Scholar]
- 31. Kaganovich D., Kopito R., Frydman J. (2008) Nature 454, 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Webb J. L., Ravikumar B., Atkins J., Skepper J. N., Rubinsztein D. C. (2003) J. Biol. Chem. 278, 25009–25013 [DOI] [PubMed] [Google Scholar]
- 33. Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B., Yuan J. (2006) J. Biol. Chem. 281, 14474–14485 [DOI] [PubMed] [Google Scholar]
- 34. Iwata A., Riley B. E., Johnston J. A., Kopito R. R. (2005) J. Biol. Chem. 280, 40282–40292 [DOI] [PubMed] [Google Scholar]
- 35. Qin Z. H., Wang Y., Kegel K. B., Kazantsev A., Apostol B. L., Thompson L. M., Yoder J., Aronin N., DiFiglia M. (2003) Hum. Mol. Genet. 12, 3231–3244 [DOI] [PubMed] [Google Scholar]
- 36. Ravikumar B., Vacher C., Berger Z., Davies J. E., Luo S., Oroz L. G., Scaravilli F., Easton D. F., Duden R., O'Kane C. J., Rubinsztein D. C. (2004) Nat. Genet. 36, 585–595 [DOI] [PubMed] [Google Scholar]
- 37. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 39. Zatloukal K., Stumptner C., Fuchsbichler A., Heid H., Schnoelzer M., Kenner L., Kleinert R., Prinz M., Aguzzi A., Denk H. (2002) Am. J. Pathol. 160, 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nagaoka U., Kim K., Jana N. R., Doi H., Maruyama M., Mitsui K., Oyama F., Nukina N. (2004) J. Neurochem. 91, 57–68 [DOI] [PubMed] [Google Scholar]
- 41. Pandey U. B., Nie Z., Batlevi Y., McCray B. A., Ritson G. P., Nedelsky N. B., Schwartz S. L., DiProspero N. A., Knight M. A., Schuldiner O., Padmanabhan R., Hild M., Berry D. L., Garza D., Hubbert C. C., Yao T. P., Baehrecke E. H., Taylor J. P. (2007) Nature 447, 859–863 [DOI] [PubMed] [Google Scholar]
- 42. Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A., Yao T. P. (2003) Cell 115, 727–738 [DOI] [PubMed] [Google Scholar]
- 43. Teckman J. H., Perlmutter D. H. (2000) Am. J. Physiol. Gastrointest. Liver Physiol. 279, G961–G974 [DOI] [PubMed] [Google Scholar]
- 44. Teckman J. H., An J. K., Loethen S., Perlmutter D. H. (2002) Am. J. Physiol. Gastrointest. Liver Physiol. 283, G1156–G1165 [DOI] [PubMed] [Google Scholar]
- 45. Teckman J. H., An J. K., Blomenkamp K., Schmidt B., Perlmutter D. (2004) Am. J. Physiol. Gastrointest. Liver Physiol. 286, G851–G862 [DOI] [PubMed] [Google Scholar]
- 46. Perlmutter D. H. (2006) Autophagy 2, 258–263 [DOI] [PubMed] [Google Scholar]
- 47. Bernales S., McDonald K. L., Walter P. (2006) PLoS Biol. 4, e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yorimitsu T., Nair U., Yang Z., Klionsky D. J. (2006) J. Biol. Chem. 281, 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hampton R. Y., Gardner R. G., Rine J. (1996) Mol. Biol. Cell 7, 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loayza D., Tam A., Schmidt W. K., Michaelis S. (1998) Mol. Biol. Cell 9, 2767–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Elble R. (1992) BioTechniques 13, 18–20 [PubMed] [Google Scholar]
- 52. Straub M., Bredschneider M., Thumm M. (1997) J. Bacteriol. 179, 3875–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim I., Ahn J., Liu C., Tanabe K., Apodaca J., Suzuki T., Rao H. (2006) J. Cell Biol. 172, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hosomi A., Tanabe K., Hirayama H., Kim I., Rao H., Suzuki T. (2010) J. Biol. Chem. 285, 24324–24334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suzuki T., Park H., Hollingsworth N. M., Sternglanz R., Lennarz W. J. (2000) J. Cell Biol. 149, 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. (2004) J. Biol. Chem. 279, 38369–38378 [DOI] [PubMed] [Google Scholar]
- 57. Wilhovsky S., Gardner R., Hampton R. (2000) Mol. Biol. Cell 11, 1697–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gardner R. G., Shearer A. G., Hampton R. Y. (2001) Mol. Cell. Biol. 21, 4276–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rabinovich E., Kerem A., Fröhlich K. U., Diamant N., Bar-Nun S. (2002) Mol. Cell Biol. 22, 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hampton R. Y., Koning A., Wright R., Rine J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Koning A. J., Roberts C. J., Wright R. L. (1996) Mol. Biol. Cell 7, 769–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anderson R. G., Orci L., Brown M. S., Garcia-Segura L. M., Goldstein J. L. (1983) J. Cell Sci. 63, 1–20 [DOI] [PubMed] [Google Scholar]
- 63. Jingami H., Brown M. S., Goldstein J. L., Anderson R. G., Luskey K. L. (1987) J. Cell Biol. 104, 1693–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Orci L., Brown M. S., Goldstein J. L., Garcia-Segura L. M., Anderson R. G. (1984) Cell 36, 835–845 [DOI] [PubMed] [Google Scholar]
- 65. Lynch-Day M. A., Klionsky D. J. (2010) FEBS Lett. 584, 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Istvan E. S., Palnitkar M., Buchanan S. K., Deisenhofer J. (2000) EMBO J. 19, 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Takeuchi J., Chen H., Coffino P. (2007) EMBO J. 26, 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stolz A., Schweizer R. S., Schafer A., Wolf D. H. (2010) Traffic 10, 1363–1369 [DOI] [PubMed] [Google Scholar]
- 69. Okamoto K., Kondo-Okamoto N., Ohsumi Y. (2009) Dev. Cell 17, 87–97 [DOI] [PubMed] [Google Scholar]
- 70. Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. (2009) Dev. Cell 17, 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Scott S. V., Guan J., Hutchins M. U., Kim J., Klionsky D. J. (2001) Mol. Cell 7, 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mazón M. J., Eraso P., Portillo F. (2007) Mol. Microbiol. 63, 1069–1077 [DOI] [PubMed] [Google Scholar]
- 73. Brodsky J. L., Werner E. D., Dubas M. E., Goeckeler J. L., Kruse K. B., McCracken A. A. (1999) J. Biol. Chem. 274, 3453–3460 [DOI] [PubMed] [Google Scholar]
- 74. Casagrande R., Stern P., Diehn M., Shamu C., Osario M., Zúñiga M., Brown P. O., Ploegh H. (2000) Mol. Cell 5, 729–735 [DOI] [PubMed] [Google Scholar]
- 75. Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. (2001) J. Cell Biol. 153, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhao G., Zhou X., Wang L., Li G., Schindelin H., Lennarz W. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8785–8790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Katiyar S., Joshi S., Lennarz W. J. (2005) Mol. Biol. Cell 16, 4584–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shintani T., Huang W. P., Stromhaug P. E., Klionsky D. J. (2002) Dev. Cell 3, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Farré J. C., Manjithaya R., Mathewson R. D., Subramani S. (2008) Dev. Cell 14, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kraft C., Deplazes A., Sohrmann M., Peter M. (2008) Nat. Cell Biol. 10, 602–610 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.