Abstract
In the present study we demonstrate an association between mammalian myosin Va and cytoplasmic P bodies, microscopic ribonucleoprotein granules that contain components of the 5′–3′ mRNA degradation machinery. Myosin Va colocalizes with several P body markers and its RNAi-mediated knockdown results in the disassembly of P bodies. Overexpression of a dominant-negative mutant of myosin Va reduced the motility of P bodies in living cells. Co-immunoprecipitation experiments demonstrate that myosin Va physically associates with eIF4E, an mRNA binding protein that localizes to P bodies. In contrast, we find that myosin Va does not play a role in stress granule formation. Stress granules are ribonucleoprotein structures that are involved in translational silencing and are spatially, functionally, and compositionally linked to P bodies. Myosin Va is found adjacent to stress granules in stressed cells but displays minimal localization within stress granules, and myosin Va knockdown has no effect on stress granule assembly or disassembly. Combined with recently published reports demonstrating a role for Drosophila and mammalian class V myosins in mRNA transport and the involvement of the yeast myosin V orthologue Myo2p in P body assembly, our results provide further evidence that the class V myosins serve an important role in the transport and turnover of mRNA.
Keywords: Molecular Motors, mRNA, Myosin, RNA Transport, RNA Turnover, P Bodies, Stress Granules
Introduction
Class V myosins are actin-based motor proteins composed of two identical heavy chains and several associated light chains. The amino-terminal head domain binds to ATP and actin filaments and provides the motive force for the complex. The neck region contains six calmodulin binding IQ motifs and acts as a lever arm, whereas the carboxyl-terminal tail domain mediates binding to cargo (1). Yeast have two class V myosins, flies and worms have one, whereas mammals have three (Va, Vb, and Vc). Myosin Va (MyoVa)2 is widely expressed but is enriched in brain, testes, and skin. It has been implicated in a diverse range of cellular roles, including synaptic vesicle transport (2), insulin secretion (3), myelination (4), and long term potentiation (5). People who lack functional myosin Va suffer from a rare autosomal recessive disorder called Griscelli syndrome, which is characterized by hypopigmentation and severe neurological defects (6).
It is well established that the yeast class V myosin Myo4p actively transports Ash1 mRNA to the bud tip of dividing cells (7), but it is now emerging that class V myosins from higher eukaryotes also play roles in RNA transport. Myosin Va has been found associated with several RNA binding proteins in a messenger ribonucleoprotein (mRNP) complex precipitated from a mouse brain extract (8). The intracellular distribution of RNA is dramatically altered in primary cells derived from dilute-lethal (myosin Va-null) mice (9). Yoshimura et al. (10) have shown that myosin Va mediates the translocation of TLS (translated in liposarcoma) and its target RNA, Nd1-L, into dendritic spines, and myosin V is involved in targeting oskar mRNA to the posterior pole of the Drosophila oocyte (11).
Recently, the other Saccharomyces cerevisiae myosin V orthologue, Myo2p, was found to be associated with hundreds of mRNA transcripts and to partially colocalize with P bodies (12). P bodies are microscopic structures composed of enzymes involved in mRNA turnover. They are believed to perform a number of functions, including storage of translationally inactive mRNP complexes and the decapping and degradation of unwanted mRNA (13). Chang et al. (14) suggest that Myo2p facilitates the release of mRNA transcripts from P bodies. P bodies also contain components of the miRNA- and siRNA-mediated translational repression pathways. In the present study, we identified a pool of mammalian myosin Va that localizes to P bodies. We found that it physically associates with the mRNA cap binding protein eIF4E and showed that the siRNA-mediated depletion of myosin Va affects the assembly of P bodies but has no effect on the formation of the closely related stress granules.
EXPERIMENTAL PROCEDURES
Cell Lines and Plasmids
HeLa and S91-6 cells were obtained from the European Collection of Cell Cultures, and B16-F10 cells were a kind gift from E. Sviderskaya (Wellcome Trust Functional Genomics Cell Bank, St. George's Hospital Medical School, London, UK). Cells were maintained in DMEM (BioWhittaker) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm glutamine at 5% CO2. mCherry-C1 MyoVa(1100–1853) was constructed by subcloning the open reading frame from pGADGH MyoVa(1100–1853) into the EcoRI/SalI sites of pmCherry-C1. Dcp1a-GFP and mCherry were kind gifts from R. Parker (15) and R. Tsien (16), respectively.
Antibodies
Rabbit polyclonal antibodies to myosin Va (LF-18) and Dcp1a (HPA013202), mouse monoclonal anti-α tubulin, and chicken anti-Lsm1 were from Sigma-Aldrich. Rabbit polyclonal anti-myosin Vb has been described elsewhere (17). Mouse monoclonal anti-eIF4E (product P-2) and goat anti-TIA-1 (product C-20) were from Santa Cruz Biotechnology. The mouse monoclonal anti-FMRP (1C3) was obtained from Chemicon. Two anti-GFP antibodies from Abcam were used; mouse anti-GFP (catalog no. 1218) for co-immunoprecipitations and rabbit anti-GFP (catalog no. 290) for Western blots. Secondary antibodies for immunofluorescence were from Molecular Probes and included Alexa Fluor 488 goat anti-mouse, Alexa Fluor 594 goat anti-rabbit, Alexa Fluor 488 goat anti-chicken and Alexa Fluor 488 donkey anti-goat. Western blots were performed on a LiCor Odyssey Infrared Imaging System using the following secondary antibodies from Rockland: IRDye 700DX goat anti-chicken, IRDye 800 donkey anti-goat, IRDye 800 goat anti-mouse, and IRDye800 goat anti-rabbit.
RNAi
HeLa cells seeded the day previously were transfected with siLam A/C (5′-AACUGGACUUCCAGAAGAACA-3′), siMyoVa1 (5′-AACUGACUACCUGAAUGAUGA-3′), siMyoVa2 (5′-CGAAACAACUGGAACUCGA-3′), and siLuc (5′-CGUACGCGGAAUACUUCGA-3′) for 72 h using Oligofectamine (Invitrogen) according to the manufacturer's instructions. To deplete MyoVa in the B16-F10 mouse melanoma cell lines, the HuSH 29-mer myosin Va shRNA vectors were purchased from Origene (shMyoVa1, 5′-CAGGTACAATGTCAGTCAACTGGAAGAAT-3′ and shMyoVa2, 5′-GTCAATCAGGCTCTCCATTCTGCTGTCAA-3′). The same parental plasmid expressing a noneffective GFP shRNA was used as a negative control. To visualize cells transfected with the shRNA the HuSH vectors were co-transfected with pEGFP-C1 (Clontech) empty vector using Lipofectamine 2000. The cells were processed for immunofluorescence 72 h post-transfection.
Immunofluorescence and Confocal Microscopy
Cells seeded on 10-mm glass coverslips at least 48 h previously were fixed with 3% paraformaldehyde for 15 min at room temperature and quenched with 50 mm NH4Cl. After washing, the cells were permeabilized with 0.1% Triton X-100 and labeled for 1 h in a humid chamber with the indicated primary antibodies. The coverslips were washed and incubated for a further 1 h with secondary antibodies, washed again, and mounted on microscope slides using Mowiol. When chicken anti-Lsm1 was used the cells were permeabilized in PP buffer (0.05% saponin; 80 mm Pipes, pH 6.8, 1 mm MgCl2, 5 mm EGTA) plus 20 units/ml RNasin for 3 min at room temperature prior to fixation. Fluorescence images were acquired on a Zeiss LSM 510 Meta confocal microscope and processed with Image Examiner software (Zeiss). Figures were created in Adobe Illustrator (version 10). The number of P bodies per cell was counted manually using the Cell Counter plug-in of ImageJ software (U. S. National Institutes of Health).
Live Cell Microscopy
HeLa cells were seeded on 35-mm glass-bottomed dishes (Iwaki, Japan). On the following day, cells were transfected with Dcp1a-GFP and either mCherry-C1 or mCherry-MyoVa(1100–1853) using Lipofectamine 2000. 24 h post-transfection, the cells were washed with PBS, and fresh medium was added. Confocal videomicroscopy time lapse images were acquired on a Leica DMIRE2 inverted microscope equipped with a CSUXI spinning disk head (Yokogawa), and temperature and CO2 controller (Life Imaging Services). GFP and mCherry were excited with 491- and 561-nm laser lines (MAG Biosystems), respectively. Images were captured on a QuantEM 512SC camera (Photometrics). A z stack of seven planes was acquired every 2 s for a total duration of 2 min. Movies were generated by compiling three-dimensional maximum intensity projections. P body tracks were generated using the Object Tracking journal in MetaMorph (version 7, Molecular Devices). Calculations and graphs were performed in Excel 2007 (Microsoft).
Immunoprecipitations
The immunoprecipitating antibody was prebound to anti-mouse IgG magnetic beads (Dynal) in TKN Buffer (20 mm Tris, pH 7.4, 50 mm KCl, 0.5% Nonidet P-40) for 1 h. Meanwhile, HeLa cells seeded in 100-mm dishes were washed with cold PBS and harvested in TKN, including protease inhibitors, plus 20 units/ml RNasin (Promega) and rotated for ∼30 min in a cold room. The lysate was passed six times through a 26-gauge needle and spun at 1000 × g for 5 min. The resulting supernatant was used as the input material and incubated overnight with the antibody-bead complexes. The next day, the beads were washed three times with TKN plus 20 units/ml RNasin and resuspended after the final wash in sample buffer. For the Lsm1 co-immunoprecipitations, anti-Lsm1 was bound to anti-chicken IgY conjugated to Sepharose (Davids Biotechnologie).
Fractionation
For fractionation experiments, HeLa cells seeded in 100-mm dishes were stressed as indicated, washed with cold PBS, harvested in TKN plus protease inhibitors plus RNasin, and rotated in a cold room for 30 min. The lysates were then sonicated for 2 × 2 min in a water-bath sonicator prior to centrifugation at 14,000 × g for 25 min at 4 °C. The soluble fraction (supernatant) was removed and the pellet (detergent-resistant fraction) was resuspended in an equal volume of TKN plus 2% SDS, heated to 95 °C for 5 min, and sonicated.
RESULTS
Myosin Va Is Necessary for PB Assembly
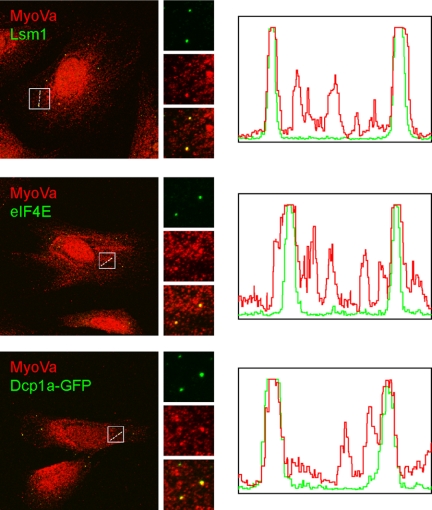
Examination of the intracellular distribution of endogenous myosin Va in HeLa cells using an isoform-specific antibody revealed that, in addition to the expected vesicular pattern, MyoVa also labeled several bright “foci” in the cytoplasm. These foci did not belong to elements of the endocytic or secretory pathways as evidenced by the lack of colocalization with various markers of these pathways (data not shown). Instead, these myosin Va-positive foci were identified as P bodies. P bodies are large foci located in the cytoplasm that contain translation repression and mRNA degradation machinery and accumulate translationally inactive mRNA transcripts (18). Myosin Va colocalized with the P body markers Lsm1, eIF4E, and Dcp1a-GFP (Fig. 1). The majority of P bodies examined were positive for myosin Va. This correlates with the recent observation that the yeast myosin V orthologue Myo2p plays a role in P body function in budding yeast (12). In contrast, endogenous myosin Vb was not found on P bodies (supplemental Fig. S1A).
FIGURE 1.
Myosin Va localizes to P bodies. HeLa cells grown on glass coverslips for 48 h were fixed and labeled for MyoVa, eIF4E, and Lsm1. To visualize Dcp1a, cells were transfected with Dcp1a-GFP for 18 h prior to fixation. Scale bar, 10 μm. Fluorescence intensity distribution along dashed white lines is shown in the histograms.
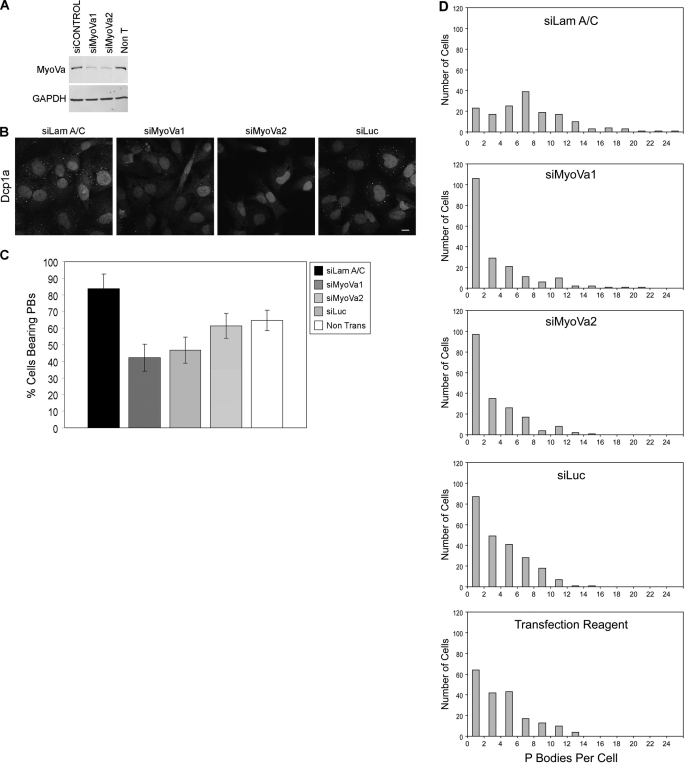
To further investigate the role of mammalian myosin Va in P body function, the effect of its knockdown on P body numbers and structure was determined. Two different siRNA oligonucleotides directed against myosin Va were used, both of which reduced the level of myosin Va protein by ∼85% after 72 h of transfection (Fig. 2A). The effect of myosin Va knockdown on the proportion of cells displaying P bodies and the number of P bodies in each cell were compared with three different control conditions; (1) cells transfected with a “targeting” siRNA to lamin A/C (siLam A/C), (2) cells transfected with a “nontargeting” siRNA to firefly luciferase (siLuc), and (3) cells treated with the transfection reagent alone. 72 h post-transfection the cells were fixed and P bodies were labeled with an antibody to Dcp1a. Knockdown of myosin Va resulted in an almost 2-fold reduction in the number of cells displaying P bodies, when compared with the targeting control siRNA, siLamA/C (Fig. 2, B and C). A similar level of reduction in the number of P bodies was consistently observed for both myosin Va targeting siRNA oligonucleotides. More than 80% of cells transfected with siLamA/C displayed P bodies, and this was reduced to between 40 and 50% in cells transfected with the myosin Va siRNA oligonucleotides. It should be noted that only ∼60% of cells transfected with the nontargeting siLuc, or treated with the transfection reagent alone, displayed P bodies. This is consistent with reports from a number of groups, which demonstrate that activation of RNAi and miRNA pathways leads to the accumulation of a pool of nontranslating mRNP complexes that can nucleate the assembly of P bodies (19–21). Also, Jagannath et al. (22) have found that transfection of cells with targeting siRNA oligonucleotides results in up-regulated expression of the P body components GW182 and Ago2. Because both siMyoVa1 and siMyoVa2 are targeting oligonucleotides, we have compared them with a targeting control siRNA, siLamA/C. siLuc does not have a target mRNA in HeLa cells and therefore does not induce the formation of P bodies as the RNAi pathway is not activated.
FIGURE 2.
Myosin Va knockdown results in P body disassembly. A, the degree of silencing of siMyoVa1 and siMyoVa2 was assessed by Western blot analysis. Non T, nontransfected. B, HeLa cells were transfected with the indicated siRNA oligonucleotides for 72 h and then immunostained with an antibody that detects endogenous Dcp1a. Nuclei were stained with DAPI. Scale bars, 10 μm. C, shown is the quantification of the percentage of cells displaying P bodies after 72 h of transfection with the indicated siRNA oligonucleotides. At least 200 cells were counted for each condition from three independent experiments. Data are shown as mean ± S.E. D, shown are distributions of the number of P bodies per cell in cells transfected with the indicated siRNAs for 72 h. The number of P bodies in >150 cells for each condition was counted from three independent experiments.
A closer inspection of the 40–50% of cells that retained P bodies when transfected with siRNA oligonucleotides to myosin Va revealed that the average number of PBs in these cells was four, in comparison with an average of eight in siLamA/C-transfected cells (Fig. 2D). Overall, these results indicate that a pool of myosin Va localizes to P bodies and this motor protein is required for their assembly and/or the maintenance of their structure in the cytoplasm.
Myosin Va Complexes with eIF4E
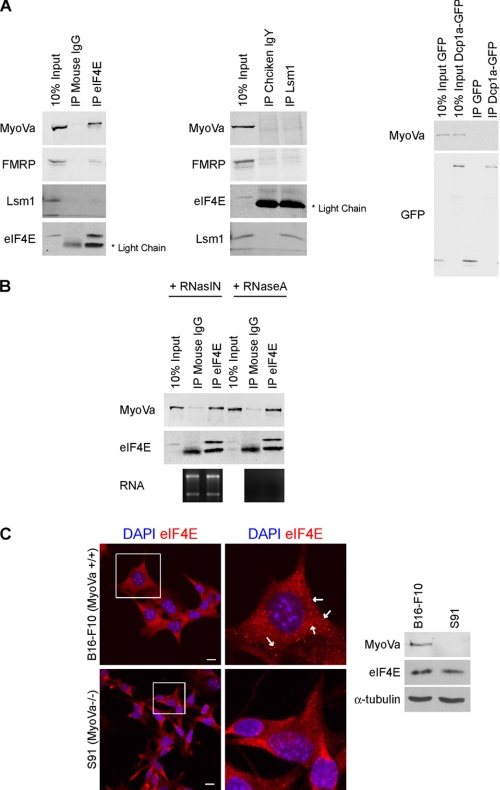
Co-immunoprecipitation assays were used to determine whether myosin Va physically associates with P body components. Endogenous eIF4E and Lsm1 were immunoprecipitated from HeLa cell lysates with specific antibodies. Nonimmune IgGs from the same species as the primary antibody were used as negative controls. In the case of Dcp1a, HeLa cells were transfected for 24 h with a Dcp1a-GFP construct, and the fusion protein was immunoprecipitated with an anti-GFP antibody. Cells expressing GFP alone were used as a negative control. These assays revealed that myosin Va associates with eIF4E but not with Lsm1 or Dcp1a (Fig. 3A). This contradicts data from the Mooseker laboratory, which shows that Myo2p co-immunoprecipitates with Lsm1p (12); however, we cannot rule out the possibility that our antibody to endogenous human Lsm1 masks its myosin Va binding domain. eIF4E (eukaryotic translation initiation factor 4E) is a translation initiation factor that binds the m7GpppN cap present at the 5′ end of all nuclear transcribed mRNAs. In conjunction with the eIF4F complex, it is involved in directing ribosomes to the m7G cap structure (23). eIF4E also localizes to P bodies, and it has been postulated to play a role in mRNP remodeling events that target mRNAs to P bodies for degradation (24). Lsm1 and eIF4E did not associate with each other, but interestingly, fragile X mental retardation protein (FMRP), an RNA binding protein previously shown to associate with myosin Va (8), was also pulled down in the eIF4E immunoprecipitation. FMRP colocalizes with myosin Va in HeLa cells (supplemental Fig. S1B). The association between myosin Va and eIF4E is not mediated via the binding of both proteins to the same mRNA transcript as the interaction is resistant to RNase A treatment (Fig. 3B). The localization of eIF4E was next examined in S91 cells, a mouse melanoma cell line derived from a dilute (MyoVa-null) mouse (25). In these cells, eIF4E displayed a punctate pattern distributed throughout the cytoplasm but did not localize to characteristic P bodies, whereas in B16-F10 melanoma cells (wild-type for myosin Va), eIF4E labeled several P bodies in each cell (Fig. 3C). There was no difference in the expression levels of eIF4E in both cell lines as determined by Western blot. Down-regulation of myosin Va expression in the B16-F10 cells using small hairpin RNAs resulted in a reduction in the number of cells displaying eIF4E-labeled P bodies (supplemental Fig. S2A). This corresponds to the reduction observed in HeLa cells transfected with siRNA oligonucleotides targeting myosin Va. Given the established role that myosin Va plays in the transport of mRNP complexes in a number of model systems (10, 11, 26), our data suggests that myosin Va is involved in the targeting of mRNA to P bodies, or the sites of P body nucleation, for storage and/or degradation.
FIGURE 3.
Myosin Va physically associates with eIF4E. A, PB components from HeLa cell lysates were immunoprecipitated with antibodies to eIF4E, Lsm1, and Dcp1a-GFP, and the immunoprecipitates were probed by immunoblotting with the indicated antibodies. Nonimmune IgG from the same species as the immunoprecipitating antibody was used as a control. B, the MyoVa-eIF4E interaction is not mediated via binding to the same mRNA transcripts. eIF4E was immunoprecipitated (IP) from the HeLa cells lysates in the presence of RNaseA or an RNase inhibitor (RNasin). To control for the activity of RNase A, total RNA was purified from the nonbound fraction and 28 S and 18 S rRNA was visualized on an agarose gel. C, eIF4E does not localize to P bodies in MyoVa-null cells. B16-F10 (MyoVa+/+) and S91 (MyoVa−/−) mouse melanoma cells were grown on glass coverslips for 48 h, fixed, and labeled with DAPI and anti-eIF4E. Arrows indicate P bodies. Scale bars, 10 μm. A Western blot of B16-F10 and S91 cell lysates probed with anti-MyoVa, anti-eIF4E, and anti-α tubulin (loading control) is also shown.
Overexpression of a Dominant-negative Mutant of Myosin Va Inhibits Motility of P Bodies
P bodies display a spatially restricted, microtubule-dependent, pattern of motility (27). Myosin Va has been reported to track the plus ends of microtubules in association with EB1 (28), and several reports have shown that class V myosins function together with kinesin microtubule motor proteins to transport cargo (29–31). Moreover, Krauss et al. (11) have demonstrated that myosin V and kinesin interact directly with each other in Drosophila oocytes and suggest that myosin V acts as a counterbalance to kinesin to prevent ectopic expression of oskar mRNA in the cytoplasm. We next examined whether myosin Va plays a role in the motility of P bodies. HeLa cells were co-transfected with a green fluorescent protein-tagged Dcp1a construct and a vector that expresses the tail region of myosin Va (amino acids 1100–1853) fused to mCherry. The tail regions of the class V myosins are commonly used as dominant-negative mutants (5, 32). The movement of the Dcp1a-labeled P bodies was recorded by live-cell spinning disk confocal microscopy. The use of a dominant-negative mutant was employed rather than an siRNA approach as we could never be sure that myosin Va was sufficiently depleted to have an effect in the cells that retained P bodies.
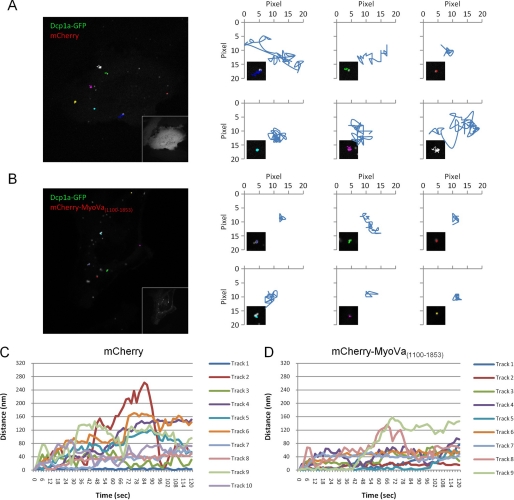
In cells expressing Dcp1a-GFP and mCherry alone, P body motility patterns similar to those reported by Aizer et al. (27) were observed. Tracking of these movements revealed some P bodies that were stationary for the duration of the movie, but the majority of P bodies displayed a highly dynamic, but restricted, motility pattern (Fig. 4, A and C, and supplemental Movie 1). These motile P bodies could travel up to 300 nm from their point of origin during the 2-min imaging period. In contrast, in cells that expressed medium to high levels of mCherry-MyoVa(1100–1853), the proportion of immobile P bodies was greatly increased (Fig. 4, B and D, and supplemental Movie 2), with only 39.4% (of a total of 33 tracks measured) of P bodies traveling >80 nm from their point of origin, in comparison with 59.6% (of a total of 47 tracks measured) traveling >80 nm in cells expressing mCherry alone (supplemental Fig. S3). Aizer et al. (27) observed that disruption of the microtubule network with the depolymerizing drug nocodazole reduces P body motility, whereas treatment with the microtubule-stabilizing drug taxol has no effect on P body movement (supplemental Figs. S4 and S5). The reduction in motility upon overexpression of mCherry-MyoVa(1100–1853) may be due to the uncoupling of P bodies from microtubules by this dominant-negative mutant. These results suggest that myosin Va plays a role in the dynamics of P bodies.
FIGURE 4.
A dominant-negative mutant of myosin Va reduces the motility of P bodies. A, Dcp1a-GFP in cells co-expressing mCherry were imaged in live HeLa cells (60 frames, total 2 min). The last acquired frame is represented with the tracks of six P bodies from the previous frames overlaid onto the image. The inset contains the corresponding mCherry image. Each of the six tracks is plotted on an XY graph, and the inset contains a zoomed image of the represented track. B, Dcp1a-GFP in cells co-expressing mCherry-MyoVa(1100–1853) were imaged in live HeLa cells (60 frames, total 2 min). The last acquired frame is represented with the tracks of six P bodies from the previous frames overlaid onto the image. The inset contains the corresponding mCherry-MyoVa(1100–1853) image. Each of the six tracks is plotted on an XY graph, and the inset contains an enlarged image of the represented track. C, line plot of all of the P body tracks measured in the cell depicted in A. The x axis represents time (s) and the y axis represented the distance (nm) from the origin (location of the P body at time 0). D, line plot of all of the P body tracks measured in the cell represented in B. The x axis represents time (s), and the y axis represented the distance (nm) from the origin (location of the P body at time 0).
Myosin Va Is Not Involved in Stress Granule Assembly
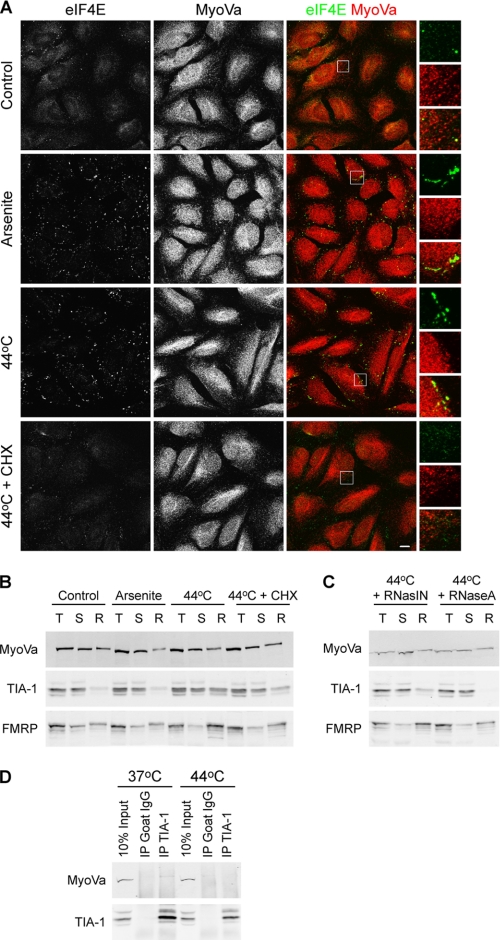
Stress granules are large nonmembranous cytoplasmic aggregates (from 0.1 to 2 μm in size). They form when a cell is exposed to a variety of environmental stresses and selectively accumulate nontranslating mRNPs containing transcripts encoding housekeeping genes but not those encoding stress-induced genes such as HSP70 (33). This allows the cell to reprogram its translational machinery to facilitate the synthesis of proteins required to deal with the stress. P bodies and stress granules are spatially, functionally, and compositionally linked (34). A number of proteins, including eIF4E, translocate from P bodies to SGs in response to stress. FMRP also localizes to stress granules in stressed cells (35). To determine whether myosin Va also translocates to stress granules, HeLa cells were exposed to oxidative or heat stress for 30 min prior to fixation and antibody labeling (Fig. 5A). Under these conditions, eIF4E was clearly found in stress granule aggregates; however, myosin Va did not undergo a similar translocation and remained in adjacent P bodies. Only rarely was myosin Va found to localize within stress granules. As reported previously, cycloheximide inhibited the formation of stress granules (36). This minimal association of myosin Va with stress granules was confirmed biochemically. Endogenous stress granule components partition into a detergent insoluble fraction in cells that have been exposed to stress (37). Control and stress-induced HeLa cells were harvested in an Nonidet P-40 containing buffer, sonicated, and fractionated into detergent-soluble and -insoluble fractions by centrifugation. Both TIA-1, an RNA binding protein that promotes stress granule assembly (37), and FMRP are enriched in the detergent-resistant fraction from stress-induced cells. Cycloheximide treatment releases these proteins back into the soluble fraction (Fig. 5B). In contrast, a pool of myosin Va consistently partitions into the detergent-resistant fraction, even under control, and unstressed, conditions and exposure to stress does not promote an increase in the proportion of myosin Va in the detergent-resistant fraction. RNaseA releases TIA-1 and some FMRP into the soluble fraction; however, it had no effect on the insoluble myosin Va pool (Fig. 5C). It is likely that this insoluble pool of myosin Va is associated with an actin fraction and not stress granules as β-actin is also consistently present in the detergent-resistant fraction under all conditions (data not shown). Co-immunoprecipitation assays revealed that myosin Va does not associate with TIA-1 under control or stress conditions (Fig. 5D).
FIGURE 5.
Myosin Va does not localize to stress granules. A, HeLa cells seeded on glass coverslips were either incubated at 37 °C in fresh medium or stressed by treatment with 500 μm sodium arsenite, or incubation at 44 °C plus or minus 10 μg/ml cycloheximide (CHX) for 30 min prior to fixation and immunostaining with anti-MyoVa and anti-eIF4E. Enlarged images of the boxed regions are displayed on the right. Scale bar, 10 μm. B, HeLa cells placed under the indicated stress conditions were fractionated into total (T), detergent-soluble (S) and -resistant (R) fractions. Fractions were analyzed by Western blotting. C, HeLa cells incubated at 44 °C were fractionated into total, detergent-soluble, and -resistant fractions in the presence of RNase A or an RNase inhibitor. Fractions were analyzed by Western blotting. D, myosin Va does not interact with TIA-1. TIA-1 was immunoprecipitated from HeLa cell lysates harvested from cells that had been maintained at 37 °C or incubated at 44 °C for 1 h. Immunoprecipitates were immunoblotted with anti-MyoVa and anti-TIA-1.
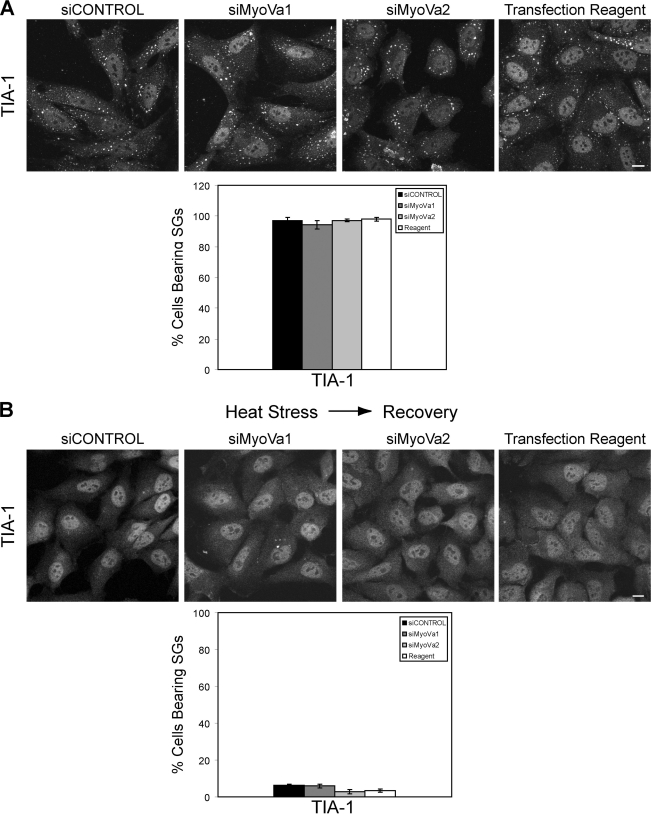
Myosin Va depletion has no effect on stress granule assembly. HeLa cells transfected with siCONTROL, siMyoVa1, siMyoVa2, or treated with the transfection reagent alone for 72 h were subjected to heat stress (44 °C for 1 h) prior to fixation and labeling with anti-TIA-1. Myosin Va knockdown had no effect on the percentage of cells bearing stress granules. Close to 100% of cells transfected with the MyoVa targeting, or control, siRNAs displayed TIA-1 positive stress granules (Fig. 6A). Myosin Va knockdown also had no effect on the TIA-1 localization pattern in unstressed cells (data not shown). The number and size of stress granules per cell was unaffected by myosin Va depletion. Also, myosin Va knockdown had no effect on the localization of eIF4E to stress granules (supplemental Fig. S2B). It has recently been reported that a dynein motor protein is involved in the formation of stress granules, whereas kinesin is required for stress granule disassembly (38). To determine whether myosin Va is required for SG disassembly, myosin Va-depleted HeLa cells were exposed to heat stress and then allowed to recover at 37 °C for 60 min. The cells were then fixed, and SGs were labeled with anti-TIA-1. Depletion of myosin Va did not affect the disassembly of SGs once the stress had been lifted. In both control and myosin Va knockdown cells <10% displayed stress granules (Fig. 6B). Thus, from these data, we can conclude that myosin Va does not play a role in the formation or dissolution of stress granules.
FIGURE 6.
Myosin Va depletion does not affect the assembly or disassembly of stress granules. A, HeLa cells were transfected with the indicated siRNA oligonucleotides for 72 h and placed at 44 °C for 1 h in fresh medium. The cells were fixed, and SGs were visualized by labeling with anti-TIA1. Scale bar, 10 μm. The histogram represents the percentage of cells bearing SGs. At least 300 cells were counted for each condition from three independent experiments. Data are shown as mean ± S.E. B, HeLa cells were transfected with the indicated siRNA oligonucleotides for 72 h and placed at 44 °C for 1 h in fresh medium. The medium was replaced, and the cells were allowed to recover at 37 °C for 1 h prior to fixation fixed and labeling with anti-TIA1. Scale bar, 10 μm. The histogram represents the percentage of cells bearing SGs. At least 300 cells were counted for each condition from three independent experiments. Data are shown as mean ± S.E.
DISCUSSION
The results presented here demonstrate that a pool of mammalian myosin Va localizes to P bodies, and our knockdown experiments indicate that this actin-based motor protein is required for the assembly and/or maintenance of these P body foci in the cytoplasm. We have found that myosin Va physically associates with eIF4E, a translation initiation factor that binds the 5′ cap on all nuclear transcribed mRNAs. eIF4E localizes to P bodies but translocates to stress granules upon exposure to environmental stresses. However, myosin Va did not translocate to stress granules along with eIF4E, and myosin Va knockdown had no effect on SG assembly.
The interaction between myosin Va and eIF4E is resistant to RNase A, suggesting that it is mediated by a protein-protein interaction, either direct or indirect. It is unlikely that myosin Va binds directly to mRNA transcripts as it does not possess classical RNA binding motifs such as KH and RRM domains. We believe that myosin Va mediates its role in RNA transport through interaction with RNA binding proteins such as eIF4E.
In live-cell studies, we have demonstrated that overexpression of the tail region of myosin Va results in a reduction of P body motility. It can be hypothesized that if a complex composed of myosin Va and kinesin is involved in tethering P bodies to the microtubule network, then the overexpression of a dominant-negative mutant of myosin Va would uncouple the P bodies from microtubules, resulting in the observed reduction of motility.
P bodies and stress granules are dynamic structures that are linked to the microtubule cytoskeleton (27, 38, 39). The retrograde and anterograde microtubule-based motor proteins dynein and kinesin-1 (KIF5B) have recently been shown to have antagonistic effects on P body enlargement and stress granule formation (38). The authors of this study demonstrate that dynein heavy chain 1 and bicaudal D1 are required for the formation of stress granules and the growth of P bodies upon stress and that KIF5B is required for the disassembly of SGs. They propose that a balance between dynein- and kinesin-driven transport regulates stress granule and P body growth.
Myosin Va is well established to move along microtubules in association with kinesin (28, 31, 40, 41). In fact, Krauss et al. (11) have demonstrated that myosin V acts in conjunction with kinesin to transport oskar mRNA to the posterior pole of the Drosophila oocyte. We postulate that myosin Va mediates the active transport of mRNP complexes along the cytoskeleton to P bodies. A common feature of P bodies from yeast to humans is the enhancement of their formation in the presence of increased concentrations of nontranslating RNA or mRNAs undergoing decapping (24, 42–45). We hypothesize that myosin Va plays a key role in the delivery of RNA that has exited the translation cycle to the sites of P body nucleation. The decline of P body numbers observed in cells depleted of myosin Va can thus be explained by the inhibition of this transport pathway and the failure of the cell to deliver nontranslating RNA to these P body nucleation sites.
There is a distinction between the cytoskeleton-dependent transport of nontranslating RNA to P bodies and the rapid exchange of certain P body components such as Dcp1a between the cytosol and PBs, which is not dependent on an intact cytoskeleton (27). Initial fluorescence recovery after photobleaching experiments in MyoVa-depleted cells suggest that myosin Va does not play a role in the exchange of Dcp1a between the cytosol and PBs (data not shown). This would explain the absence of an interaction between myosin Va and Dcp1a or Lsm1.
Cytoplasmic mRNAs and their associated RNA binding proteins cycle between polysomes, P bodies, and stress granules, and it is emerging that motor proteins play important roles in this process. Interestingly, Kwon et al. (46) have demonstrated that disruption of the actin cytoskeleton with cytochalasin D and latrunculin B does not inhibit the formation of SGs. This agrees with our observations that myosin Va, an actin-based motor protein, is not required for the formation or dissolution of SGs. It is attractive to speculate that the opposite is true for P bodies. We are currently investigating whether actin is associated with P bodies and plays a role in tethering the myosin Va-kinesin-mRNP complexes to these structures.
An unresolved issue in the field is why mRNPs aggregate into these large P bodies and stress granules. Aggregation of mRNPs into P bodies is not required for mRNA decapping or stress-induced translation repression in yeast (47). Also, disruption of P bodies in metazoans does not appear to inhibit miRNA-mediated repression or degradation of transcripts (20, 48). Perhaps these functions can take place in granules that are below the level of detection of the light microscope. Given the fact that the aggregation of mRNAs into large mRNP granules is a conserved feature of eukaryotic cells, it is very likely that P bodies and SGs have an important role (18).
In summary, given the evolutionarily conserved role played by class V myosins in mRNA transport from yeast to mammals (8, 10, 11, 49), we propose that mammalian myosin Va also plays an important role in the transport of mRNP complexes to P bodies.
Supplementary Material
Acknowledgments
We thank Elena Sviderskaya, Roger Tsien, and Roy Parker for generous gifts of reagents. We also acknowledge the PICT-IBiSA Imaging Platform and Bruno Goud at the Institut Curie.
This work was supported by Health Research Board, Ireland Career Development Fellowship PD/2005/25 and Joint Health Research Board/Marie-Curie Postdoctoral Mobility Fellowship HRB MCPD/2009/6 (to A. J. L.) and SFI Programme Grant 05/IN.3/B859 (to M. W. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Movies 1 and 2.
- MyoVa
- myosin Va
- FMRP
- fragile X mental retardation protein
- mRNP
- messenger ribonucleoprotein particle
- PB
- processing body
- SG
- stress granule
- Luc
- luciferase.
REFERENCES
- 1. Trybus K. M. (2008) Cell Mol. Life Sci. 65, 1378–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prekeris R., Terrian D. M. (1997) J. Cell Biol. 137, 1589–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Varadi A., Tsuboi T., Rutter G. A. (2005) Mol. Biol. Cell 16, 2670–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sloane J. A., Vartanian T. K. (2007) J. Neurosci. 27, 11366–11375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Correia S. S., Bassani S., Brown T. C., Lisé M. F., Backos D. S., El-Husseini A., Passafaro M., Esteban J. A. (2008) Nat. Neurosci. 11, 457–466 [DOI] [PubMed] [Google Scholar]
- 6. Pastural E., Barrat F. J., Dufourcq-Lagelouse R., Certain S., Sanal O., Jabado N., Seger R., Griscelli C., Fischer A., de Saint Basile G. (1997) Nat. Genet. 16, 289–292 [DOI] [PubMed] [Google Scholar]
- 7. Paquin N., Chartrand P. (2008) Trends Cell Biol. 18, 105–111 [DOI] [PubMed] [Google Scholar]
- 8. Ohashi S., Koike K., Omori A., Ichinose S., Ohara S., Kobayashi S., Sato T. A., Anzai K. (2002) J. Biol. Chem. 277, 37804–37810 [DOI] [PubMed] [Google Scholar]
- 9. Salerno V. P., Calliari A., Provance D. W., Jr., Sotelo-Silveira J. R., Sotelo J. R., Mercer J. A. (2008) Cell Motil. Cytoskeleton 65, 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoshimura A., Fujii R., Watanabe Y., Okabe S., Fukui K., Takumi T. (2006) Curr. Biol. 16, 2345–2351 [DOI] [PubMed] [Google Scholar]
- 11. Krauss J., López de Quinto S., Nüsslein-Volhard C., Ephrussi A. (2009) Curr. Biol. 19, 1058–1063 [DOI] [PubMed] [Google Scholar]
- 12. Chang W., Zaarour R. F., Reck-Peterson S., Rinn J., Singer R. H., Snyder M., Novick P., Mooseker M. S. (2008) RNA 14, 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parker R., Sheth U. (2007) Mol. Cell 25, 635–646 [DOI] [PubMed] [Google Scholar]
- 14. Anderson P. (2005) Dev. Cell 9, 311–312 [DOI] [PubMed] [Google Scholar]
- 15. Lykke-Andersen J. (2002) Mol. Cell. Biol. 22, 8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 17. Lindsay A. J., McCaffrey M. W. (2009) Cell Motil. Cytoskeleton 66, 1057–1072 [DOI] [PubMed] [Google Scholar]
- 18. Balagopal V., Parker R. (2009) Curr. Opin. Cell Biol. 21, 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pauley K. M., Eystathioy T., Jakymiw A., Hamel J. C., Fritzler M. J., Chan E. K. (2006) EMBO Rep. 7, 904–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. (2007) Mol. Cell. Biol. 27, 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lian S., Fritzler M. J., Katz J., Hamazaki T., Terada N., Satoh M., Chan E. K. (2007) Mol. Biol. Cell 18, 3375–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jagannath A., Wood M. J. (2009) Mol. Biol. Cell 20, 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sonenberg N. (2008) Biochem. Cell Biol. 86, 178–183 [DOI] [PubMed] [Google Scholar]
- 24. Andrei M. A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Lührmann R. (2005) RNA 11, 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seperack P. K., Mercer J. A., Strobel M. C., Copeland N. G., Jenkins N. A. (1995) EMBO J. 14, 2326–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gonsalvez G. B., Little J. L., Long R. M. (2004) J. Biol. Chem. 279, 46286–46294 [DOI] [PubMed] [Google Scholar]
- 27. Aizer A., Brody Y., Ler L. W., Sonenberg N., Singer R. H., Shav-Tal Y. (2008) Mol. Biol. Cell 19, 4154–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu X. S., Tsan G. L., Hammer J. A., 3rd (2005) J. Cell Biol. 171, 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ali M. Y., Lu H., Bookwalter C. S., Warshaw D. M., Trybus K. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4691–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hodges A. R., Bookwalter C. S., Krementsova E. B., Trybus K. M. (2009) Curr. Biol. 19, 2121–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang J. D., Brady S. T., Richards B. W., Stenolen D., Resau J. H., Copeland N. G., Jenkins N. A. (1999) Nature 397, 267–270 [DOI] [PubMed] [Google Scholar]
- 32. Hales C. M., Vaerman J. P., Goldenring J. R. (2002) J. Biol. Chem. 277, 50415–50421 [DOI] [PubMed] [Google Scholar]
- 33. Anderson P., Kedersha N. (2009) Curr. Biol. 19, R397–398 [DOI] [PubMed] [Google Scholar]
- 34. Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. (2005) J. Cell Biol. 169, 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mazroui R., Huot M. E., Tremblay S., Filion C., Labelle Y., Khandjian E. W. (2002) Hum. Mol. Genet. 11, 3007–3017 [DOI] [PubMed] [Google Scholar]
- 36. Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E., Anderson P. (2000) J. Cell Biol. 151, 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M., Anderson P. (2004) Mol. Biol. Cell 15, 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loschi M., Leishman C. C., Berardone N., Boccaccio G. L. (2009) J. Cell Sci. 122, 3973–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sweet T. J., Boyer B., Hu W., Baker K. E., Coller J. (2007) RNA 13, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ali M. Y., Krementsova E. B., Kennedy G. G., Mahaffy R., Pollard T. D., Trybus K. M., Warshaw D. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4332–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cao T. T., Chang W., Masters S. E., Mooseker M. S. (2004) Mol. Biol. Cell 15, 151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brengues M., Teixeira D., Parker R. (2005) Science 310, 486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cougot N., Babajko S., Séraphin B. (2004) J. Cell Biol. 165, 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sheth U., Parker R. (2003) Science 300, 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R. (2005) RNA 11, 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kwon S., Zhang Y., Matthias P. (2007) Genes Dev. 21, 3381–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Decker C. J., Teixeira D., Parker R. (2007) J. Cell Biol. 179, 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chu C. Y., Rana T. M. (2006) PLoS Biol 4, e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bobola N., Jansen R. P., Shin T. H., Nasmyth K. (1996) Cell 84, 699–709 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.