Abstract
The molecular basis for retention of integral membrane proteins in the endoplasmic reticulum (ER) is not well understood. We recently discovered a novel ER molecular chaperone termed Cosmc, which is essential for folding and normal activity of the Golgi enzyme T-synthase. Cosmc, a type II single-pass transmembrane protein, lacks any known ER retrieval/retention motifs. To explore specific ER localization determinants in Cosmc we generated a series of Cosmc mutants along with chimeras of Cosmc with a non-ER resident type II protein, the human transferrin receptor. Here we show that the 18 amino acid transmembrane domain (TMD) of Cosmc is essential for ER localization and confers ER retention to select chimeras. Moreover, mutations of a single Cys residue within the TMD of Cosmc prevent formation of disulfide-bonded dimers of Cosmc and eliminate ER retention. These studies reveal that Cosmc has a unique ER-retention motif within its TMD and provide new insights into the molecular mechanisms by which TMDs of resident ER proteins contribute to ER localization.
Keywords: Endoplasmic Reticulum (ER), Glycosylation, Golgi, Plasma Membrane, Protein Chimeras, Protein Folding, Protein Sorting, Cosmc, Localization, T-synthase
Introduction
After translocation into the endoplasmic reticulum (ER),2 both soluble and membrane proteins move by default toward the plasma membrane unless specific primary or structural motifs are present. Such motifs include targeting, retention, and/or retrieval signals that ultimately determine the localization of proteins in specific organelles (1). However, the molecular basis of retrieval/retention of specific integral membrane protein in different organelles along the secretory pathway of eukaryotic cells, and especially in the ER, is not well understood. One of the best studied mechanisms of ER localization involves retrieval motifs, and the best understood of those is the tetrapeptide H/KDEL lumenal sequence at the C terminus of ER lumenal proteins (2, 3). Recognition of H/KDEL by the ERD2-like receptor in post-ER compartments leads to formation of COPI-coated vesicles and eventual transport of proteins with the H/KDEL sequence back to the ER (4, 5). Some ER resident proteins carry a di-lysine motif (K(X)KXX) in the C-terminal cytosolic tails of type I membrane proteins (6), that promotes their interaction with COPI vesicle machinery and redistribution to the ER (7–9). Similarly, di-arginine motifs, consecutive Arg residues within the N terminus of a type II membrane protein, as seen for MHC class II-associated invariant chain (10, 11), can also promote ER localization by COPI mechanisms. Retrieval motifs are sometimes identified through Golgi-related glycosylation changes in the glycoprotein (9). Both cytosolic di-arginine and lumenal determinants may be important for ER localization of some proteins such as Arabidopsis glucosidase I (12). Additional motifs include the di-phenylalanine (FF) motif (13, 14), as seen in type I proteins like the p24 family, which are also associated with COPI coat protein binding.
Other less well understood mechanisms of ER retrieval/retention have also been observed. For example, the transmembrane domain (TMD) of resident ER proteins may also be important (15–17), as seen for the type II membrane protein Sec12p, where the cytosolic tail is required for retention and the TMD is required for recycling (18). Finally, protein oligomerization within large complexes within the ER may also contribute to ER localization (19–23).
We recently discovered a novel ER-localized molecular chaperone termed Cosmc, which assist in the folding and prevention of oligomerization of the key enzyme involved in mucin-type O-glycosylation, the core 1 β1,3-galactosyltransferase (T-synthase) (24–28). The T-synthase is a Golgi enzyme that adds a galactose residue from the donor UDP-Gal to glycoproteins entering the Golgi that have the GalNAcα1-Ser/Thr (Tn antigen) on mucin-type sequences to generate the core 1 disaccharide Galβ1–3GalNAcα1-Ser/Thr (T antigen) (29). Cosmc is a type II transmembrane protein (∼36 kDa) with a short cytoplasmic N-terminal domain, and a large ER lumenal domain that is not efficiently N-glycosylated and is the functional chaperone domain (24, 25, 28).
The mechanism of retention of Cosmc in the ER is not known, because Cosmc lacks any canonical retrieval/retention motifs. We reported earlier that transferrin N-terminal signal sequence fused with the Cosmc lumenal domain (soluble Cosmc) leads to its secretion into culture media, indicating that retrieval/retention signals may be associated with the transmembrane and/or cytosolic domains (28). Consistent with this was the finding that soluble Cosmc engineered to contain a KDEL tag at its C terminus is retained in the ER and functions as efficiently as wild-type (WT) Cosmc (28).
To better understand the retrieval/retention signals in Cosmc for ER localization and explore whether Cosmc has novel signals, we generated mutants of Cosmc, as well as chimeric constructs of Cosmc with a non-ER glycoprotein, the human transferrin receptor (TfR). The TfR is also a single pass type II membrane protein (30–32). The TfR is localized to the plasma membrane and endosomal vesicles in all animal cells (33). Our studies show that the TMD of Cosmc is essential for ER localization and that residues within the TMD are critical for disulfide-bonded dimerization of Cosmc.
EXPERIMENTAL PROCEDURES
Reagents
African Green Monkey SV40-transfected kidney fibroblast cell line COS7 was obtained from American Type Culture Collection. Restriction enzymes were obtained from New England Biolabs, Inc. (Ipswich, MA). Fugene6 and TaqDNA polymerase were obtained from Roche Diagnostics (Mannheim, Germany). TNM-FH and EX-Cell 405 media were purchased from BD Biosciences (San Jose, CA). Rabbit anti-human giantin mAb (IgG1) was purchased from Abcam, Inc. (Cambridge, MA). Rabbit anti-human calnexin antiserum and mouse anti-KDEL (GRP78 and GRP94) mAb (10C3) were purchased from Assay Designs (Ann Arbor, MI). Alexa Fluor-labeled secondary antibodies, Vector pcDNA3.1(+), PCR TOPO4 cloning kit, and SDS-PAGE gels were purchased from Invitrogen (Carlsbad, CA). Proteasome inhibitor MG-132 was purchased from EMD Chemicals (Gibbstown, NJ). QIAquick Gel Extraction kits were obtained from Qiagen (Valencia, CA). Chemiluminescent Substrate and BCA protein assay kit were purchased from Thermo Fisher Scientific (Pittsburgh, PA).
Preparation of Expression Constructs
A construct encoding C-terminal HPC4-tagged Cosmc (Cosmc-HPC4) was made by introducing the HPC4 epitope (KGDILRPDVQDE) into wild-type Cosmc at its C terminus by PCR. The product was cloned into PCR3.1. The insert was cut with BamHI (partially)-XbaI and cloned into pcDNA3.1(+). A construct encoding C-terminal HPC4-tagged TfR (TfR-HPC4) was made using a similar strategy to Cosmc-HPC4. The HPC4 epitope tag was introduced into the C terminus of TfR by PCR. The product for TfR-HPC4 and the products from the asymmetric PCR as prepared below were cloned into pCR4-TOPO. The insert was cut with BamHI-XbaI and cloned into pcDNA3.1(+). The PCR primers are listed in supplemental Table S1.
The six chimeric constructs were prepared using the strategy outlined in supplemental Fig. S1A. Plasmids (A) (Cosmc) and (B) (TfR) are color-coded and shown as two paired strands. Synthetic oligonucleotide primers are also color-coded and shown as single strands, with half arrowheads indicating the direction (34). The intermediate PCR-amplified products (C) and (D) derived from plasmid (A) and (B), are also shown as two paired strands and color-coded corresponding to the plasmid template and primers, respectively (34). In the first PCR, Plasmid (A) was amplified by high-fidelity PCR employing Pfu polymerase and synthetic oligonucleotide primers a and b (34). Plasmid (B) was amplified by high-fidelity PCR employing Pfu polymerase and synthetic oligonucleotide primers c and d. The conditions for the high-fidelity PCR amplification were: 1 cycle at 98 °C for 30 s, 35 cycles of PCR at 98 °C for 10 s, 62 °C for 30 s, and 72 °C for 3–4 min (30 s per kilobase DNA fragment to be amplified) in a volume of 50 μl with 5 ng of plasmid DNA template, 1 unit of Pfu polymerase, 10 pmol of each primer, 5 μl of mixed dNTP at 2.5 mm concentration, and 10 μl of 5× Pfu buffer (containing 1.0 mm Mg2+), and 1 cycle at 72 °C for 5 min. Then the products from each individual PCR were analyzed on agarose gels and purified, mixed in an asymmetric ratio for a second PCR; the (C) strand at its 3′-end and the (D) strand at its 5′-ends have overlapping regions to pair with each other. Extension of this overlap by DNA polymerase created the full-length chimeric molecule (E) (34). The new strand (E) then acted as a template to make the final PCR product (F) by high-fidelity PCR employing Pfu polymerase and synthetic oligonucleotide primers a and d (34). The optimal conditions for the second PCR were: 1 μl of each product from the first PCR by the high-fidelity PCR amplification mixed with 10 pmol of synthetic oligonucleotide primers. The mixture was subjected to a PCR in a volume of 50 μl containing 1 unit of Pfu polymerase, 5 μl of mixed dNTP at 2.5 mm concentration, and 10 μl 5× Pfu buffer (containing 1.0 mm Mg2+) (34). PCR cycling parameters were the same as in the first PCR.
Site-directed Mutagenesis
Wild-type Cosmc was used as template to make mutations either from cysteine to alanine or from cysteine to serine using QuickChangeTM site-directed mutagenesis kit (Stratagene) following the manufacturer's protocol. For Cys-to-Ala site-directed mutagenesis, we used forward primer: 5′-GGAAGCATTTTCGCTGCTTTGATC-3′ and reverse primer: 5′-GATCAAAGCAGCGAAAATGCTTCC-3′. For Cys-to-Ser site directed mutagenesis, we used forward primer: 5′-GGAAGCATTTTCAGTGCTTTGATC-3′ and reverse primer: 5′-GATCAAAGCACTGAAAATGCTTCC-3′. The recombinant plasmids containing the desired mutations were confirmed by DNA sequencing.
Cell Culture and Transfection
Monolayer COS7 cells were cultured at 37 °C in 5% CO2 in DMEM supplemented with 10% FBS, penicillin, and streptomycin at 100 μg/ml. One day prior to transfection, the cells were seeded into a 10-cm dish and cultured in complete media overnight to reach 50–60% confluency. Cells were transfected with Fugene6 transfection reagent according to manufacturer's instructions.
Immunofluorescent Staining of COS7 Cells
COS7 cells were cultured on a chambered slide and transiently transfected with the expression constructs using Fugene6 transfection reagent according to the manufacturer's protocol. At 48 h post transfection, cells were washed with TBS and fixed with 4% paraformaldehyde on ice for 1h and permeabilized with 0.1% Triton X-100 for 45 min on ice. After blocking with 1% BSA in TBS for 1 h at room temperature, the cells were incubated with primary antibodies for 1 h at room temperature. The cells were washed with TBS three times and incubated with Alexa Fluor-labeled secondary antibodies at room temperature for 1 h. Cells were then washed four times with TBS and mounted with Prolong Antifade Media (Invitrogen). After drying at room temperature for 12–16 h, cells were visualized by confocal microscopy (TCS NT; Leica) at room temperature under 40× Plan Fluotar 1.0 NA oil immersion or 100× Plan APO 1.4 NA oil immersion objective lenses. The images were maximum projection collected with a pinhole of 1 using 0.5-μm step size. Images were analyzed using the TCS and Volocity software (Leica).
Subcellular Fractionation
COS7 cells grown to 80% confluence in 10 cm2 dishes and transiently transfected with the expression constructs for 48 h were harvested and washed with cold PBS. Cells were homogenized in 25 mm HEPES, pH7.5 containing 250 mm sucrose. Then post-nuclear supernatants (PNS) were made by centrifugation at 20,000 × g for 30min. The concentration of sucrose in PNS was adjusted to 40% (w/v) and loaded on 60, 50, 30, and 20% sucrose gradient. After centrifugation at 100,000 × g for 20 h, 18 fractions were collected from the bottom of the tube (from top to bottom: fraction No. 1 to 18). The fractions were analyzed by Western blotting with the indicated antibodies.
Preparation of Cell Extracts
Transfected cells were resuspended in an appropriate volume of TBS buffer including 1 mm CaCl2 and proteinase inhibitor mixture (Roche Applied Science, Indianapolis, IN) and homogenized by sonication on an ice-bath for 5 s 4 times. In some extractions, as noted, we included 250 mm iodoacetamide to modify free Cys residues and limit artifactual dimerization. The PNS were obtained by centrifugation of homogenate at 700 × g for 10min, and the extracts were obtained by adding 0.5% Triton X-100 (final concentration) to the supernatant and solubilizing on ice for 30 min.
Assay of Glycosyltransferases
T-synthase and β4-Gal-T were assayed as previously described (28, 69).
Western Blot
Western blot with HPC4-tagged proteins as well as ER marker calnexin and Golgi protein giantin were performed as previously described (28).
Proteasome Inhibition
About 1 × 106 COS7 cells were seeded in T75 flasks and transiently transfected with the expression constructs using Fugene 6 transfection reagent according to the manufacturer's protocol. At 48 h after transfection, the cells were split into two plates, one was treated with 10 μm MG-132 (dissolved in 100% DMSO at 2 mm stock) the other with 0.5% DMSO in complete media for 12–14 h,. The cells were harvested for Western blot analysis.
Immunoprecipitation of Cosmc
Cell extracts or the subcellular fractions were incubated with monoclonal antibody HPC4-conjugated AffiGel10 (HPC4-Beads) overnight while rotating at 4 °C. The beads were collected using bench centrifugation and washed five times with 400 μl of TBS containing 1 mm CaCl2. The bead-bound material was eluted five times with 20 μl of elution buffer (50 mm TBS pH8.0, 10 mm EDTA). The fractions were pooled for endoglycosidase treatment and Western blot analysis.
RESULTS
Generation of Cosmc-TfR Chimeras
To examine the role(s) of the cytoplasmic domain (CD), the transmembrane domain (TMD), and the lumenal domain (LD) of Cosmc in the secretory pathway, a series of chimeric cDNAs were constructed by recombinant DNA techniques. Cosmc was fused to the equivalent region from a cell surface membrane protein, transferrin receptor (TfR), which, like Cosmc, it is a type II transmembrane protein, with a relatively short CD, a single TMD, and a large extracellular domain. Six constructs for expression of chimeras were generated (supplemental Fig. S1B). Chimera 1 contains the TfR-CD and Cosmc-TMD and -LD (#1, TfR/Cosmc/Cosmc). Chimera 2 contains the Cosmc-CD and TfR-TMD and -LD (#2, Cosmc/TfR/TfR). Chimera 3 contains the Cosmc-LD and the TfR-CD and -TMD (#3, TfR/TfR/Cosmc). Chimera 4 contains the TfR-LD and the Cosmc-CD and -TMD (#4, Cosmc/Cosmc/TfR). Chimera 5 contains the Cosmc-TMD and the TfR-CD and -LD (#5, TfR/Cosmc/TfR). Chimera 6 contains the TfR-TMD and the Cosmc-CD and -LD (6, Cosmc/TfR/Cosmc). Each construct has a C-terminal HPC4 epitope tag (35, 36).
wtCosmc Localizes to the ER
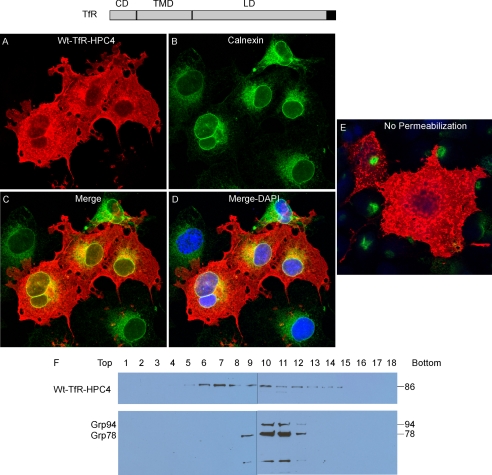
COS7 cells, transiently transfected with full-length (FL), HPC4-epitope tagged wild-type Cosmc (wtCosmc), were stained with anti-HPC4 mAb by immunofluorescence. Cosmc (red) was expressed in a perinuclear pattern in the transfected cells, with a very similar pattern to the ER marker Calnexin (green), which stained all cells (Fig. 1, A–C). A merge of the stained images shows a yellow/orange color in the cells expressing Cosmc, indicating co-localization of Cosmc and Calnexin. These results are consistent with prior studies showing ER localization of Cosmc (28, 37). Subcellular fractionation by sucrose gradient centrifugation showed that wtCosmc was recovered primarily in fractions 9–11, corresponding to fractions 9–12 containing the ER marker GRP78/94 (Fig. 1D). By contrast, T-synthase activity, used as a Golgi marker, was mainly recovered in fractions 6–8 (Fig. 1E), which also correspond to the fractions containing the activity of the Golgi protein β4-Gal-T (data not shown). Overall, the immunofluorescence staining and subcellular fractionation results are in agreement and indicate that wtCosmc mainly localizes in the ER of COS7 cells.
FIGURE 1.
Localization of wtCosmc. A–C, immunofluorescent staining of wtCosmc. Cells were stained with mouse anti-HPC4 (red) antibody and rabbit anti-Calnexin (green) antibody. Merge, yellow. DAPI, blue. D, sucrose gradient subcellular fractionation. The postnuclear supernatant (PNS) was applied to a sucrose gradient and 18 fractions (top to bottom) were obtained after ultracentrifugation. Proteins from each fraction were analyzed on Western blot with anti-HPC4 and anti-KDEL antibodies and (E) measured for T-synthase activity.
wtTfR Localizes in the Plasma Membrane
COS7 cells expressing the wild-type TfR (wtTfR) were immunofluorescently stained with anti-HPC4 and anti-Calnexin antibodies in the presence and absence of detergent Triton X-100 for membrane permeabilization. The wtTfR (red) was observed as a bright plasma membrane staining pattern under both permeable and non-permeable conditions (Fig. 2, A–E). By contrast, Calnexin (green) was apparent only when cells were permeabilized by Triton X-100, consistent with its intracellular localization. Minor staining in the ER was also observed for wtTfR, which could be due to the high expression of this protein in COS7 cells or overall fluorescence in many membranes including endosomes. Subcellular fractionation by sucrose gradient centrifugation showed that the wtTfR-HPC4 was recovered in fractions 5–15 with major bands in fractions 6∼10, while GRP78/94 as markers of the ER were mainly found in fractions 8∼12, as shown by Western blot (Fig. 2F). Because the wtTfR is synthesized in the ER and sorted to the plasma membrane through the secretory pathway, it is likely that some protein will be found in all membranes in this pathway. Overall, the immunofluorescence staining and subcellular fractionation results are in agreement and indicate that wtTfR is present on plasma membranes of COS7 cells. Thus, the localization of recombinant forms of wtCosmc and wtTfR in COS7 cells are clearly different and validate the potential of using chimeric constructs of the two proteins to explore localization determinants.
FIGURE 2.
Localization of wtTfR. A–D, immunofluorescent staining of wtTfR. Cells were stained with anti-HPC4 (red) antibody and anti-Calnexin (green). Merge, yellow. DAPI, blue. E, transfected cells not treated with Triton X-100 were stained with anti-HPC4 (red) antibody and anti-calnexin (green). F, sucrose gradient subcellular fractionation. The PNS was applied to a sucrose gradient and 18 fractions (top to bottom) were obtained after ultracentrifugation. Proteins from each fraction were analyzed on Western blot with anti-HPC4 and anti-KDEL antibodies.
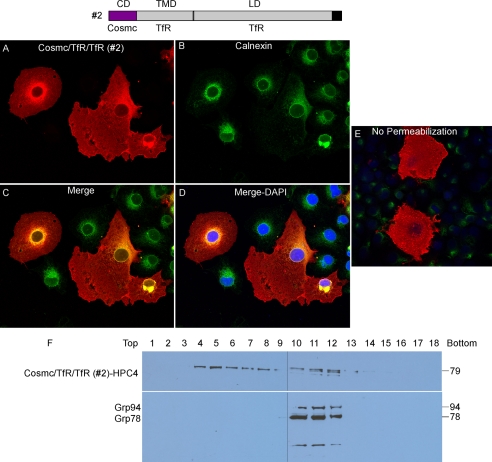
Cosmc/TfR/TfR (Construct #2) Localizes in the Plasma Membrane
COS7 cells, transiently transfected with the construct expressing chimera Cosmc/TfR/TfR (Construct #2), were examined by immunofluorescence with anti-HPC4 and anti-Calnexin antibody (Fig. 3, A–D). Cosmc/TfR/TfR (red) was displayed on the cell surface along the clear edges of the plasma membrane, similar to what was observed for wtTfR staining (Fig. 2, A–E), which was also observed as perinuclear and punctate pattern in some cells. These results indicate that Cosmc/TfR/TfR was mainly localized on the plasma membrane, and partially retained in the ER and Golgi apparatus and behaved similarly to wtTfR. The merged image with Calnexin (green) confirmed the partial co-localization of Cosmc/TfR/TfR with the ER marker, and the partial ER localization of this fusion protein (Fig. 3, A–D). To further explore its plasma membrane localization, the transfected COS7 cells were also stained with anti-HPC4 under non-permeable conditions. The Cosmc/TfR/TfR was similarly stained on the whole cell with a pattern similar to that observed in the permeabilized cells, except for the perinuclear staining (Fig. 3E). Another line of evidence showing that Cosmc/TfR/TfR was mainly plasma membrane-localized was obtained from subcellular fractionation/Western blotting. Cosmc/TfR/TfR-HPC4 from transfected COS7 cells was recovered in fractions 4–13, similar to the wtTfR, whereas GRP78/94 were mainly in fractions 10–12 (Fig. 3F). The plasma membrane and Golgi fractions are represented in fractions 5–7 and 6–9, respectively. These data show that Cosmc/TfR/TfR was present on the plasma membrane, and indicate that the CD of Cosmc is not sufficient to retain the TfR in the ER.
FIGURE 3.
Localization of Cosmc/TfR/TfR (construct #2). A–D, immunofluorescent staining of Cosmc/TfR/TfR. Cells were stained with anti-HPC4 (red) antibody and anti-calnexin (green). Merge, yellow. DAPI, blue. E, transfected cells not treated with Triton X-100 were stained with anti-HPC4 (red) antibody and anti-calnexin (green). F, sucrose gradient subcellular fractionation. The PNS was applied to a sucrose gradient and 18 fractions (top to bottom) were obtained after ultracentrifugation. Proteins from each fraction were analyzed on Western blot with anti-HPC4 and anti-KDEL antibodies.
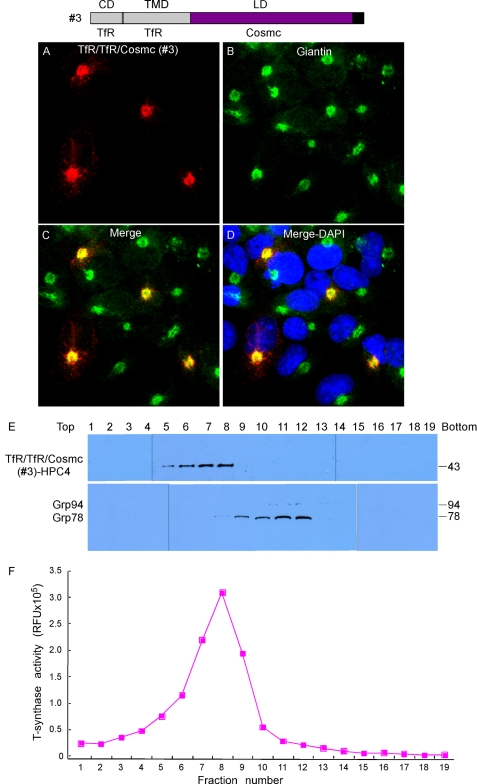
TfR/TfR/Cosmc (Construct #3) Localizes in the Golgi
To test whether the lumenal domain (LD) of Cosmc is important in ER localization, we generated a construct expressing the chimeric protein TfR/TfR/Cosmc (Construct #3). COS7 cells were transiently transfected with the construct, and the cellular localization was examined by immunofluorescence with anti-HPC4 antibody (Fig. 4, A–D). The Golgi was visualized by antibody to a Golgi-resident protein Giantin. The TfR/TfR/Cosmc (red) was expressed in a punctate pattern in many cells that was coincident with the localization of Giantin (green), which was visualized in every cell. A merge of the stained cell images showed a yellow color in every TfR/TfR/Cosmc stained cell, indicating the co-localization of TfR/TfR/Cosmc with Giantin. Subcellular fractionation by sucrose gradient centrifugation showed that TfR/TfR/Cosmc was present in fractions 5–8 as detected by anti-HPC on Western blot (Fig. 4E), which is different from fractions 9–12 containing the ER marker Grp78/94. By contrast, T-synthase activity was mainly recovered in fractions 7–9, corresponding to the Golgi fractions (Fig. 4F). This result is consistent with the immunofluorescent staining data and indicates that TfR/TfR/Cosmc localizes to the Golgi. These results indicate that the LD of Cosmc is not sufficient to retain the TfR in the ER. Interestingly, the LD of Cosmc has the ability to retain Construct #3 in the Golgi apparatus and efficiently prevent its movement to the plasma membrane. This interesting mechanism will be studied in the future using Golgi-associated chimeric constructs.
FIGURE 4.
Localization of TfR/TfR/Cosmc (construct #3). A–D, immunofluorescent staining of TfR/TfR/Cosmc. Cells were stained with anti-HPC4 (red) antibody and anti-Giantin (green) antibodies. Merge, yellow. DAPI, blue. E, sucrose gradient subcellular fractionation. The PNS was applied to a sucrose gradient and 19 fractions (top to bottom) were obtained after ultracentrifugation. Proteins from each fraction were analyzed on Western blot with anti-HPC4 and anti-KDEL antibodies and (F) measured for T-synthase activity.
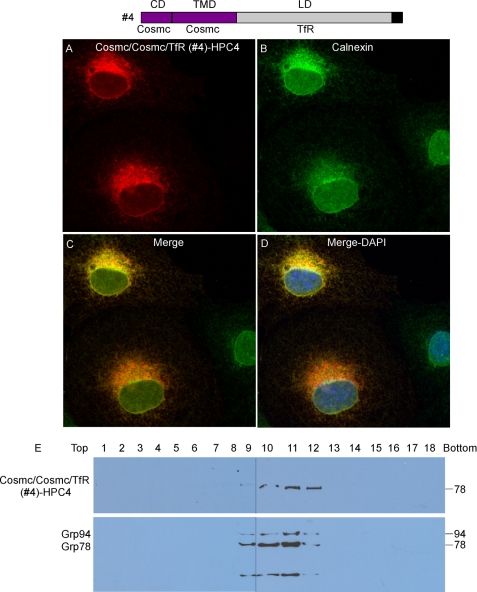
Cosmc/Cosmc/TfR (Construct #4) Localizes in the ER
COS7 cells, transiently transfected with the construct expressing Cosmc/Cosmc/TfR (Construct #4), were examined by immunofluorescence with anti-HPC4 and anti-Calnexin antibodies (Fig. 5, A–D). Cosmc/Cosmc/TfR (red) was observed in a perinuclear pattern in some cells, similar to staining with Calnexin (green), which stained every cell. The merge of the stained cell images shows a yellow color in the Cosmc/Cosmc/TfR-stained cells, indicating their co-localization. Subcellular fractionation by sucrose gradient centrifugation showed that Cosmc/Cosmc/TfR-HPC4 was recovered primarily in fractions 9–12 as detected by anti-HPC4 on Western blot, corresponding to fractions 9–12 containing the ER markers GRP78 and GRP94 (Fig. 5E). This result is consistent with the immunofluorescent staining data, and indicates that Chimera #4 exhibits an ER-localization. These results show that the CD and TMD of Cosmc are sufficient to retain the TfR lumenal domain in the ER.
FIGURE 5.
Localization of Cosmc/Cosmc/TfR (construct #4). A–D, immunofluorescent staining of Cosmc/Cosmc/TfR fusion proteins. Cells were stained with anti-HPC4 (red) antibody and anti-calnexin (green). Merge, yellow. DAPI, blue. E, sucrose gradient subcellular fractionation. The PNS was applied to a sucrose gradient and 18 fractions (top to bottom) were obtained after ultracentrifugation. Proteins from each fraction were analyzed on Western blot with anti-HPC4 and anti-KDEL antibodies.
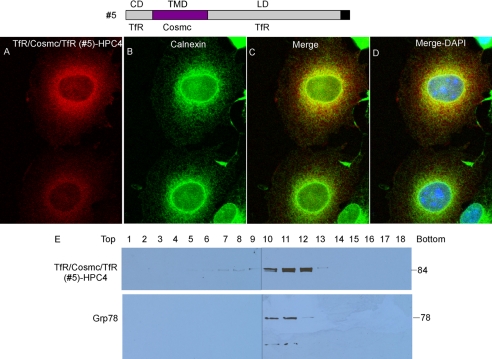
TfR/Cosmc/TfR (Construct #5) Localizes in the ER
To test whether the TMD of Cosmc is the ER-localization determinant for Cosmc, we generated a construct expressing the chimera protein of TfR/Cosmc/TfR (Construct #5). COS7 cells were transiently transfected with the construct, and cellular localization was examined by immunofluorescence with anti-HPC4 and anti-Calnexin antibodies (Fig. 6, A–D). TfR/Cosmc/TfR (red) was observed in a perinuclear pattern in some cells, similar to staining with Calnexin (green), which stained every cell. Merge of the stained cell images shows a yellow color in the TfR/Cosmc/TfR-stained cells, indicating the co-localization of those two proteins. Subcellular fractionation by sucrose gradient centrifugation showed that TfR/Cosmc/TfR-HPC4 was recovered primarily in fractions 10–12 as detected by anti-HPC4 on Western blot, corresponding to fractions 10–12 containing the ER markers GRP78 and GRP94 (Fig. 6E). This result is consistent with the immunofluorescent staining data and further confirms the ER localization of Chimera #5. Thus, these results show that the TMD alone of Cosmc is sufficient to retain the TfR cytoplasmic and lumenal domains in the ER, suggesting that the TMD of Cosmc is the primary ER localization determinant.
FIGURE 6.
Localization of TfR/Cosmc/TfR (construct #5). A–D, immunofluorescent staining of TfR/Cosmc/TfR. Cells were stained with anti-HPC4 (red) antibody and anti-calnexin (green). Merge, yellow. E, sucrose gradient subcellular fractionation. The PNS was applied to a sucrose gradient and 18 fractions (top to bottom) were obtained after ultracentrifugation. Proteins from each fraction were analyzed on Western blot with anti-HPC4 and anti-KDEL antibodies.
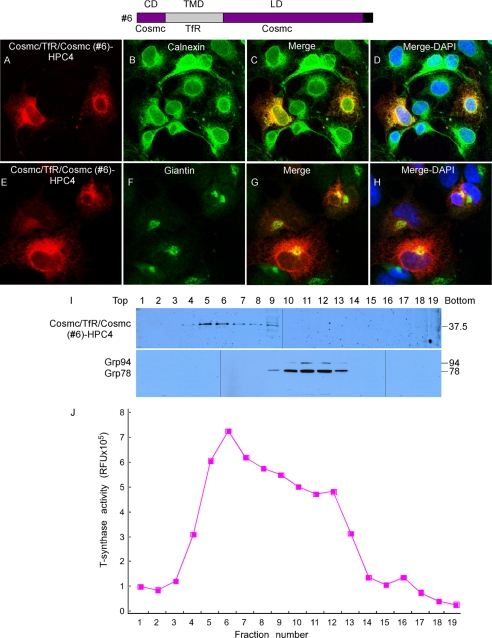
Cosmc/TfR/Cosmc (Construct #6) Mainly Localizes in the Golgi, but Partially in the ER
To further explore the role of the TMD in Cosmc for ER localization, we generated a chimera of Cosmc in which its TMD was replaced by that of the TfR. COS7 cells were transiently transfected with the construct expressing Cosmc/TfR/Cosmc (Construct #6), and cellular localization was examined by immunofluorescence with anti-HPC4, anti-Calnexin, and anti-Giantin antibodies (Fig. 7, A–H). Cosmc/TfR/Cosmc (red) was observed in both a perinuclear localization similar to staining with the ER marker Calnexin (green) (Fig. 7, A–D) but it showed a punctate pattern that was also coincident with the localization of the Golgi marker Giantin (green) (Fig. 7, E–H). The merge of the stained cell images shows a yellow color in the Cosmc/TfR/Cosmc-stained cells, indicating the co-localization of those two proteins. Subcellular fractionation by sucrose gradient centrifugation showed that Cosmc/TfR/Cosmc was observed in fractions 4–9 with major bands in fractions 5–7 as detected by anti-HPC4 antibody on Western blot; this is different from fractions 10–13 containing the ER markers GRP78 and GRP94 (Fig. 7I). T-synthase activity was mainly recovered in fractions 5–8, corresponding to the Golgi fractions (Fig. 7J). The immunofluorescence imaging indicates that Cosmc/TfR/Cosmc may localize in both ER and Golgi, whereas the subcellular fractionation studies suggest that it more localized with the peak activity of the Golgi marker T-synthase. In any case, in the absence of the TMD of Cosmc, the other domains of Cosmc are not sufficient to cause ER retention and prevent movement to the Golgi apparatus.
FIGURE 7.
Localization of Cosmc/TfR/Cosmc (construct #6). A–H, immunofluorescent staining of Cosmc/TfR/Cosmc. Cells were stained with anti-HPC4 (red) antibody and anti-calnexin (green) (A–D), or anti-giantin (green) (E–H) antibodies. Merge, yellow. DAPI, blue. I, sucrose gradient subcellular fractionation. The PNS was applied to a sucrose gradient and 19 fractions (top to bottom) were obtained after ultracentrifugation. Proteins from each fraction were analyzed on Western blot with anti-HPC4 and anti-KDEL antibodies and (J) measured for T-synthase activity.
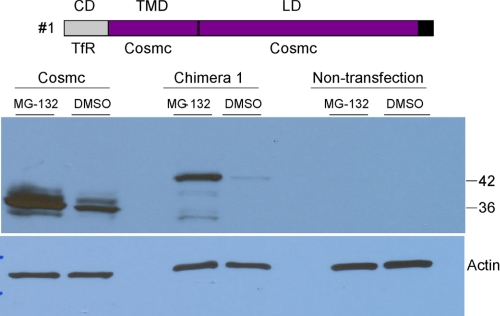
TfR/Cosmc/Cosmc (Construct #1) Is Degraded by Proteasomal Pathway
We also generated the interesting construct TfR/Cosmc/Cosmc (Construct #1). COS7 cells were transiently transfected with wtCosmc and with TfR/Cosmc/Cosmc (Construct #1), however, unlike expression of other constructs, there was very little expression of TfR/Cosmc/Cosmc in any cell. This result suggested that the recombinant protein might be degraded. To test this prediction, after 48 h transfection, both transfected and non-transfected COS7 cells were equally split into two plates. One was treated with proteasome inhibitor MG-132, while the other was treated with DMSO overnight as a negative control. After collecting the cells, we examined expression of Construct #1 using the anti-HPC4 antibody in Western blotting. There were no significant bands present in non-transfected cells. Interestingly, there was very little expression of Construct #1 in the absence of MG-132, but expression was significantly enhanced in the presence of the inhibitor (Fig. 8). In addition, while wtCosmc was expressed in the absence of MG-132, there was somewhat enhanced expression in the presence of the inhibitor, indicating that some of the wtCosmc is also being degraded in cells by a proteasomal pathway. These data demonstrate that TfR/Cosmc/Cosmc was degraded through the proteasomal pathway and thus, it is not possible to examine its localization by confocal imaging.
FIGURE 8.
Degradation of TfR/Cosmc/Cosmc (construct #1) through the proteasome pathway. COS-7 cells were treated with 10 μm MG-132 or DMSO and cell extracts were analyzed on SDS-PAGE by Western blot with mouse anti-HPC4 antibody.
Sensitivity of Chimeras to Endoglycosidase Treatment
TfR has both N- and O-glycans (38–41). The three N-glycosylation sites of the TfR occur in the C-terminal lumenal domain, and during maturation from the ER through the Golgi apparatus, some N-glycans remain high mannose-type and others are converted to complex-type chains (42, 43). These complex-type N-glycans are sensitive to Peptide:N-glycosidase F (PNGase F), but resistant to endoglycosidase H (Endo H), which is specific for high mannose/hybrid-type N-glycans. We considered that if the chimeric proteins containing the C-terminal domain of TfR remain in the ER they might contain only high mannose- or hybrid-type N-glycans, whereas if they move out of the ER into Golgi compartment, and then are retrieved to the ER, they may be more resistant to Endo H. Transiently transfected COS7 cells were harvested and cell extracts were prepared. Anti-HPC4 antibody was used for immunoprecipitation to pull down chimeric proteins, as well as wtTfR, followed by enzyme treatments and Western blots to probe for changes in glycosylation. TfR and the chimera Cosmc/TfR/TfR (construct #2) were sensitive to PNGase F, but partially resistant to Endo H (Fig. 9). However, chimeras Cosmc/Cosmc/TfR (construct #4) and TfR/Cosmc/TfR (construct #5) were sensitive to both PNGase F and Endo H. The results suggest that TfR and Chimera 2 traffic through the Golgi apparatus, while chimeras Cosmc/Cosmc/TfR and TfR/Cosmc/TfR lack complex-type N-glycans, and thus remain in the ER. These results are consistent with the localization data shown in Figs. 5 and 6, showing ER localization for these chimeras.
FIGURE 9.
COS7 cells were transfected to express wild-type TfR and chimeras 2, 4, and 5, all as HPC4-epitope-tagged proteins. The proteins were immunoprecipitated with anti-HPC4, and either non-treated, treated with Endo H, or treated with PNGase F. The samples were then analyzed on SDS-PAGE followed by Western blot with mouse anti-HPC4 antibody.
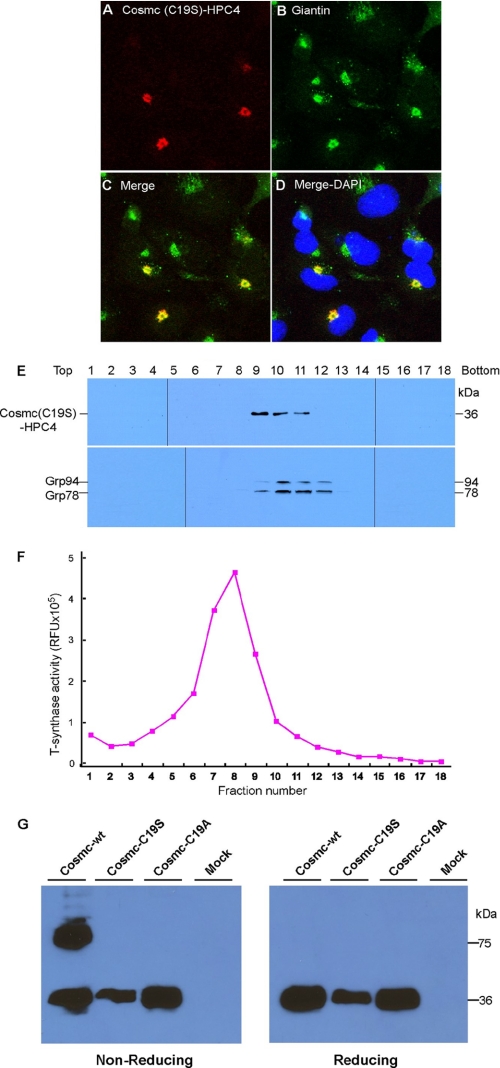
Cysteine within the TMD of Cosmc Is Required for Retention of Full-length Cosmc in the ER
While the above results show that the TMD of Cosmc is essential for its ER localization, the TMD does not contain any known ER retention motif. We noted that there is a single residue of cysteine (Cys-19) within the TMD, which led us to explore whether this residue might contribute to the ER localization function of the TMD. The 18 amino acid sequence of the TMD of human Cosmc is predicted to be -G-V-M-L-G-S-I-F-C19-A-L-I-T-M-L-G-H-I-. To explore the potential contribution of the Cys-19 residue in wtCosmc, the Cys was mutated to either Ala (C19A) or Ser (C19S) by site-directed mutagenesis. COS7 cells were transfected with these two new constructs individually and the cellular localization of the constructs was assayed. Unexpectedly, the results from immunofluorescence showed that both mutant Cosmc forms exhibited Golgi localization. Shown in Fig. 10, A–D is the localization of the C19S Cosmc mutant (red), which colocalized with the Golgi marker, Giantin (green), in a punctate pattern. The results are clearly in contrast to those in Fig. 1, A–C, where Cosmc is co-localized with the ER marker calnexin. These results demonstrate that the TMD of Cosmc is responsible for ER retention and that mutation of the single Cys residue in the TMD to Ser causes the TMD to lose its ER localization function and the mutated proteins accumulate in the Golgi apparatus.
FIGURE 10.
Localization of cysteine mutant Cosmc. A–D, immunofluorescent staining of mutant Cosmc (C19S). Cells were stained with anti-HPC4 (red) antibody and anti-giantin (green) antibodies. Merge, yellow. DAPI, blue. E, sucrose gradient subcellular fractionation. The PNS was applied to a sucrose gradient and 18 fractions (top to bottom) were obtained after ultracentrifugation. Proteins from each fraction were analyzed on Western blot with anti-HPC4 and anti-KDEL antibodies and (F) measured for T-synthase activity. G, COS-7 cells transiently expressing wild-type Cosmc (FL) or mutant Cosmc (C19S and C19A) were extracted by the given protocol and included 250 mm iodoacetamide in the extraction buffer to limit artifactual disulfide-bonded dimerization. After treatment, the cell extracts were analyzed on SDS-PAGE with or without β-ME by Western blot with anti-HPC4 antibody.
To further explore the localization of the C19S mutant, we performed subcellular fractionation on sucrose gradients of COS7 cells expressing mutant Cosmc-HPC4. The C19S mutant was present in fractions 9–11 with the major band in fraction 9 as detected by anti-HPC4 on Western blot (Fig. 10E). Similar results were observed for the C19A mutant. The ER marker Grp78/94 was found in fractions 9–12 with the major band in fraction 10 (Fig. 10E). By contrast, T-synthase activity was mainly recovered in fractions 7–9, corresponding to the Golgi fractions (Fig. 10F). These results indicate that a fraction of the C19S mutant is localized to the Golgi, but a portion of the C19S mutant may also be present in pre-Golgi/post-ER compartments.
In preliminary studies we noted that wtCosmc behaved partly as a disulfide-bonded dimeric protein on non-reducing SDS gels. Thus, we tested whether this Cys19 residue within the TMD of Cosmc might contribute to potential intermolecular disulfide formation. To this end, both FL Cosmc and the two cysteine mutants of Cosmc (C19S and C19A) expressed in COS7 cells were analyzed by reducing and non-reducing gel electrophoresis. Cell extracts were prepared in the presence of 250 mm iodoacetamide to limit artifactual disulfide-bonded dimerization. We observed both dimeric forms and monomeric forms of FL wtCosmc in non-reducing gels, and only monomeric forms in reducing gels. Interestingly, both C19S and C19A mutants of Cosmc behaved as monomeric forms in both reducing and non-reducing gels (Fig. 10G). These results demonstrate that the mutation of the single Cys residue in the TMD to either Ala or Ser affects the ability of Cosmc to form dimers.
DISCUSSION
The results presented here show that the single TMD within the type II integral membrane structure of Cosmc is sufficient to promote its localization to the ER and that substitution or mutation within the TMD leads to loss of ER retention (summarized in Table 1). The evidence here also suggests that the TMD of Cosmc is a retention rather than a retrieval signal. The ER retention mechanism for Cosmc is unusual and may be novel among the myriad of mechanisms observed to date for retrieval/retention of resident ER proteins.
TABLE 1.
Summary of the localization results with wild-type Cosmc and TfR and chimeras #1–6
| Construct (CD/TMD/LD) | Primary localization results |
|---|---|
| Cosmc (full-length) | ER |
| TfR (full-length) | Plasma membrane |
| #1-TfR/Cosmc/Cosmc | Degraded |
| #2-Cosmc/TfR/TfR | Plasma membrane |
| #3-TfR/TfR/Cosmc | Golgi |
| #4-Cosmc/Cosmc/TfR | ER |
| #5-TfR/Cosmc/TfR | ER |
| #6-Cosmc/TfR/Cosmc | Mainly Golgi |
The two major pathways of localizing proteins to the ER are retrieval from distal compartments back to ER by retrograde transport or retention through active exclusion from vesicles that exit the ER (6), and some proteins, such as the ER chaperone calreticulin (44), may utilize both retention and retrieval. ER retrieval systems include the C-terminal H/KDEL lumenal sequence (2, 3), which is recognized by the ERD2-like receptor in post-ER compartments, leading to formation of COPI-coated vesicles and eventual retrotranslocation to the ER (4, 5). Some ER-resident proteins carry other specific sequence motifs that interact with COPI complexes, such as di-lysine (K(X)KXX) (45, 46), di-arginine (RR or RXR) (10, 11), and di-phenylalanine (FF) (13, 14) that promote recruitment into COPI vesicle machinery for retrieval to the ER. Although relatively few type II resident membrane proteins, such as Cosmc, have been identified to date, some of these are known to be retained in the ER by the di-arginine motif located at the cytosolic N terminus, as seen for the MHC class II-associated invariant chain (10, 11). Using similar approaches to those we have used, it was shown that introduction of the di-arginine motif into the human TfR and N-acetylglucosaminyltransferase I (GlcNAc-TI) type II caused both to be efficiently localized in the ER (47, 48). Other retrieval systems have been noted, such as those involving the resident ER protein Rer1p, which physically recognizes the TMD of clients, such as Sec12p, and returns them to the ER via COPI vesicles (49). Other proteins, which lack canonical ER retrieval motifs, may reside in the ER by complexation with other proteins containing such motifs, as has been seen for ribophorins I and II, that may associate with the oligosaccharyltransferase complex, which contains proteins bearing di-lysine motifs (50–52).
Such retrieval pathways do not appear to be operative for Cosmc, since Cosmc lacks identifiable retrieval motifs. Furthermore, retrieval systems can often be identified by changes in glycosylation observed by proteins that may reach distal secretory compartments, where the N-glycans are trimmed and modified (9, 53). Cosmc is not efficiently N-glycosylated although it has a glycosylation sequon close to its C terminus (28), but our results show that the N-glycans of chimeras Cosmc/Cosmc/TfR (Construct #4) and TfR/Cosmc/TfR (Construct #5), which are ER localized, are Endo-H sensitive, in contrast to other chimeras that acquire Endo-H resistant N-glycans. Thus, it is reasonable to conclude that Cosmc is retained in the ER and inefficiently exits to post-ER compartments. However, in future experiments we plan to explore the association of both the T-synthase and possibly Cosmc and some of the Cosmc chimeras described here with COPI vesicles, which should provide further insight into the specific trafficking of T-synthase versus Cosmc. The T-synthase is a resident Golgi enzyme that relies on Cosmc for efficient folding and maturation, both in vivo and in vitro (24, 25, 27, 28), but the mechanisms for T-synthase targeting and separation from Cosmc in the ER are not known.
The localization of proteins to the ER by retention and exclusion from transport vesicles is poorly understood, but may involve oligomerization and complexes that are excluded from budding vesicles (reviewed by Teasdale and Jackson (6)). However, while the specific roles of the TMD within resident ER proteins are unclear, the length and hydrophobic nature of the TMD has been shown to be important for ER retention (15–17), as exemplified recently for the E protein of dengue virus (54). In this example, mutation of residues within the TMD of the E protein to increase its hydrophobicity leads to increased surface expression. This mechanism may relate to earlier studies on the α chain of the T cell receptor (TCR), which is retained in the ER through its single TMD (55, 56), and retention involves specific basic amino acids within the TMD (57).
Other less well understood mechanisms of ER retrieval/retention have also been observed. For example, the TMD of resident ER proteins may also be important, as seen for the type II membrane protein Sec12p, where the cytosolic tail is required for retention, and the TMD is required for recycling (18). Some glycoproteins may be retained in the ER by interactions with lectins within the quality control system of the ER (58–60), but since Cosmc is not glycosylated it is unlikely to involve this pathway. Finally, protein oligomerization within large complexes within the ER may also contribute to ER localization (19–23). In this regard our preliminary studies indicate that Cosmc occurs as an oligomer, and it is possible that oligomerization may be important for its ER retention, but further detailed biochemical and molecular studies are underway to explore that possibility.
Additionally, unpaired Cys residues on ER proteins may lead to their retention by thiol-dependent mechanisms, as observed for unassembled Ig light chains (61) and the ERp44-mediated localization of Ero1α (53). Such processes involve lumenal thiol residues, rather than those in the TMD, in contrast to our results with Cosmc, where the Cys-19 residue in the TMD of Cosmc is crucial for its ER retention. Furthermore, loss of the Cys-19 residue in the Cosmc TMD causes decreased disulfide dimer formation of Cosmc, suggesting that Cys-19 is normally involved in disulfide formation. However, it is certainly possible that thiol residues may be involved in Cosmc localization in some way, and future experiments will explore that possibility, along with formally demonstrating the potential of Cys-19 to form a disulfide bond.
Various post-translational modifications can also contribute to ER retrieval/retention. An example is reversible S-palmitoylation in which the 16-carbon fatty acid is covalently attached to a protein through a thio-ester linkage (62–64). The demonstration of palmitoylation required for lysosomal enzyme-sorting of mannose-6-phosphate receptor has reinforced the hypothesis of the role of palmitoylation for normal trafficking and localization (65, 66). There is also emerging evidence that palmitoylation of Cys residues in the TMD of resident ER proteins may influence the ER retention. For example, the lipoprotein receptor-related protein 6 (LRP6) is involved in canonical Wnt signaling (67), and palmitoylation of membrane Cys residues is required for its exit from the ER; consequently mutation of Cys residues leads to ER retention (68). However, this mechanism is not likely to be involved in Cosmc retention in the ER since the Cys-19 residue in the TMD is required for disulfide-bond formation and loss of Cys leads to movement out of the ER. Nevertheless, in future studies, we intent to examine whether reversible or transient palmitoylation of Cosmc or adjacent ER proteins may be important for Cosmc retention or exit from the ER.
In summary, our results show the important role of the TMD of Cosmc for its ER retention, but additional studies will need to be performed to identify the role of Cosmc oligomerization or association with other ER proteins, and the roles of Cys and other aspects of Cosmc structure for its ER retention. Our studies extend the range of resident ER proteins and contribute to our overall understanding of the complex mechanism(s) for ER retention, and suggest that the TMD of Cosmc may be a valuable tool to study ER retention and function of proteins in the secretory pathway.
Supplementary Material
Acknowledgments
We thank the Emory Winship Cancer Institute Cell Imaging and Microscopy Core at Emory University School of Medicine for help with confocal imaging. We thank Dr. Jamie Heimburg-Molinaro of Emory University for manuscript editing and review, and Raj Aryal, Dr. Anthony Luyai, and Dr. Yingchun Wang for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM068559 (to R. D. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
- ER
- endoplasmic reticulum
- TMD
- transmembrane domain
- CD
- cytoplasmic domain
- LD
- lumenal domain
- TfR
- transferrin receptor
- DMSO
- dimethyl sulfoxide
- PNS
- postnuclear supernatant.
REFERENCES
- 1. Pfeffer S. R., Rothman J. E. (1987) Annu. Rev. Biochem. 56, 829–852 [DOI] [PubMed] [Google Scholar]
- 2. Gomord V., Wee E., Faye L. (1999) Biochimie 81, 607–618 [DOI] [PubMed] [Google Scholar]
- 3. Pelham H. R. (1990) Trends Biochem. Sci 15, 483–486 [DOI] [PubMed] [Google Scholar]
- 4. Lewis M. J., Pelham H. R. (1992) Cell 68, 353–364 [DOI] [PubMed] [Google Scholar]
- 5. Wieland F., Harter C. (1999) Curr. Opin. Cell Biol. 11, 440–446 [DOI] [PubMed] [Google Scholar]
- 6. Teasdale R. D., Jackson M. R. (1996) Annu. Rev. Cell Dev. Biol. 12, 27–54 [DOI] [PubMed] [Google Scholar]
- 7. Gaynor E. C., te Heesen S., Graham T. R., Aebi M., Emr S. D. (1994) J. Cell Biol. 127, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jackson M. R., Nilsson T., Peterson P. A. (1993) J. Cell Biol. 121, 317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Townsley F. M., Pelham H. R. (1994) Eur. J. Cell Biol. 64, 211–216 [PubMed] [Google Scholar]
- 10. Bakke O., Dobberstein B. (1990) Cell 63, 707–716 [DOI] [PubMed] [Google Scholar]
- 11. Lotteau V., Teyton L., Peleraux A., Nilsson T., Karlsson L., Schmid S. L., Quaranta V., Peterson P. A. (1990) Nature 348, 600–605 [DOI] [PubMed] [Google Scholar]
- 12. Boulaflous A., Saint-Jore-Dupas C., Herranz-Gordo M. C., Pagny-Salehabadi S., Plasson C., Garidou F., Kiefer-Meyer M. C., Ritzenthaler C., Faye L., Gomord V. (2009) BMC Plant Biol. 9, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Contreras I., Ortiz-Zapater E., Aniento F. (2004) Plant J. 38, 685–698 [DOI] [PubMed] [Google Scholar]
- 14. Sohn K., Orci L., Ravazzola M., Amherdt M., Bremser M., Lottspeich F., Fiedler K., Helms J. B., Wieland F. T. (1996) J. Cell Biol. 135, 1239–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Honsho M., Mitoma J. Y., Ito A. (1998) J. Biol. Chem. 273, 20860–20866 [DOI] [PubMed] [Google Scholar]
- 16. Rayner J. C., Pelham H. R. (1997) EMBO J. 16, 1832–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang M., Ellenberg J., Bonifacino J. S., Weissman A. M. (1997) J. Biol. Chem. 272, 1970–1975 [DOI] [PubMed] [Google Scholar]
- 18. Sato M., Sato K., Nakano A. (1996) J. Cell Biol. 134, 279–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cocquerel L., Duvet S., Meunier J. C., Pillez A., Cacan R., Wychowski C., Dubuisson J. (1999) J. Virol. 73, 2641–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cocquerel L., Meunier J. C., Pillez A., Wychowski C., Dubuisson J. (1998) J. Virol. 72, 2183–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cocquerel L., Wychowski C., Minner F., Penin F., Dubuisson J. (2000) J. Virol. 74, 3623–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hardt B., Kalz-Fuller B., Aparicio R., Volker C., Bause E. (2003) Glycobiology 13, 159–168 [DOI] [PubMed] [Google Scholar]
- 23. Opat A. S., Houghton F., Gleeson P. A. (2000) J. Biol. Chem. 275, 11836–11845 [DOI] [PubMed] [Google Scholar]
- 24. Aryal R. P., Ju T., Cummings R. D. (2010) J. Biol. Chem. 285, 2456–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ju T., Cummings R. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16613–16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ju T., Cummings R. D. (2005) Nature 437, 1252. [DOI] [PubMed] [Google Scholar]
- 27. Wang Y., Ju T., Ding X., Xia B., Wang W., Xia L., He M., Cummings R. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 9228–9233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ju T., Aryal R. P., Stowell C. J., Cummings R. D. (2008) J. Cell Biol. 182, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ju T., Otto V. I., Cummings R. D. (2011) Angewandte Chemie 50, 2–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ponka P., Lok C. N. (1999) Int. J. Biochem. Cell Biol. 31, 1111–1137 [DOI] [PubMed] [Google Scholar]
- 31. Trowbridge I. S., Omary M. B. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 3039–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trowbridge I. S., Shackelford D. A. (1986) Biochem. Soc. Symp. 51, 117–129 [PubMed] [Google Scholar]
- 33. Rutledge E. A., Gaston I., Root B. J., McGraw T. E., Enns C. A. (1998) J. Biol. Chem. 273, 12169–12175 [DOI] [PubMed] [Google Scholar]
- 34. Fang G., Weiser B., Visosky A., Moran T., Burger H. (2002) in PCR-Mediated Recombination: A General Method Applied to Construct Chimeric Infectious Molecular Clones, PCR Cloning Protocols (Chen B.-Y., Janes H. W. eds) Springer, Secaucus, NJ: [DOI] [PubMed] [Google Scholar]
- 35. Rezaie A. R., Fiore M. M., Neuenschwander P. F., Esmon C. T., Morrissey J. H. (1992) Protein Expr. Purif. 3, 453–460 [DOI] [PubMed] [Google Scholar]
- 36. Stearns D. J., Kurosawa S., Sims P. J., Esmon N. L., Esmon C. T. (1988) J. Biol. Chem. 263, 826–832 [PubMed] [Google Scholar]
- 37. Narimatsu Y., Ikehara Y., Iwasaki H., Nonomura C., Sato T., Nakanishi H., Narimatsu H. (2008) Biochem. Biophys. Res. Commun. 366, 199–205 [DOI] [PubMed] [Google Scholar]
- 38. Do S. I., Enns C., Cummings R. D. (1990) J. Biol. Chem. 265, 114–125 [PubMed] [Google Scholar]
- 39. Omary M. B., Trowbridge I. S. (1981) J. Biol. Chem. 256, 12888–12892 [PubMed] [Google Scholar]
- 40. Do S. I., Cummings R. D. (1992) Glycobiology 2, 345–353 [DOI] [PubMed] [Google Scholar]
- 41. Hayes G. R., Enns C. A., Lucas J. J. (1992) Glycobiology 2, 355–359 [DOI] [PubMed] [Google Scholar]
- 42. Hayes G. R., Williams A. M., Lucas J. J., Enns C. A. (1997) Biochemistry 36, 5276–5284 [DOI] [PubMed] [Google Scholar]
- 43. Schneider C., Sutherland R., Newman R., Greaves M. (1982) J. Biol. Chem. 257, 8516–8522 [PubMed] [Google Scholar]
- 44. Sönnichsen B., Füllekrug J., Nguyen Van P., Diekmann W., Robinson D. G., Mieskes G. (1994) J. Cell Sci. 107, 2705–2717 [DOI] [PubMed] [Google Scholar]
- 45. Harter C., Pavel J., Coccia F., Draken E., Wegehingel S., Tschochner H., Wieland F. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1902–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harter C., Wieland F. (1996) Biochim. Biophys Acta 1286, 75–93 [DOI] [PubMed] [Google Scholar]
- 47. Nilsson T., Warren G. (1994) Curr. Opin. Cell Biol. 6, 517–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schutze M. P., Peterson P. A., Jackson M. R. (1994) EMBO J. 13, 1696–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sato K., Sato M., Nakano A. (2003) Mol. Biol. Cell 14, 3605–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kelleher D. J., Kreibich G., Gilmore R. (1992) Cell 69, 55–65 [DOI] [PubMed] [Google Scholar]
- 51. Fu J., Kreibich G. (2000) J. Biol. Chem. 275, 3984–3990 [DOI] [PubMed] [Google Scholar]
- 52. Fu J., Pirozzi G., Sanjay A., Levy R., Chen Y., De Lemos-Chiarandini C., Sabatini D., Kreibich G. (2000) Eur. J. Cell Biol. 79, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pedrazzini E., Villa A., Longhi R., Bulbarelli A., Borgese N. (2000) J. Cell Biol. 148, 899–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hsieh S. C., Tsai W. Y., Wang W. K. (2010) J. Virol. 84, 4782–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bonifacino J. S., Suzuki C. K., Klausner R. D. (1990) Science 247, 79–82 [DOI] [PubMed] [Google Scholar]
- 56. Bonifacino J. S., Cosson P., Klausner R. D. (1990) Cell 63, 503–513 [DOI] [PubMed] [Google Scholar]
- 57. Bonifacino J. S., Cosson P., Shah N., Klausner R. D. (1991) EMBO J. 10, 2783–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) Trends Biochem. Sci. 35, 74–82 [DOI] [PubMed] [Google Scholar]
- 59. D'Alessio C., Caramelo J. J., Parodi A. J. (2010) Semin. Cell Dev. Biol. 21, 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pearse B. R., Hebert D. N. (2010) Biochim. Biophys. Acta 1803, 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reddy P., Sparvoli A., Fagioli C., Fassina G., Sitia R. (1996) EMBO J. 15, 2077–2085 [PMC free article] [PubMed] [Google Scholar]
- 62. Dietrich L. E., Ungermann C. (2004) EMBO Rep. 5, 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Aoki D., Lee N., Yamaguchi N., Dubois C., Fukuda M. N. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4319–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mitchell D. A., Vasudevan A., Linder M. E., Deschenes R. J. (2006) J. Lipid Res. 47, 1118–1127 [DOI] [PubMed] [Google Scholar]
- 65. Schweizer A., Kornfeld S., Rohrer J. (1996) J. Cell Biol. 132, 577–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stöckli J., Rohrer J. (2004) Mol. Biol. Cell 15, 2617–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Clevers H. (2006) Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 68. Abrami L., Kunz B., Iacovache I., van der Goot F. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5384–5389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ju T., Xia B., Aryal R. P., Wang W., Wang Y., Ding X., Mi R., He M., Cummings R. D. (2011) Glycobiology, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.