Abstract
Actin-related proteins (Arps) are a highly conserved family of proteins that have extensive sequence and structural similarity to actin. All characterized Arps are components of large multimeric complexes associated with chromatin or the cytoskeleton. In addition, the human genome encodes five conserved but largely uncharacterized “orphan” Arps, which appear to be mostly testis-specific. Here we show that Arp7A, which has 43% sequence identity with β-actin, forms a complex with the cytoskeletal proteins Tes and Mena in the subacrosomal layer of round spermatids. The N-terminal 65-residue extension to the actin-like fold of Arp7A interacts directly with Tes. The crystal structure of the 1–65Arp7A·LIM2–3Tes·EVH1Mena complex reveals that residues 28–49 of Arp7A contact the LIM2–3 domains of Tes. Two alanine residues from Arp7A that occupy equivalent apolar pockets in both LIM domains as well as an intervening GPAK linker that binds the LIM2–3 junction are critical for the Arp7A-Tes interaction. Equivalent occupied apolar pockets are also seen in the tandem LIM domain structures of LMO4 and Lhx3 bound to unrelated ligands. Our results indicate that apolar pocket interactions are a common feature of tandem LIM domain interactions, but ligand specificity is principally determined by the linker sequence.
Keywords: Actin, Cytoskeleton, Protein Domains, Protein Structure, Protein-Protein Interactions, Spermatogenesis, Actin-related Protein, LIM Domain, Testis
Introduction
Actin, one of the major components of the eukaryotic cytoskeleton, is an evolutionarily conserved protein that has structural and functional homologues in prokaryotes (1–4). In addition to actin, eukaryotes also contain a conserved family of actin-related proteins (Arps)4 that have 17–45% sequence identity to actin and are predicted to adopt a similar structural fold (5–8).
Arp1, the first Arp identified, is most closely related to actin and is an integral component of the dynactin complex, which also contains a single molecule of Arp11 (9). The dynactin complex together with the dynein motor play an essential role in transporting a variety of cellular cargoes in a retrograde fashion along microtubules (9–11). In contrast to Arp1/11, Arp2 and -3, which form a stable complex with five unrelated proteins (ARPC1–5), are associated with the actin cytoskeleton (12–14). The Arp2/3 complex, which is conserved in all eukaryotes, plays an essential role in promoting actin polymerization during a variety of different cellular processes, including cell motility and endocytosis (12–14). In recent years, it has become clear that a number of evolutionarily conserved Arps (Arp4, -5, -6, and -8) have no apparent association with the cytoskeleton but are in fact found in the nucleus as components of chromatin-remodeling complexes (6, 8, 15). In most cases, the exact in vivo function of these Arp-containing chromatin-remodeling complexes still remains to be established. The function of some of these complexes may, however, involve actin, which is constantly shuttling in and out of the nucleus and appears to have additional nuclear functions beyond its role in the cytoplasm (6, 8).
One common theme that has emerged is that Arps are often present in pairs in large multimeric protein complexes, which frequently also contain actin. It is possible that “orphan” Arps, whose cellular function remains to be established, may also be components of large actin-containing complexes. A number of these orphan Arps (Arp-T1, Arp-T2, Arp7A, Arp7B, ArpM1) appear to be testis-specific (16–20). Recently, Arp7A, which is also known as ACTL7A, T-actin 2, and Tact2, was identified in a yeast two-hybrid screen as a potential interacting partner for Tes (21).
Tes is a putative human tumor suppressor, which is frequently down-regulated in a variety of tumor cell lines as well as primary breast tumors and glioblastomas (22–26). Targeted deletion of the Tes gene in mice leads to an increased susceptibility to carcinogenic drug-induced gastric cancer (24). In contrast, overexpression of Tes suppresses cell growth and significantly reduces the tumorigenic potential of T47D (ductal breast carcinoma) and MES-SA (uterine sarcoma) tumor cell lines in nude mice (25, 27). Tes, which contains three tandemly arranged LIM domains in its C-terminal half, interacts with a variety of cytoskeletal proteins including actin, α-actinin, Mena, paxillin, talin, and zyxin (21, 27, 28). LIM domains define protein interaction motifs that bind a wide range of different proteins (29). Tes is recruited to focal adhesions via a direct interaction between its LIM1 domain and zyxin (27). Moreover, Tes is able to regulate Mena-dependent cell migration by virtue of the ability of its C-terminal LIM3 domain to compete with FPPPP-containing proteins for binding to the EVH1 domain of Mena (28).
The potential association of Tes with the actin cytoskeleton is highly indicative of a possible cytoskeletal function for Arp7A. Consistent with this, the sequence of Arp7A has no major deletions or insertions within its predicted actin fold and has 43% sequence identity to β-actin, which is higher than that of Arp3 (5, 6, 16, 19). Arp7A does, however, have a unique 65-amino acid extension at its N terminus (16). In this study, we sought to examine whether Tes is indeed complexed with Arp7A in testis, and if so, to understand the molecular basis of the interaction between the two proteins.
EXPERIMENTAL PROCEDURES
Antibodies and Immunofluorescence Analysis
The Arp7A antibody was produced by immunization of rabbits with a peptide corresponding to residues 1–65 of human Arp7A. Arp7A antibodies were affinity-purified on a 1–65-peptide column according to the manufacturer's instructions (SulfoKink kit, Pierce). Polyclonal antibodies against actin, Mena, profilin I, and VASP were obtained from Cytoskeleton Inc., Drs. Walter Witke, Roger Karlsson, and Frank Gertler, respectively. Monoclonal antibodies against actin (AC74 and AC40 Sigma; MAB1501R Chemicon International); α-actinin (MAB1682 Chemicon International); GFP (3E1) and Tes (1A9, 7F6, and 5E6) (Cancer Research UK); GM130 (2C10, Abcam); Mena (21) and paxillin (m349) (BD Biosciences), and zyxin (164D4) (SySy) were used. FITC-PNA was obtained from Sigma. Adult mouse testes were fixed in 4% paraformaldehyde in PBS for 3 h at 4 °C and impregnated in 20% sucrose in PBS for 6 h at 4 °C before being embedded in Tissue-Tek (Sakura) prior to cryostat sectioning. Immunofluorescence analysis was performed as described previously (28).
Mammalian Expression Vectors, Pulldown, and Immunoprecipitation Assays
Arp7A (GenBankTM: NM_006687) was amplified by PCR from a human testis Marathon-Ready cDNA library (Clontech) and cloned into the NotI-EcoRI sites of CB6-N-GFP (30) to generate GFP-Arp7A for expression in mammalian cells. GFP-tagged 1–65 and Arp7A-Δ1–65 were generated by PCR. All CB6-N-GFP Tes expression vectors have been previously described (27). Mammalian cell extracts containing the different GFP-tagged proteins were prepared and incubated with His- or GST-tagged protein resins as described previously (27, 28). Rat testes were homogenized in 50 mm Tris, pH 7.5, 150 mm NaCl, 0,1% Triton, and the homogenate was centrifuged at 80,000 × g at 4 °C for 1 h. The resulting supernatant was incubated for 2 h at 4 °C with Tes antibody for immunoprecipitation and processed as described previously (28).
Bacterial Expression Vectors and Interaction Assays
His-Tes and GST-3C-EVH1Mena expression vectors have been described (27, 28). Residues 1–65 of human Arp7A (1-65Arp7A) and the LIM2–3 domains of human Tes (LIM2–3Tes, residues 296–421) were amplified by PCR and cloned into the NotI-EcoRI sites of pMW172-GST-3C and pMW172-His-3C (28). All 1–65Arp7A deletion mutants were generated by PCR.
GST-tagged 1–65Arp7A and EVH1Mena were produced at 30 °C in BL21(DE3) Rosetta. Bacterial cell pellets were sonicated in 50 mm Tris, pH 8, 500 mm NaCl, 1 mm DTT, 10 mm benzamidine, and 1 mm PMSF. Cleared supernatants were mixed for 90 min with glutathione-Sepharose 4B beads, which were then washed with 20 mm Tris, pH 8, 100 mm NaCl, and 1 mm DTT. 1–65Arp7A and EVH1Mena proteins were released from the beads by overnight incubation with PreScission protease (GE Healthcare) at 4 °C. GST-LIM2–3Tes was prepared as above, but before adding the PreScission protease, the protein-bound resin was incubated at 4 °C for 1 h with a solution of 15 mm ATP, 30 mm MgCl2, 5 mm β-octyl glucoside containing bacterial extracts dissolved in 8 m urea to remove heat shock proteins.
For direct binding assays, GST-tagged proteins were incubated with His-Tes or His-LIM2–3 resin 1 h at 4 °C and processed as described previously (27, 28). The ternary complex of 1–65Arp7A·LIM2–3Tes·EVH1Mena for crystallization was prepared by adding purified 1–65Arp7A and EVH1Mena to GST-tagged LIM2–3Tes immobilized on glutathione-Sepharose 4B resin and incubated at 4 °C with PreScission protease for 18 h. The released 1–65Arp7A·LIM2–3Tes·EVH1Mena ternary complex was further purified by Superdex-75 size exclusion chromatography.
Crystallization, Data Collection, and Structure Solution
The ternary complex of 1–65Arp7A·LIM2–3Tes·EVH1Mena was crystallized at 22 °C from sitting drops containing 2 μl of protein solution (37.5 mg/ml in 20 mm Tris-HCl, pH 8.0, 100 mm NaCl) and 1 μl of well solution (17.5% PEG 6000, 0.1 m Tris, pH 8.5, 0.15 m KSCN) over a 500 μl well. Crystals appeared after ∼10 days; for data collection, they were flash-frozen in a stream of nitrogen at 100 K using paratone-N as cryo-protectant.
Diffraction data were collected in-house using CuKα radiation from a Rigaku MicroMax-007HF and a Mar345 image plate; an initial dataset to 3.0 Å resolution was superseded by one at 2.6 Å, which was used for final refinement (Table 1). Data were integrated with mosflm (31) and scaled with scala (32). Other programs from the CCP4 suite (33) were used for general crystallographic computations, and PyMOL (34) was used to prepare structural figures.
TABLE 1.
Data processing and refinement statistics
| Diffraction data | |
|---|---|
| Space group | P212121 |
| Cell a, b, c (Å) | a = 37.83, b = 86.79, c = 115.27 |
| Zab | 1 |
| Wavelength (Å) | 1.5418 |
| Resolution (Å) | 25.56–2.60 (2.74–2.60)a |
| Completeness (%) | 96.9 (96.9) |
| Multiplicity | 4.2 (4.0) |
| Rmeas (%)c | 10.7 (49.4) |
| Rp.i.m. (%)d | 6.8 (32.1) |
| I/σ(I) | 14.2 (2.9) |
| Refinement | |
| R | 21.8 |
| Rfree | 26.8 |
| Reflections | 11,400 |
| No. of protein atoms | 2040 |
| No. of zinc ions | 5 |
| No. of solvent atoms | 49 |
| Wilson B factor | 56.2 |
| Mean B factor (Å2) Protein chains A, M, T | 53.0, 45.5, 42.1 |
| r.m.s.d. bonds (Å), angles (°) | 0.018, 1.888 |
| Ramachandran plot (%) (preferred, allowed, outliers) | 90.62, 7.42, 1.95 |
a Numbers in parentheses refer to the highest resolution shell.
b Za is the number of molecules of the complex in the asymmetric unit.
c Rmeas = ∑hkl|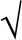 (N/(N − 1))∑i|Ii(hkl) − <I(hkl)>|/∑hkl∑iIi(hkl).
(N/(N − 1))∑i|Ii(hkl) − <I(hkl)>|/∑hkl∑iIi(hkl).
d Rp.i.m. = ∑hkl|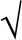 (1/(N − 1))∑i|Ii(hkl) − <I(hkl)>|/∑hkl∑iIi(hkl).
(1/(N − 1))∑i|Ii(hkl) − <I(hkl)>|/∑hkl∑iIi(hkl).
The structure was solved by molecular replacement. First, the EVH1 component of the ternary complex 1-65Arp7A·LIM2–3Tes·EVH1Mena was located using the program molrep (35) using the mouse EVH1 structure (Protein Data Bank (PDB) code 1EVH) as a search model and using a preliminary dataset at 3.0Å resolution. A second round of molecular replacement used the LIM3Tes·EVH1Mena complex (PDB code 2IYB) as search model. Having placed these two chains, a search for LIM2Tes used a model based on LIM3Tes generated by the program chainsaw (36). When the 2.6 Å data became available, refinement continued with refmac (37), alternating with model adjustment with coot (38).
Protein Interfaces and Comparisons
Protein-protein interfaces in the ternary complex were analyzed using the PISA server (39) (supplemental Table S1). Structural comparisons of LIM domains were calculated using SSM via the PDBeFold server at the European Bioinformatics Institute (EBI) The PDB entry most similar to the tandem LIM2–3Tes taken together is the Lhx3 LIM domains 1 and 2 in complex with the binding domain of Isl1 (PDB code 2RGT; r.m.s.d. 3.89 Å for 105 Cα atoms).
ITC Calorimetry and Peptide Arrays
Isothermal calorimetry (ITC) calorimetry was performed using a VP-ITC 200 microcalorimeter (MicroCal LLC). To minimize protein aggregation, the concentration of LIM2–3Tes or LIM2Tes was kept below 50 μg/ml and was dialyzed into 20 mm Tris, pH 8, 100 mm NaCl, 0.2 mm tris(2-carboxyethyl)phosphine buffer prior to the ITC experiment. It was then concentrated to 0.6 mg/ml immediately before use and placed in the ITC cell compartment. A 7 mg/ml solution of the various wild type and mutant Arp7A peptides was prepared in a 20 mm Tris, pH 8, 100 mm NaCl, 0.2 mm tris(2-carboxyethyl)phosphine buffer following purification of the peptide from low molecular weight contaminants on a G10 Sepharose column. The eluted Arp7A peptide peak was diluted to 0.3 mg/ml and drawn into the ITC syringe for injection. Volumes of 5–7.5 μl were injected into the ITC cell containing either LIM2–3Tes or LIM2Tes. Data analysis and curve fitting were done using the Origin 7.0 software (OriginLab).
Cellulose membranes were prepared and spotted with peptides using standard protocols. To eliminate nonspecific binding, the membrane was first blocked for 30 min using 1 mg/ml BSA in PBS containing 0.1% Tween 20 prior to the addition of GST-LIM2–3Tes at a final concentration of 0.4 μg/ml. The membrane was then washed three times in BSA buffer and then incubated with anti-GST antibody for 30 min. Finally, the membrane was washed three times for 5 min and then incubated with anti-rabbit HRP (Santa Cruz Biotechnology), washed, and developed using ECL reagent (GE Healthcare).
RESULTS
Arp7A, Tes, and Mena Co-localize in the Subacrosomal Layer of Round Spermatids
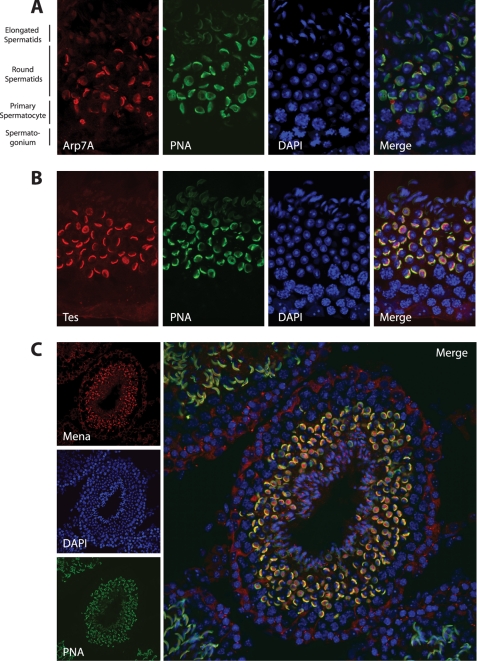
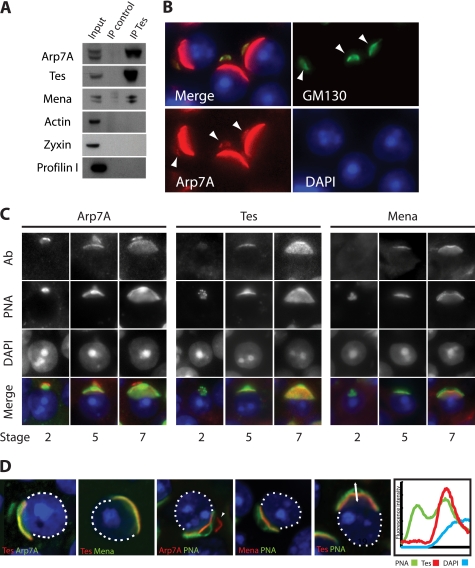
Molecular analysis in mice indicates that Arp7A is only expressed in testis (18, 19). To determine whether Tes co-localizes with Arp7A in testis, we raised an antibody against a peptide corresponding to the unique 65-amino acid extension at its N terminus (1–65). Western blot analysis on extracts prepared from HeLa cells expressing GFP-Arp7A or Escherichia coli expressing GST-Arp7A confirmed that the Arp7A antibody is specific (data not shown). Immunohistochemistry on testis sections reveals that Arp7A is associated with the acrosome of round spermatids but is absent from the surrounding cells (Fig. 1A). In addition, we found that both Tes and Mena, but not its homologue, VASP, are associated with the acrosome of round spermatids (Fig. 1, B and C, and data not shown). By contrast, we were unable to detect the Tes-interacting partners actin, α-actinin, paxillin, and zyxin on the acrosome (data not shown). Immunoprecipitations from testes extracts confirmed that Arp7A is indeed complexed with Tes and Mena (Fig. 2A). Consistent with our immunofluorescence analysis, we were unable to detect actin or zyxin within this complex (Fig. 2A). We also could not detect the Mena-binding protein, profilin I, in the Arp7A·Tes·Mena complex (Fig. 2A).
FIGURE 1.
Arp7A, Tes, and Mena associate with the acrosome of round spermatids. A, immunofluorescence analysis of testis sections reveals that Arp7A co-localizes with the acrosomal marker peanut agglutinin (PNA) on round spermatids. B and C, Tes (B) and Mena (C) have a similar localization to Arp7A.
FIGURE 2.
Arp7A, Tes, and Mena are components of the subacrosomal layer. A, immunoblot analysis of Tes immunoprecipitations (IP) with the indicated antibodies reveals that Arp7A, Tes, and Mena are complexed to each other in rat testes extracts. B, immunofluorescence analysis of testis sections reveals that Arp7A co-localizes with GM130 on the Golgi apparatus of round spermatids (white arrowhead). C, temporal localization of Arp7A, Tes, and Mena during acrosome formation. PNA labels the developing acrosome, and the stage of spermatogenesis is indicated at the bottom. Ab, antibody. D, confocal immunofluorescence images reveal that Arp7A, Tes, and Mena co-localize between the acrosome (PNA-positive) and the nucleus, in the subacrosomal layer. The fluorescence intensity profile distribution of Tes, PNA, and DNA in the region indicated by the double-headed arrow highlights the subacrosomal localization of Tes (right panel).
Closer examination of the testis sections reveals that that Arp7A is also associated with an additional structure, which was identified as the Golgi apparatus based on its labeling with the Golgi-specific protein GM130 (Fig. 2B). Arp7A is associated with the Golgi apparatus prior to and throughout the biogenesis of the acrosome (Fig. 2, B and C). In contrast, Tes and Mena are never observed on the Golgi apparatus, suggesting that they only interact with Arp7A as the developing acrosome associates with the nuclear envelope (Fig. 2C). Moreover, closer examination of the localization of Arp7A, Tes, and Mena reveals that all three proteins are actually localized in the subacrosomal layer that links the acrosome to the nuclear envelope (Fig. 2D).
Residues 1–65 of Arp7A Bind Directly to the LIM2 Domain of Tes
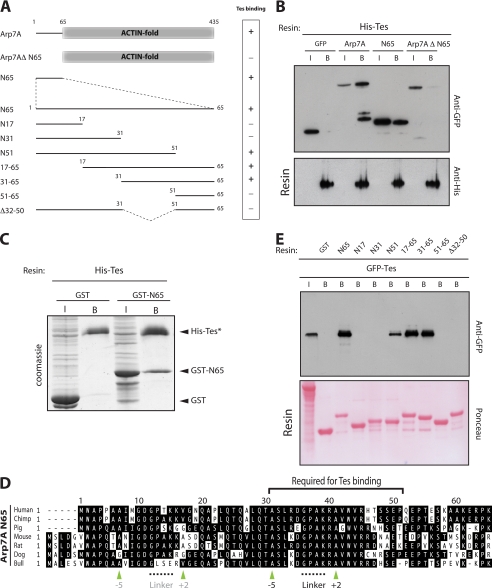
Arp7A was identified as a Tes-interacting protein in a yeast two-hybrid screen to identify potential Tes binding partners (21). To validate and extend this yeast two-hybrid interaction, we performed pulldown assays on extracts prepared from 293 cells expressing GFP-tagged Arp7A using His-Tes produced in E. coli. We found that the His-Tes resin retains GFP-tagged Arp7A, as well as its first 65 residues (N65), but not the core actin fold of the protein (Fig. 3, A and B). Our previous results have shown that Mena and zyxin are capable of binding directly to Tes (27, 28). Pulldown assays using bacterially expressed proteins demonstrated that Arp7A represents the third direct binding partner for Tes (Fig. 3C). Furthermore, Arp7A, like Mena, can bind full-length Tes. This is in contrast to zyxin, which can only interact with the isolated LIM1 domain or C-terminal half of Tes, presumably due to a masking of the binding site in a “closed” conformation of the full-length Tes (27).
FIGURE 3.
Tes binds directly to the N-terminal 65 residues of Arp7A. A, schematic representation of the Arp7A mutants together with their Tes binding capacity. B, immunoblot analysis reveals that a His-Tes resin retains GFP-tagged Arp7A and 1–65, but not Arp7A lacking the first 65 residues (Arp7AΔ1–65) or GFP. The input (I) and bound (B) samples of the respective GFP-tagged proteins are indicated above the top panel. All samples contain equivalent amounts of His-Tes resin (bottom panel). C, Coomassie Brilliant Blue-stained gel reveals that a His-Tes resin (*) binds directly to GST-1–65 but not to GST. D, sequence alignment of the N-terminal extension to the actin-like fold of Arp7A from placental mammals. Apolar pocket alanine residues at −5 and +2 (green arrowheads) relative the GPAK linker in the Tes-binding region are highlighted below the alignment. A second putative tandem LIM-binding site near the N terminus of Arp7A is indicated in gray. E, immunoblot analysis of GST pulldowns using the 1–65 deletion mutants in A demonstrates that residues 31–51 of Arp7A are required to bind GFP-Tes. The Ponceau staining demonstrates that all samples contained equivalent amounts of each GST resin.
The sequence of the first 65 residues of Arp7A is highly conserved among placental mammals (60–98% identity) (Fig. 3D). To obtain insights into which region is responsible for interacting with Tes, we performed pulldown assays using a series of deletion mutants produced in E. coli (Fig. 3, A and E). We found that the sequence between residues 31 and 51 of Arp7A is required to retain GFP-Tes from HeLa cell extracts (Fig. 3, A and E). Sequence alignment of available Arp7A orthologues from placental mammals reveals that residues 31–51 correspond to the most conserved region of the N-terminal extension (Fig. 3D). Database searches, however, reveal that these residues have no obvious sequence homology to any other protein, suggesting that the interaction between Arp7A and Tes is unique.
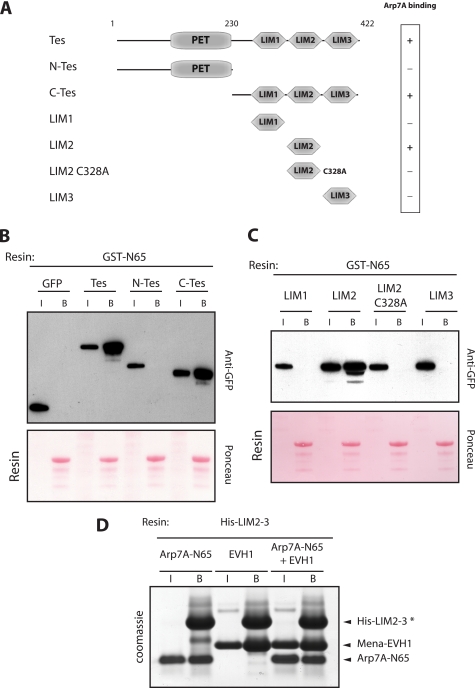
Previous observations have shown that zyxin and Mena interact directly with the LIM1 and LIM3 domains of Tes, respectively (27, 28). To identify the Arp7A-binding site in Tes, we performed pulldown assays on cell extracts containing GFP-tagged Tes and its domains using a GST-N65 resin. We found that the GST-N65 resin binds GFP-Tes and its C-terminal half but not the N-terminal half of the protein (Fig. 4, A and B). Pulldown assays with the individual Tes LIM domains reveal that GST-N65 only binds to the second LIM domain (Fig. 4, A and C). Disruption of the LIM2 domain by the introduction of a single point mutation (C328A, a zinc-coordinating residue) (27), completely abolished its interaction with Arp7A (Fig. 4C). Interestingly, pulldown assays using protein produced in E. coli demonstrate that the EVH1 domain of Mena and the N-terminal 65 residues of Arp7A are able to form a tripartite complex with the LIM2–3 domain of Tes (Fig. 4D).
FIGURE 4.
Arp7A binds the LIM2 domain of Tes. A, schematic representation of the different Tes domains together with their ability to bind 1–65Arp7A. B, immunoblot analysis reveals that a GST-1–65 resin retains GFP-tagged Tes and C-Tes but not N-Tes or GFP. The input (I) and bound (B) samples with the respective GFP-tagged protein are indicated. The Ponceau staining demonstrates that all samples contain equivalent amounts of GST-1–65 resin. C, immunoblot analysis reveals that the GST-1–65 resin binds GFP-tagged LIM2 but not the LIM1, LIM2-C328A, or LIM3 domains of Tes. D, Coomassie Blue-stained gel reveals that a His-LIM2–3 resin (*) can simultaneously bind 1–65Arp7A and EVH1Mena.
Structure of the 1–65Arp7A·LIM2–3Tes·EVH1Mena Complex
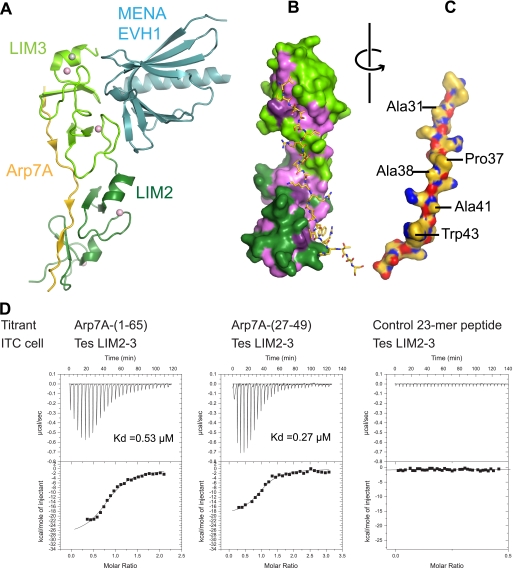
To obtain molecular insights into this tripartite complex, we determined the structure of 1–65Arp7A·LIM2–3Tes·EVH1Mena at 2.6 Å (Fig. 5A; supplemental Fig. S1A and Table 1) (PDB code 2XQN). There is a single ternary complex present in the asymmetric unit. All residues within the LIM2–3Tes and EVH1Mena domains are well defined in the structure. Both LIM domains are very similar (r.m.s.d. of 2.65 Å for 52 matched Cα atoms out of 62/63 in the two domains), and each binds two zinc ions. A fifth zinc ion is also found between symmetry contacts within the crystal lattice (supplemental Fig. S1B). The LIM3Tes·EVH1Mena interface in the tripartite complex is reduced to 599 Å2 from the 759 Å2 observed in the LIM3Tes-EVH1Mena binary complex (PDB code 2IYB) (supplemental Fig. S1C). The EVH1 domain in both complexes is very similar (r.m.s.d. of 0.56 Å over 107 Cα atoms), as are contacts between EVH1Mena and the first zinc finger motif of LIM3Tes. The second zinc finger motif is, however, displaced by ∼10° (supplemental Fig. S1C).
FIGURE 5.
Structure of the 1–65Arp7A·LIM2–3Tes·EVH1Mena complex. A, a schematic of the ternary complex. LIM2–3Tes is shown in green (darker for LIM2 and lighter for LIM3), the EVH1Mena is in pale cyan, and 28–49Arp7A is in gold. The four canonical zinc ions bound to the tandem LIM domains are shown as pink spheres, and an additional zinc ion associated with a crystal contact is shown in gray. B, a surface rendering of LIM2–3Tes is colored as in A but with hydrophobic/aromatic residues highlighted in purple. C, the 28–49Arp7A peptide has been rotated 180° from its position in B to expose buried side chains. D, isothermal calorimetry data show that Arp7A-(1–65) and Arp7A-(27–49) have a similar binding affinity for LIM2–3Tes. A control peptide from talin shows no specific binding to LIM2–3Tes.
In contrast to LIM2–3Tes and EVH1Mena, only residues 28–49 of 1–65Arp7A are ordered (Fig. 5, A–C, and supplemental Fig. S1A). Residues 28–49 of 1–65Arp7A (referred to as 28–49Arp7A) map closely to the minimal Tes-binding fragment identified by pulldowns (Fig. 3, A and E). Overall, the LIM2–3Tes·1–65Arp7A interface buries an appreciable surface area of 1216 Å2. ITC indicated that a 27–49Arp7A peptide has essentially the same binding affinity for LIM2–3Tes as 1–65Arp7A (Kd of 0.27 as compared with 0.53 μm) (Fig. 5D), confirming that 27–49Arp7A contains the major LIM2–3Tes-binding epitope. Consistent with our pulldown assays (Fig. 4C), ITC experiments reveal that LIM2Tes binds independently to 27–49Arp7A with a Kd of ∼1.4 μm.
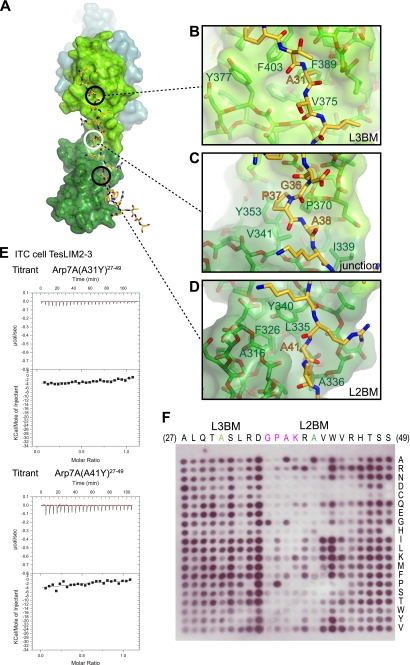
Arp7A binds in an extended conformation (Fig. 5A). The majority of contacts are made by antiparallel β-strand main chain-main chain interactions and a limited number of hydrophobic interactions (Fig. 5, B and C; supplemental Table S1). A striking feature is the equivalent “apolar” pockets between the two zinc fingers of each LIM domain, which engage a single LIM-binding motif that contains a central alanine residue (Fig. 6A). The reverse orientation of the 28–49Arp7A peptide chain relative to LIM2–3Tes positions Ala-31 (L3BM) and Ala-41 (L2BM) of Arp7A to occupy the apolar pockets of LIM3 and LIM2, respectively (Fig. 6, B and D). Consistent with our pulldown assays, the EVH1Mena makes no contacts to 28–49Arp7A as it binds to the opposite surface on LIM3Tes (Fig. 5A).
FIGURE 6.
Molecular and biochemical analysis of the Arp7A-Tes interaction. A, surface rendering of LIM2–3Tes with the 28–49Arp7A peptide in stick representation. Circles highlight three areas of contact between Arp7A and Tes, which are shown in detail in B–D. The pockets for Ala-31 and Ala-41 of Arp7A are located in equivalent sites of the LIM3 and LIM2 domains, respectively (black circles), whereas the binding region of the GPAK motif of Arp7A binds the junction of the two LIM domains (white circle). Orange and green labels mark Arp7A and Tes residues, respectively. E, calorimetry data for the indicated Arp7A (27–49) peptides reveal that mutation of Ala-31 and Ala-41 side chains ablates LIM2–3Tes binding. F, far Western analysis of an Arp7A (27–49) peptide array indicates that critical residues in the linker region are required for LIM2–3Tes binding.
A third hydrophobic interaction is contributed by a linker sequence, Gly-Pro-Ala-Lys (residues 36–39), that connects L2BM and L3BM (Fig. 6A). Within this linker motif, Gly-36 has a positive φ torsion angle and Pro-37 makes hydrophobic contacts with Pro-370 and Val-341, whereas Ala-38 occupies a shallow hydrophobic pocket between LIM2 and LIM3 (Fig. 6C). This third interaction effectively fixes the relative orientation between the two LIM domains, which have a relatively small interaction surface.
Critical Tes Binding Determinants within 27–49Arp7A
To investigate the interaction of Arp7A with the LIM2–3 domain in more detail, we mutated Ala-31 or Ala-41 individually to a bulkier tyrosine side chain and assessed changes in binding affinities by ITC. Either mutation ablated interaction with LIM2–3Tes (Fig. 6E), confirming the importance of binding the apolar pockets in the two LIM domains. We then undertook a more systematic approach to identify essential LIM2–3Tes-binding residues within 27–49Arp7A by using a far Western analysis of a 23-mer peptide array (Fig. 6F). The arrayed peptides were chosen to systematically screen the effect of having all 20 amino acids at each position. Consistent with the structure of the complex, we found that sequence substitutions within the Gly-Pro-Ala-Lys linker and L2BM resulted in a loss or reduction in LIM2–3Tes binding (Fig. 6F). The importance of the L2BM-LIM2 interaction is consistent with our findings from pulldown experiments (Fig. 4C).
DISCUSSION
Previous observations have shown that Arp7A is expressed in testis and is associated with the nucleus of spermatids (18, 19). We have now extended these earlier studies and shown that Arp7A is actually localized in the subacrosomal layer, which is also known as the acroplaxome (40). The acroplaxome is a morphologically distinct junctional complex that is thought to play an important role in anchoring the acrosome to the nuclear envelope of round spermatids (41). During acrosome biogenesis, pro-acrosomal vesicles derived from the trans-Golgi interact with the acroplaxome and fuse into a single acrosomal sac that is anchored to the nuclear envelope by the acroplaxome. Fusion of additional pro-acrosomal vesicles continues as the acrosome increases in size until it spreads over the anterior portion of the nucleus.
Although we have a good morphological understanding of acrosome biogenesis, we still lack a complete understanding of the protein composition and molecular organization of the acroplaxome (41, 42). The association of Arp7A with the Golgi apparatus, as well as the earliest stages of the developing acrosome, suggests that Arp7A is likely to play an important role during the assembly and function of the acroplaxome. Morphologically, the acroplaxome appears to contain actin-like filaments (40, 41). The margin of the acroplaxome of isolated round spermatids also stains with phalloidin and β-actin antibodies (40, 43). Our observation that Tes and Mena, which are known to interact with actin and associate with focal adhesions (21, 27), are present in the acroplaxome also points to an important role for actin in the assembly and maintenance of this junctional complex. Consistent with this notion, it has recently been reported that the transmembrane protein vezatin, which is associated with adherens junctions and the actin cytoskeleton, is also found in the acroplaxome (44). Moreover, the appearance of vezatin, which parallels that of Tes and Mena, coincides both temporally and spatially with acrosome formation (44). Based on our observations, we think it is highly likely that the Arp7A·Tes·Mena complex is an integral component of the acroplaxome that together with vezatin is responsible for anchoring the acrosome to the nuclear envelope, possibly by interacting with actin filaments. At this stage, however, we cannot rule out that this complex may actually promote actin filament assembly. However, in contrast to mature spermatids or isolated round spermatids, we failed to detect actin in the subacrosomal layer of round spermatids on testis sections using phalloidin or a panel of anti-actin antibodies (data not shown). This raises the question whether these actin-like filaments are in fact largely composed of Arps rather than just actin.
Previous work has shown that at least Arp1, which exists as a short actin-like filament in the dynactin complex, is capable of assembling into filaments when expressed in cells (9). In contrast, when expressed in HeLa cells, GFP-Arp7A is cytoplasmic and does not assemble into filaments or co-localize with and/or affect the localization of Tes and Mena at focal adhesions (data not shown). This suggests that recruitment of Arp7A to the acroplaxome also involves additional components besides Tes, consistent with the idea that it is part of a larger multimeric protein complex. Given that Arps appear to come in pairs, it is feasible that one or more of Arp-T1, Arp-T2, Arp7B, or ArpM1, which appear to be largely testis-specific, are also components of the same complex. Indeed, ArpM1, which has 46% identity with β-actin, has recently been shown to be localized in a region that appears to correspond to the acroplaxome (20). Moreover, ArpM1 interacts with profilin III, which is also localized in the acroplaxome (20, 45). In addition, we have found that Arp7B, whose N-terminal extension is distinct from that of Arp7A (16), is also strongly enriched in the acroplaxome.5
Arp7A was originally identified as a potential binding partner of Tes in a yeast two-hybrid screen (21). In this study, we sought to understand the molecular basis of the interaction between Arp7A and Tes. Our initial biochemical analysis demonstrated that the N-terminal extension to the actin-like fold of Arp7A interacts directly with the LIM2 domain of Tes. Finer mapping of the interaction site by deletion analysis reveals that residues 28–49 of Arp7A are required to interact with Tes. Our crystallographic evidence, however, shows that resides 28–49 actually interact with the LIM2 and LIM3 domains of Tes. ITC experiments confirmed that Arp7A has a stronger interaction with LIM2–3 than LIM2 alone.
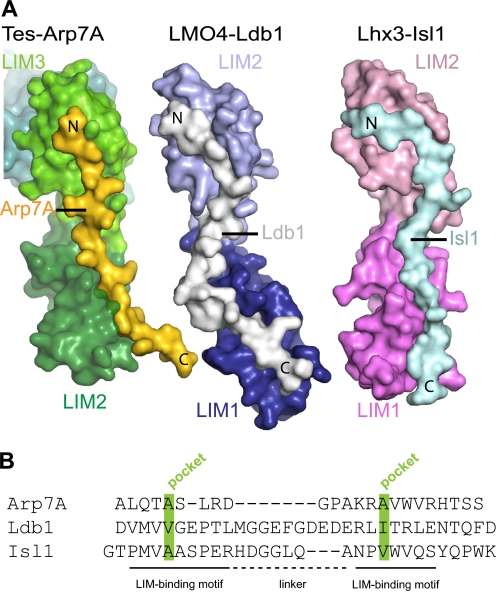
Arp7A contributes three clusters of hydrophobic residues to the LIM2–3 interaction, two of which contact the LIM domains, whereas the third bridges across the interface between both LIM domains. The Tes-binding site on Arp7A shares no apparent sequence similarity with other tandem LIM domain ligands (Fig. 7, A and B). Nevertheless, the 1–65Arp7A-LIM2–3Tes interaction is reminiscent of the LIDIsl1·LIM1–2Lhx3, LIDldb1·LIM1–2LMO4, and LIDldb1·LIM1–2LMO2 complexes (46–48). These binary complexes contain tandem LIM domains from members of the LMO and LIM-HD subgroup of LIM domain proteins bound to a LIM-interacting domain (LID) from a partner protein (Isl1 or Ldb1) (46–48). As observed for Arp7A, these tandem LIM domain ligands bind in an antiparallel orientation and engage each LIM domain apolar pocket, suggesting that pocket occupancy is a common feature of tandem LIM domain ligands (Fig. 7, A and B). Arp7A residues 28–49 follow a straight trajectory (29.7 Å distance between the Cα atoms of Arp7A Ala-31 and Ala-41) rather than the meandering S-shaped path of the other two tandem LIM ligands (for example, Ldb1 has an equivalent distance of 38.9 Å between Val-304 and Ile-322) (Fig. 7A). Consistent with this, 28–49Arp7A is significantly shorter than the LID peptides bound to LMO4 and Lhx3 (Fig. 7B). Residues 28–49 of Arp7A possibly approach the minimum length capable of simultaneously engaging both LIM apolar pockets. Previous alanine-scanning mutational studies demonstrated that hydrophobic residues in Ldb1 and Isl1, equivalent to the apolar pocket-binding alanine side chains of Arp7A, are important in tandem LIM interactions (49). The contribution of these hydrophobic residues, however, may have been underestimated because alanine-scanning was used to assess their importance (49). In addition, these previous studies did not implicate the linker connecting the LIM-binding regions of Isl1 or Ldb1 as contributing to tandem LIM binding (47, 49). In contrast, we found that the Arp7A GPAK linker makes crucial contacts with a shallow hydrophobic pocket between the LIM2 and LIM3 domains of Tes (Fig. 6). The strict sequence constraints we observe within this short linker are consistent with its important role in binding Tes.
FIGURE 7.
Structural comparison of tandem LIM domain ligands. A, comparison of 1–65Arp7A·LIM2–3Tes with the LIDLdb1·LIM1–2LMO4 (PDB code 1RUT) and LIDIsl1·LIM1–2Lhx3 (PDB code 2RGT) complexes, respectively. All three ligands have distinctive trajectories and run antiparallel to the LIM domains. B, structure-based alignment of the three tandem LIM domain ligands indicating the apolar pocket interacting residues (green) in the LIM-binding motifs.
Studies on tandem LIM domains in the absence of a ligand suggest that no fixed orientation exists (50). When bound to their respective ligands, an appreciable but small interface is formed between the tandem LIM domains. This interface in Tes and Lhx3 has a polar character and involves several hydrogen-bonding contacts. In contrast, the LMO4 interface has both hydrophobic contacts and water molecules but lacks hydrogen bonds. Using a simple metric based on the four common zinc ions in the tandem LIM structures (two per LIM domain, labeled Zn1–Zn4), we can measure the Zn1–Zn4 distance of 41.5 Å for LIM2–3Tes, whereas in LMO4·Ldb1 (1RUT) and Lxh3·Isl1 (2RGT) (molecule B), it is 58.9 and 53.1 Å, respectively. Comparing the torsion angle defined by Zn1-Zn2-Zn3-Zn4 indicates the different rotational orientations of the tandem domains, being −27° in LIM2–3Tes, −178° in 1RUT, and −99°, −17° in the two copies in 2RGT (supplemental Fig. S2). Clearly, the relative orientation of the two LIM domains is determined by the length and precise trajectory of the ligand, given that the latter is anchored to the individual LIM domains through its apolar pocket interactions.
LIM domains are well established as independently folded protein interaction modules, but the potential complexity of partner engagement in multi-LIM domain proteins is poorly understood (29). The identification of Arp7A as a new tandem LIM domain ligand that is unrelated to Ldb1 and Isl1 highlights the difficulties in predicting de novo binding partners for tandem/multiple LIM domains. Many of the contacts of Arp7A with Tes are sequence-independent as they rely on main chain-main chain interactions. However, our structure has revealed features shared with other tandem LIM ligands, namely two small hydrophobic side chains occupying an apolar pocket in each LIM domain separated by a variable linker. Apolar pocket occupancy is likely to contribute toward the affinity of the interaction, whereas the specificity of individual ligand-tandem LIM domain interactions is driven by the ligand linker. Although at present it is not possible to predict new LIM domain ligands in silico, it may be possible in the future as more structures of tandem LIM·ligand complexes are solved.
Interestingly, there is an additional GPAK motif flanked by hydrophobic residues at −5 and +2 position close to the amino terminus of Arp7A in a number of different species (Fig. 3D). The presence of these sequences points to a possible additional interaction of Arp7A with the LIM1 domain and the junction between LIM1 and LIM2 (Fig. 4). The antiparallel arrangement of Arp7A and LIM2–3Tes would, however, place the additional LIM-binding motif close to the LIM3 domain at the C terminus of Tes (Fig. 5A). Moreover, an interaction of Arp7A with the LIM1 domain and junction between LIM1 and LIM2 would compete with LIM2–3 binding. Alternatively, if the motif is functional, then Arp7A would be capable of interacting with two Tes molecules or an additional tandem LIM domain-containing protein.
In conclusion, we have defined the molecular basis of the interaction between Arp7A with Tes and shown that both proteins together with Mena are components of the acroplaxome. Our work highlights the diversity of tandem LIM domain ligand size and sequence. Future studies will be required to establish the role of the Arp7A·Tes·Mena complex in the formation and function of the acroplaxome and whether it binds to and/or induces actin filament polymerization.
Supplementary Material
Acknowledgments
We thank Drs. Frank Gertler (Massachusetts Institute of Technology, Boston, MA), Roger Karlsson (Department of Cell Biology, Stockholm University, Sweden), and Walter Witke (University of Bonn, Germany) for antibodies. We thank George Elia and Nicola O'Reilly (Cancer Research UK, London Research Institute) for cutting testis sections and for providing peptides/spot membranes, respectively.
The atomic coordinates and structure factors (code 2XQN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
B. Boëda, unpublished results.
- Arp
- actin-related protein
- PNA
- peanut agglutinin
- ITC
- isothermal titration calorimetry
- LID
- LIM-interacting domain
- r.m.s.d.
- root mean square deviation
- Mena
- mammalian enabled
- LIM domain
- Lin-11, Isl1 and Mec-3 domain
- LMO
- LIM-only protein
- LIM-HD
- LIM-homeodomain
- VASP
- vasodilator-stimulated phosphoprotein.
REFERENCES
- 1. Shih Y. L., Rothfield L. (2006) Microbiol. Mol. Biol. Rev. 70, 729–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Michie K. A., Löwe J. (2006) Annu. Rev. Biochem. 75, 467–492 [DOI] [PubMed] [Google Scholar]
- 3. Erickson H. P. (2007) Bioessays 29, 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Derman A. I., Becker E. C., Truong B. D., Fujioka A., Tucey T. M., Erb M. L., Patterson P. C., Pogliano J. (2009) Mol. Microbiol. 73, 534–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goodson H. V., Hawse W. F. (2002) J. Cell Sci. 115, 2619–2622 [DOI] [PubMed] [Google Scholar]
- 6. Blessing C. A., Ugrinova G. T., Goodson H. V. (2004) Trends Cell Biol. 14, 435–442 [DOI] [PubMed] [Google Scholar]
- 7. Muller J., Oma Y., Vallar L., Friederich E., Poch O., Winsor B. (2005) Mol. Biol. Cell 16, 5736–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen M., Shen X. (2007) Curr. Opin. Cell Biol. 19, 326–330 [DOI] [PubMed] [Google Scholar]
- 9. Schroer T. A. (2004) Annu. Rev. Cell Dev. Biol. 20, 759–779 [DOI] [PubMed] [Google Scholar]
- 10. Vallee R. B., Williams J. C., Varma D., Barnhart L. E. (2004) J. Neurobiol. 58, 189–200 [DOI] [PubMed] [Google Scholar]
- 11. Kardon J. R., Vale R. D. (2009) Nat. Rev. Mol. Cell Biol. 10, 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goley E. D., Welch M. D. (2006) Nat. Rev. Mol. Cell Biol. 7, 713–726 [DOI] [PubMed] [Google Scholar]
- 13. Pollard T. D. (2007) Annu. Rev. Biophys. Biomol. Struct. 36, 451–477 [DOI] [PubMed] [Google Scholar]
- 14. Chhabra E. S., Higgs H. N. (2007) Nat. Cell Biol. 9, 1110–1121 [DOI] [PubMed] [Google Scholar]
- 15. Meagher R. B., Kandasamy M. K., McKinney E. C., Roy E. (2009) Int. Rev. Cell Mol. Biol. 277, 157–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chadwick B. P., Mull J., Helbling L. A., Gill S., Leyne M., Robbins C. M., Pinkett H. W., Makalowska I., Maayan C., Blumenfeld A., Axelrod F. B., Brownstein M., Gusella J. F., Slaugenhaupt S. A. (1999) Genomics 58, 302–309 [DOI] [PubMed] [Google Scholar]
- 17. Heid H., Figge U., Winter S., Kuhn C., Zimbelmann R., Franke W. (2002) Exp. Cell Res. 279, 177–187 [DOI] [PubMed] [Google Scholar]
- 18. Hisano M., Yamada S., Tanaka H., Nishimune Y., Nozaki M. (2003) Mol. Reprod. Dev. 65, 148–156 [DOI] [PubMed] [Google Scholar]
- 19. Tanaka H., Iguchi N., Egydio de Carvalho C., Tadokoro Y., Yomogida K., Nishimune Y. (2003) Biol. Reprod. 69, 475–482 [DOI] [PubMed] [Google Scholar]
- 20. Hara Y., Yamagata K., Oguchi K., Baba T. (2008) FEBS Lett. 582, 2998–3004 [DOI] [PubMed] [Google Scholar]
- 21. Coutts A. S., MacKenzie E., Griffith E., Black D. M. (2003) J. Cell Sci. 116, 897–906 [DOI] [PubMed] [Google Scholar]
- 22. Tatarelli C., Linnenbach A., Mimori K., Croce C. M. (2000) Genomics 68, 1–12 [DOI] [PubMed] [Google Scholar]
- 23. Tobias E. S., Hurlstone A. F., MacKenzie E., McFarlane R., Black D. M. (2001) Oncogene 20, 2844–2853 [DOI] [PubMed] [Google Scholar]
- 24. Drusco A., Zanesi N., Roldo C., Trapasso F., Farber J. L., Fong L. Y., Croce C. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10947–10951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sarti M., Sevignani C., Calin G. A., Aqeilan R., Shimizu M., Pentimalli F., Picchio M. C., Godwin A., Rosenberg A., Drusco A., Negrini M., Croce C. M. (2005) Clin. Cancer Res. 11, 806–813 [PubMed] [Google Scholar]
- 26. Mueller W., Nutt C. L., Ehrich M., Riemenschneider M. J., von Deimling A., van den Boom D., Louis D. N. (2007) Oncogene 26, 583–593 [DOI] [PubMed] [Google Scholar]
- 27. Garvalov B. K., Higgins T. E., Sutherland J. D., Zettl M., Scaplehorn N., Köcher T., Piddini E., Griffiths G., Way M. (2003) J. Cell Biol. 161, 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boëda B., Briggs D. C., Higgins T., Garvalov B. K., Fadden A. J., McDonald N. Q., Way M. (2007) Mol. Cell 28, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 29. Kadrmas J. L., Beckerle M. C. (2004) Nat. Rev. Mol. Cell Biol. 5, 920–931 [DOI] [PubMed] [Google Scholar]
- 30. Rietdorf J., Ploubidou A., Reckmann I., Holmström A., Frischknecht F., Zettl M., Zimmermann T., Way M. (2001) Nature Cell Biol. 3, 992–1000 [DOI] [PubMed] [Google Scholar]
- 31. Leslie A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26 [Google Scholar]
- 32. Evans P. R. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 33. Collaborative Computational Project, Number Four (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 34. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 35. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 36. Stein N. (2008) J. Appl. Crystallogr. 41, 641–643 [Google Scholar]
- 37. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 38. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 40. Kierszenbaum A. L., Rivkin E., Tres L. L. (2003) Mol. Biol. Cell 14, 4628–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kierszenbaum A. L., Rivkin E., Tres L. L. (2007) Soc. Reprod. Fertil. Suppl. 65, 33–43 [PubMed] [Google Scholar]
- 42. Moreno R. D., Alvarado C. P. (2006) Mol. Reprod. Dev. 73, 1430–1434 [DOI] [PubMed] [Google Scholar]
- 43. Kierszenbaum A. L., Rivkin E., Tres L. L. (2003) Cytogenet. Genome Res. 103, 337–344 [DOI] [PubMed] [Google Scholar]
- 44. Hyenne V., Harf J. C., Latz M., Maro B., Wolfrum U., Simmler M. C. (2007) Reproduction 133, 563–574 [DOI] [PubMed] [Google Scholar]
- 45. Behnen M., Murk K., Kursula P., Cappallo-Obermann H., Rothkegel M., Kierszenbaum A. L., Kirchhoff C. (2009) BMC Cell Biol. 10, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matthews J. M., Bhati M., Craig V. J., Deane J. E., Jeffries C., Lee C., Nancarrow A. L., Ryan D. P., Sunde M. (2008) Biochem. Soc. Trans. 36, 1393–1397 [DOI] [PubMed] [Google Scholar]
- 47. Deane J. E., Ryan D. P., Sunde M., Maher M. J., Guss J. M., Visvader J. E., Matthews J. M. (2004) EMBO J. 23, 3589–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. El Omari K., Hoosdally S. J., Tuladhar K., Karia D., Vyas P., Patient R., Porcher C., Mancini E. J. (2011) Blood 117, 2146–2156 [DOI] [PubMed] [Google Scholar]
- 49. Bhati M., Lee C., Nancarrow A. L., Lee M., Craig V. J., Bach I., Guss J. M., Mackay J. P., Matthews J. M. (2008) EMBO J. 27, 2018–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Konrat R., Kräutler B., Weiskirchen R., Bister K. (1998) J. Biol. Chem. 273, 23233–23240 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.