Abstract
The prominent characteristics of pluripotent stem cells are their unique capacity to self-renew and pluripotency. Although pluripotent stem cell proliferation is maintained by specific intracellular phosphorylation signaling events, it has not been well characterized how the resulting phosphorylated proteins are subsequently regulated. We here report that the peptidylprolyl isomerase Pin1 is indispensable for the self-renewal and maintenance of pluripotent stem cells via the regulation of phosphorylated Oct4 and other substrates. Pin1 expression was found to be up-regulated upon the induction of induced pluripotent stem (iPS) cells, and the forced expression of Pin1 with defined reprogramming factors was observed to further enhance the frequency of iPS cell generation. The inhibition of Pin1 activity significantly suppressed colony formation and induced the aberrant differentiation of human iPS cells as well as murine ES cells. We further found that Pin1 interacts with the phosphorylated Ser12-Pro motif of Oct4 and that this in turn facilitates the stability and transcriptional activity functions of Oct4. Our current findings thus uncover an atypical role for Pin1 as a putative regulator of the induction and maintenance of pluripotency via the control of phosphorylation signaling. These data suggest that the manipulation of Pin1 function could be a potential strategy for the stable induction and proliferation of human iPS cells.
Keywords: Prolyl Isomerase, Protein Phosphorylation, Protein Stability, Stem Cell, Transcription Factors, Oct4, Pin1, iPS Cells, Pluripotency, Self-renewal
Introduction
Stem cells are characterized by their ability to self-renew through mitotic cell division and to differentiate into a diverse range of specialized cell types (1, 2). Human pluripotent stem cell proliferation is maintained through the action of several transcription factors including Oct4 (octamer 4), SOX2, Klf-4, Nanog, and c-Myc, which perform reprogramming functions under the stimulatory effects of stem cell-specific growth factors, including basic fibroblast growth factor (3–5). Basic fibroblast growth factor signaling has been shown to be essential for pluripotency as its depletion from cell culture media leads to aberrant cell differentiation and cell death (6, 7). Fibroblast growth factors produce mitogenic effects in targeted cells via signaling through cell surface receptor tyrosine kinases (8). These kinases can initiate intracellular signaling in cells, which is transmitted and diffused by tyrosine phosphorylation of the assembled proteins and of cellular substrates, including protein kinases with specificity for serine/threonine residues (8, 9). Although this intracellular phosphorylation signaling might indeed contribute to the self-renewal and pluripotency of stem cells (10, 11), it has not yet been fully determined how these phosphorylated proteins are further regulated.
Protein phosphorylation is a fundamental mode of intracellular signal transduction in a variety of key cellular processes such as cell proliferation, differentiation, and morphogenesis (12). A pivotal signaling mechanism that controls the function of phosphorylated proteins is the cis-trans isomerization of phosphorylated Ser/Thr-Pro motifs by the peptidylprolyl isomerase Pin1 (13, 14). This modification regulates multiple intracellular signaling pathways, including ErbB2/Ras, Wnt/β-catenin, and NF-κB, and thus plays an important role in the etiology of several human diseases (15–18). These include various cancers, Alzheimer disease, and immune disorders (14, 17, 18). However, the role of Pin1 in regulating the properties of pluripotent stem cells has not been adequately investigated to date.
In our current study, we investigated the role of Pin1 in the self-renewal and stemness of pluripotent stem cells. We reveal that Pin1 is induced upon cellular reprogramming and that its blockade significantly inhibits the self-renewal and maintenance of human iPS2 cells in addition to murine ES cells. We find also that Pin1 can interact with phosphorylated Oct4 at the Ser12-Pro motif in this protein. This enhances the stability and hence the transcriptional activity of Oct4. Our present data thus suggest that Pin1 is indeed a putative regulator of the self-renewal and proliferation of pluripotent stem cells.
EXPERIMENTAL PROCEDURES
Colony Formation Analysis
Human iPS cells were obtained from the RIKEN BioResource Center (clone no. 201B7) (19). Cells were cultured in human embryonic stem cell culture medium (KnockOut Dulbecco's modified Eagle's medium (Invitrogen)) supplemented with 20% KnockOut SR (Invitrogen), 1% GlutaMAX (Invitrogen), 100 μm nonessential amino acids (Invitrogen), 50 μm β-mercaptoethanol, and 10 ng/ml basic fibroblast growth factor). Murine ES cells were cultured in human embryonic stem cell culture medium (KnockOut Dulbecco's modified Eagle's medium supplemented with 15% KnockOut SR, 1% GlutaMAX (Invitrogen), 100 μm nonessential amino acids, 50 μm β-mercaptoethanol, and 1000 units/ml recombinant human leukemia inhibitory factor) (20). Colony formation was scored by counting the number of alkaline phosphatase (AP)-positive colonies as described previously (21). The number of cells per colony was determined by manually counting the number of DAPI-stained cells (21).
Cell Reprogramming
MRC5 fibroblasts were transduced with retroviral vectors encoding reprogramming factors as described previously (19). Briefly, the retroviral vector plasmids pMXs-hOct4, pMXs-hSOX2, pMXs-hKLF4, pMXs-hcMYC (Addgene), and pVSV-G were introduced into Plat-E cells using Effectene transfection reagent (Qiagen). After 48 h, virus-containing supernatants were passed through a 0.45-μm filter and supplemented with 10 μg/ml hexadimethrine bromide (polybrene). Cells were seeded at 6 × 105 cells per 60 mm dish at 24 h before incubation in the virus/polybrene-containing supernatants for 16 h. After 6 days, cells were plated on irradiated mouse embryonic fibroblasts, and culture medium was replaced with the hESC culture medium 24 h later. Cells were maintained at 37 °C and 5% CO2 for 30 days.
Construction of Expression Vectors
Oct4 cDNA was subcloned into pcDNA3-HA expression vector (Invitrogen). Expression constructs of Oct4 were as follows: pcDNA-HA-Oct4 wild-type, amino acids 1–360; pcDNA-HA-Oct4 ΔC, amino acids 1–297; pcDNA-HA-Oct4 ΔN1, amino acids 138–360; pcDNA-HA-Oct4 ΔN2, amino acids 113–360; and pcDNA-HA-Oct4 ΔN3, amino acids 34–360. pcDNA-HA-Oct4-S12A was generated by KOD-Plus Mutagenesis Kit (Toyobo, Osaka, Japan) according to the manufacturer's instructions. The primers were 5′-CGCCCCCTCCAGGTGGT-3′ (forward) and 5′-CGAAGGCAAAATCTGAAGCC-3′ (reverse).
Gene Reporter Assay
A pGL3-fgf4 reporter plasmid containing an Oct-SOX binding cassette and the firefly luciferase gene was transfected with pRL-CMV (22). The −2601/+1 (nucleotide positions indicated with respect to the +1 translation start site) genomic fragment of the Oct4 promoter upstream region was amplified by PCR from human lymphocyte genomic DNA and cloned into the KpnI/HindIII sites of the pGL4-basic reporter plasmid (Promega, Madison, WI) as described previously (23). The primer sets were as follows: 5′-CCTGGTACCAGGATGGCAAGCTGAGAAACACTG-3′ and 5′-TCGCAAGCTTGCGAAGGGACTACTCAAC-3′. Cells were transfected with reporter plasmid vectors using Effectene (Qiagen) or Xfect Stem (Clontech). One day after transfection, the cells were resuspended in passive lysis buffer (Promega) and incubated for 15 min at room temperature. Luciferase activities were measured with a Dual-Luciferase reporter assay system (Promega) in accordance with the manufacturer's instructions.
GST Pulldown Assay and Immunoprecipitation Analysis
Cells were lysed with GST pulldown buffer (50 mm HEPES (pH 7.4), 150 mm NaCl, 10% glycerol, 1% Triton X-100, 1.5 mm MgCl2, 1 mm EGTA, 100 mm NaF, 1 mm Na3VO4, 1 mm DTT, 5 μg/ml leupeptin, 1 μg/ml pepstatin, and 0.2 mm PMSF) and incubated with 30 μl of glutathione-agarose beads containing either GST-Pin1 or GST at 4 °C for 2 h. The precipitated proteins were then washed three times with lysis buffer and subjected to SDS-PAGE. For immunoprecipitation, cells were lysed with Nonidet P-40 lysis buffer (10 mm Tris HCl (pH 7.4), 100 mm NaCl, 0.5% Nonidet P-40, 1 mm Na3VO4, 100 mm NaF, 5 μg/ml leupeptin, 1 μg/ml pepstatin, and 0.2 mm PMSF). Cell lysates were incubated for 1 h with protein A/G-Sepharose·nonimmunized IgG complexes. Supernatant fractions were recovered and immunoprecipitated with 5 μg of anti-Myc antibody and 30 μl protein A/G-Sepharose. After washing three times with lysis buffer, the pellets were analyzed by SDS-PAGE.
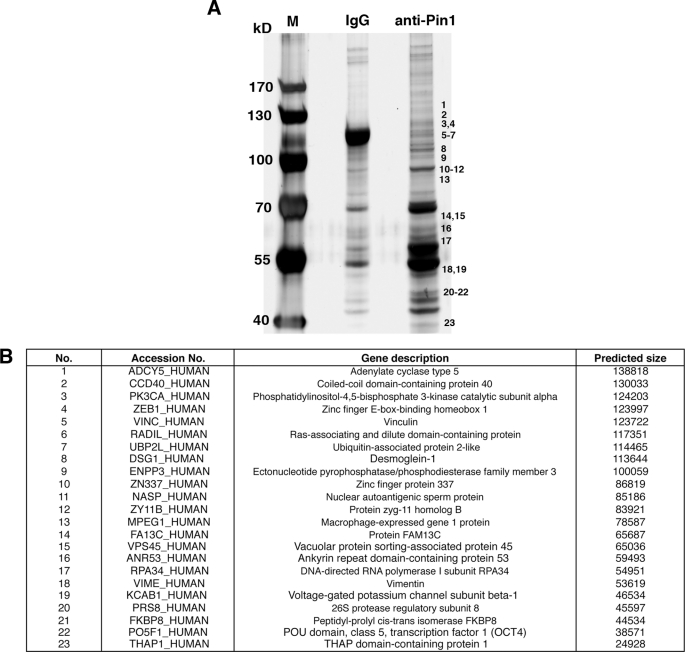
Proteomics Analysis
Human iPS cell lysates were processed for immunoprecipitation with a monoclonal anti-Pin1 antibody (clone 257417, R&D Systems) at 4 °C for 3 h followed by SDS-PAGE. Gel lanes corresponding to the region from ∼30 to 150 kDa were systematically excised, and the pieces were reduced, alkylated, and trypsinized. Peptides were analyzed by the linear ion trap Orbitrap hybrid mass spectrometer (Thermo Scientific). Protein identification was performed by peptide mass fingerprinting with the Mascot and Aldente search algorithms.
Quantitative Real-time PCR
Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's protocol. cDNA was synthesized using a cDNA synthesis kit (Toyobo, Osaka, Japan) and subjected to RT-PCR analysis with the SYBR Premix Ex gent Kit TaqII (Takara Bio, Shiga, Japan) using an Applied Biosystems 7300 real-time PCR System. The primer sets used were as follows: mOct4, 5′-CGTGTGAGGTGGAGTCTGGAGACC-3′ and 5′-ACTCGAACCACATCCTTCTCTAGCC-3′; mGAPDH, 5′-CCATGGAGAAGGCTGGGG-3′ and 5′-CAAAGTTGTCATGGATGACC-3′.
Teratoma Formation
Cells were harvested using accutase, collected into tubes, and centrifuged. The pellets were then suspended in human ESC culture medium. Fox Chase severe combined immunodeficiency mice (CREA, Tokyo, Japan) were injected with 2 × 106 cells mixed with an equal volume of Matrigel (BD Biosciences). Frozen tumor tissues embedded in optimum cutting temperature compound were sliced by cryosectioning and stained with hematoxylin and eosin.
RESULTS
Pin1 Is Induced upon Cellular Reprogramming and Enhances Generation of iPS Cells
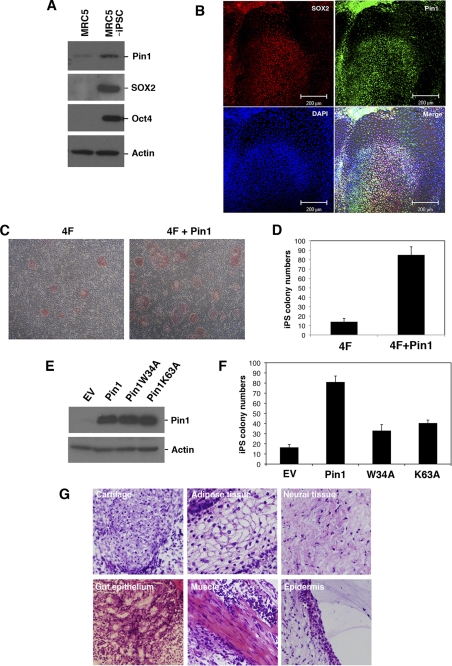
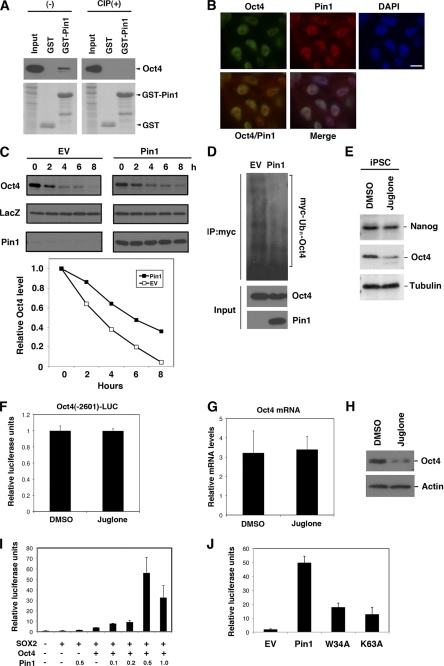
To examine the role of Pin1 in cellular reprogramming and pluripotency, we initially investigated the expression levels of this prolyl isomerase in human iPS cells. Pin1 was found to be significantly induced upon the generation of iPS cells derived from MRC5 human fibroblasts (Fig. 1A). Immunofluorescent analysis further revealed that Pin1 is selectively expressed in SOX2-positive pluripotent stem cells, whereas its expression was found to be significantly suppressed in the surrounding SOX2-negative differentiated cells (Fig. 1B). These results indicate that Pin1 is preferentially expressed in reprogramming stem cells.
FIGURE 1.
Pin1 is preferentially expressed in human iPS cells. A, immunoblotting analysis of Oct4, SOX2, and Pin1 in MRC5 and MRC5-derived iPS cells. Actin was used as a loading control. iPSC, induced pluripotent stem cells; EV, empty vector. B, immunofluorescent analysis of Pin1 and SOX2 in human iPS cells. Representative images of phase-contrast microscopy and fluorescent immunocytochemistry for SOX2 (red) and Pin1 (green) are shown. Nuclei are indicated by DAPI staining (blue). Note that Pin1 is highly expressed in SOX2-positive pluripotent stem cells. C and D, Pin1 expression enhances 4F (Oct4, SOX2, Klf4, and c-Myc)-induced iPS cell induction. MRC5 fibroblasts were infected with retrovirus vectors encoding 4F and co-infected with those encoding either empty vector or Pin1. A representative picture of colony formations stained with AP is shown (C). The numbers of AP-positive colonies were scored in three independent experiments (D). Note that the co-introduction of Pin1 with 4F increases the frequency of iPS colony formation. E and F, MRC5 fibroblasts were infected with retrovirus vectors encoding 4F and co-infected with those encoding empty vector, HA-tagged wild-type Pin1, or its W34A or K63A mutants. The expression levels of HA-Pin1 or its mutants in infected MRC5 cells were analyzed by immunoblotting analysis with anti-HA antibody (E). The number of AP-positive colonies was scored in three independent experiments (F). G, teratoma tissue derived from human iPS cells induced by 4F and Pin1. iPS cells were transplanted subcutaneously into immunodeficient mice (2 × 106/mouse). Representative images of hematoxylin and eosin stained tumor with light microscope (200×) are shown.
We next evaluated whether Pin1 affects the reprogramming of somatic cells into iPS cells. The co-infection of a Pin1-encoding retrovirus vector with those encoding four defined reprogramming factors (4F; SOX2, Oct4, Klf-4, and c-Myc) (24) notably boosted the generation of AP-positive iPS cell colonies compared with an induction of human fibroblast MRC5 cells with only four iPS factors (Fig. 1, C and D). We next performed a parallel experiment using either a WW-domain (binding domain) mutant (W34A) or a peptidyl prolyl isomerase-domain (catalytic domain) mutant (K63A) of Pin1. We confirmed the equivalent expression of each of these mutants and wild-type Pin1 (Fig. 1E). Neither of these mutants could boost iPS cell colony formation to the level seen with wild-type Pin1 (Fig. 1F), indicating that both the WW and PPIase domains are required for this function.
To test pluripotency in vivo, we transplanted 4F plus Pin1-introduced iPS cells subcutaneously into the dorsal flanks of immunodeficient mice. Nine weeks after injection, we observed teratoma formation composed of various tissues including gut-like epithelial tissues (endoderm), striated muscle (mesoderm), cartilage (mesoderm), neural tissues (ectoderm), and epidermal tissues (ectoderm) (Fig. 1G). These results indicate that the expression of Pin1 with defined reprogramming factors accelerates the frequency of iPS cell generation.
Pin1 Is Required for Pluripotent Stem Cell Self-renewal and Colony Formation
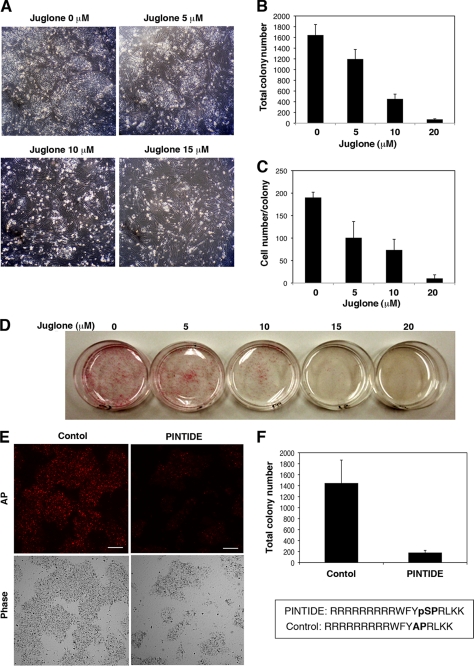
We next addressed whether Pin1 indeed plays any roles in the self-renewal of human iPS cells. iPS cells were dissociated with accutase and then plated at a clonal density in the presence of several concentrations of the selective Pin1 inhibitor juglone (5-hydroxy-1,4-naphthoquinone) (25, 26). The blockade of Pin1 by juglone considerably reduced both the numbers and size of the colonies in a dose-dependent manner (Fig. 2, A–C). It was notable also that the concentration of juglone used did not illicit nonspecific toxic effects in the feeder mouse embryonic fibroblast cells (Fig. 2A and data not shown). The effect of Pin1 inhibition upon colony formation was also confirmed in feeder-free cultures of human iPS cells by AP staining (Fig. 2D). Moreover, treatment with the Pin1 inhibitory phosphopeptide PINTIDE (27), but not a nonphosphorylated control peptide, significantly reduced the colony formation of human iPS cells (Fig. 2, E and F).
FIGURE 2.
Defective self-renewal of human iPS cells caused by Pin1 inhibition. A–C, human iPS cells were dissociated with accutase and then plated on a feeder cell layer at a clonal density in the presence of the indicated concentrations of juglone for 3 days. Colony formation was analyzed by phase-contrast microscopy (A). The number of colonies was counted at 3 days after treatment (B). The number of cells per colony was determined by manually counting the DAPI-stained cells (C). Data are the mean ± S.E. D, human iPS cells were plated at a clonal density on the feeder-free culture in the presence of the indicated concentrations of juglone followed by AP staining. E and F, human iPS cells were dissociated with accutase and then plated on feeder-free dishes at a clonal density in the presence of 50 μg/ml of the Pin1 inhibitory phosphopeptide PINTIDE (RRRRRRRRRWFYpSPRLKK) or a nonphosphorylated control peptide (RRRRRRRRRWFYAPRLKK) for 48 h (E). AP-positive colony numbers were scored (F). Data are the mean ± S.E. Scale bar, 50 μm.
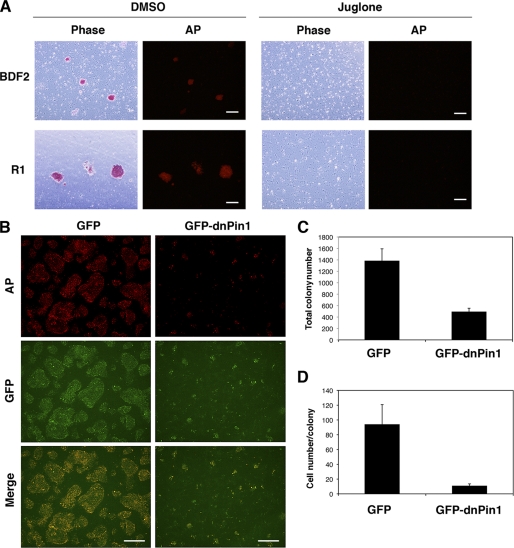
We next investigated the effects of Pin1 inhibition upon colony formation in murine ES cells. The blockade of Pin1 by juglone significantly reduced the colony numbers in two different murine ES cell types, BDF2 and R1 (Fig. 3A). The adenovirus-mediated transduction of a GFP-fused dominant-negative Pin1 (GFP-dnPin1) (28), but not a GFP control, significantly suppressed colony formation in murine ES (R1) cells manifesting as a considerable reduction in both the numbers and colony size of the murine ES cells (Fig. 3, B–D). These results together demonstrate that Pin1 is indispensable for the self-renewal and proliferation of pluripotent stem cells.
FIGURE 3.
Pin1 inhibition suppresses colony formation in murine ES cells. A, two different murine ES cell types (BDF2 and R1) were plated on gelatin-coated dishes and treated with either DMSO or juglone (10 μm). Colonies were stained with AP (red). Scale bar, 200 μm. B–D, murine ES cells (R1) were infected with an adenovirus vector encoding either GFP or GFP-dnPin1 (3000 viral particles/cell). The cells were then stained with AP (red) and DAPI and analyzed by immunofluorescent microscopy (B). Scale bar, 200 μm. The total colony number (C) and the number of cells per colony (D) were then determined. Data are the mean ± S.E.
Pin1 Functions in Maintenance of Pluripotency
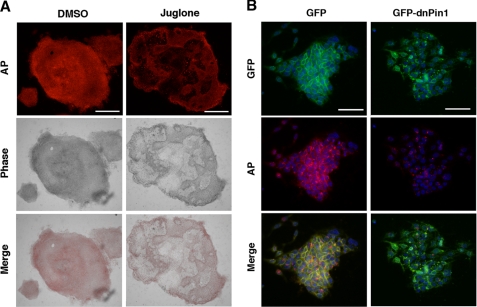
We next asked whether Pin1 has any roles in the maintenance of pluripotency in stem cells. Human iPS cells were dissociated and then cultured for 5 days to form colonies. When human iPS cells are cultured in hES medium supplemented with basic fibroblast growth factor, the overwhelming majority of the cells in the colonies are undifferentiated (Fig. 4A). However, treatment with juglone resulted in aberrant cell differentiation resulting in a “mosaic pattern” of iPS cell colonies following AP staining (Fig. 4A). Similarly, the adenovirus-mediated transduction of GFP-dnPin1, but not a GFP control, prominently reduced the number of AP-positive undifferentiated cells in murine ES cell colonies (Fig. 4B). These results together indicate that Pin1 can sustain pluripotent stem cells in an undifferentiated state in addition to the enhancement of self-renewal.
FIGURE 4.
Pin1 inhibition leads to the aberrant cell differentiation of human iPS cells. A, human iPS cells were cultured for 5 days before forming colonies and then treated with either DMSO or juglone (10 μm) for 3 days. The cells were then stained with AP (red). Representative images of phase-contrast microscopy and fluorescent immunocytochemistry are shown. Scale bar, 200 μm. B, mouse ES cells were cultured for 2 days before forming colonies and then infected with an adenovirus vector encoding either GFP or GFP-dnPin1 (3000 viral particles/cell). After 48 h, the cells were then stained with AP (red) and DAPI (blue) and analyzed by immunofluorescent microscopy. Scale bar, 50 μm.
Identification of Pin1 Binding Proteins in Human iPS Cells
Our initial data indicated that Pin1 could enhance the function of reprogramming factors during the induction and maintenance of pluripotency. We next identified the substrates targeted by Pin1 in human iPS cells. Using a monoclonal Pin1 antibody, we co-immunoprecipitated proteins from human iPS cell lysates treated with a phosphatase inhibitor mixture. These isolated immune complexes were then boiled and resolved by one-dimensional SDS-PAGE, and the proteins were visualized using silver staining. Continuous regions of the gel corresponding to proteins of ∼30 to 150 kDa in size were systematically excised (Fig. 5A), digested with trypsin, and analyzed in a linear ion trap (LTQ) Orbitrap hybrid mass spectrometer. Peptide mass fingerprinting with the Mascot and Aldente search algorithms subsequently identified 23 Pin1 interacting proteins in human iPS cells (Fig. 5B). Notably, these Pin1-binding proteins included the pluripotent transcription factor Oct4. Because Oct4 has been shown to be a master regulator of pluripotency (29), we decided to further analyze the Oct4-Pin1 interaction.
FIGURE 5.
Identification of Pin1-binding proteins in human iPS cells. A and B, lysates of human iPS cells were subjected to immunoprecipitation with either non-immunized control mouse IgG (IgG) or mouse anti-Pin1 monoclonal antibodies. Proteins bound to protein A/G-agarose beads were isolated, resolved by SDS-PAGE, and detected by silver staining (A). M indicates protein marker. Excised gel bands were digested with trypsin and analyzed on a linear ion trap (LTQ) Orbitrap hybrid mass spectrometer followed by peptide mass fingerprinting with the Mascot and Aldente search algorithms (B).
Pin1 Binds and Regulates Protein Stability of Oct4
To further characterize the Oct4-Pin1 interaction, a GST pulldown analysis was initially performed. We found that recombinant GST-Pin1, but not GST alone, binds Oct4. This association was completely abolished by pretreatment of the cell lysates with calf intestine alkaline phosphatase (Fig. 6A), indicating that Pin1 binds phosphorylated Oct4. Immunofluorescence analysis further demonstrated that Pin1 co-localizes with Oct4 in the nuclei of iPS cells (Fig. 6B). Pin1 has been shown to regulate the stability of its substrate proteins upon binding (17), and we thus addressed whether this was the case for Oct4. Cycloheximide analysis using HeLa cells transfected with Oct4 alone or co-transfected with Oct4 and Pin1 revealed that the protein half-life of Oct4 is significantly enhanced in cells co-expressing Pin1 (Fig. 6C). Moreover, immunoprecipitation analysis with cells co-transfected with Oct4 and Myc-tagged ubiquitin, with or without Pin1 co-transfection, further revealed that Pin1 overexpression significantly reduces the polyubiquitination of the Oct4 protein (Fig. 6D). Consistently, the Oct4 protein expression level was significantly reduced in human iPS cells treated with juglone as compared with control cells (Fig. 6E). These results together confirm that Pin1 enhances the protein stability of Oct4 by suppressing ubiquitin proteasome-mediated proteolysis.
FIGURE 6.
Pin1 interacts with phosphorylated Oct4 and enhances its transcriptional activity. A, human iPS cell lysates treated or untreated with calf intestine alkaline phosphatase were subjected to GST pulldown analysis with either GST or GST-Pin1, followed by immunoblotting analysis with anti-Oct4 antibody (upper panel). Coomassie staining for the GST or GST-Pin1 used in the assay is shown in the lower panel. B, human iPS cells were fixed with 4% paraformaldehyde and then co-immunostained with monoclonal antibodies against Oct4 (green) and polyclonal antibodies against Pin1 (red). Cells were then analyzed by confocal microscopy. Scale bar, 10 μm. C, HeLa cells transfected with the indicated vectors and HA-LacZ cells were treated with cycloheximide and harvested at the indicated time points. This was followed by immunoblotting analysis with Oct4, Pin1, and HA antibodies (upper panel). Quantitative data are shown in the lower panel. D, HeLa cells were transfected with Myc-tagged ubiquitin, Oct4, and co-transfected with either empty vector (EV) or Pin1. Cells were then treated with MG-132 for 12 h, and lysates were prepared and immunoprecipitated with anti-Myc antibody followed by immunoblotting analysis with anti-Oct4 antibody. Total cell lysates prior to immunoprecipitation (input) were immunoblotted with anti-Pin1 or anti-Oct4 antibody. E, human iPS cells were plated on Matrigel-coated feeder-free dishes and treated with either DMSO or juglone (20 μm) for 24 h. Cell lysates were then processed for immunoblotting analysis with anti-Nanog, anti-Oct4, or anti-tubulin antibodies. F, a plasmid containing the luciferase (LUC) gene flanked with 2601 bp of the Oct4 5′-upstream region was transfected into murine ES cells. The resulting cells were cultured in Matrigel-coated feeder-free dishes and treated with either DMSO or juglone (10 μm) for 24 h, and analyzed by gene reporter assay. G, murine ES cells were cultured in Matrigel-coated feeder-free dishes and treated with either DMSO or juglone (10 μm) for 24 h. Total RNAs were then extracted and reverse-transcribed. These preparations were then subjected to quantitative RT-PCR analysis for Oct4. The transcript levels were normalized using GAPDH. H, murine ES cells were cultured in Matrigel-coated feeder-free dishes and treated with either DMSO or juglone (10 μm) for 24 h. Cell lysates were then processed for immunoblotting analysis with either anti-Oct4 or anti-β-actin antibody. I, HeLa cells were transiently transfected with plasmids encoding Oct4, SOX2, or Pin1 and co-transfected with Oct-SOX reporter gene and pRL-CMV. At 24 h post-transfection, the cells were collected and subjected to a gene reporter assay. J, HeLa cells were transiently transfected with an Oct-SOX reporter gene and co-transfected with plasmids encoding wild-type Pin1 or its W34A or K63A mutants, together with Oct4 and SOX2. At 24 h post-transfection, the cells were collected and subjected to a gene reporter assay.
We next investigated the gene expression profile of Oct4 during the inhibition of Pin1. Murine ES cells were transfected with pGL4-Oct4-2601 promoter (harboring a genomic fragment of the Oct4 gene 5′-upstream region) and treated or not with juglone. Pin1 inhibition by juglone did not affect the transcriptional activity of the Oct4 promoter (Fig. 6F). Consistently, the results of parallel quantitative RT-PCR analysis demonstrated that the Oct4 mRNA level was not significantly altered by Pin1 inhibition (Fig. 6G), whereas the Oct4 protein level was significantly reduced by juglone treatment, as revealed by immunoblot analysis (Fig. 6H). These results together indicate that Pin1 regulates the protein stability of Oct4 but not Oct4 transcription.
We next addressed whether Pin1 enhances the transcriptional activity of the Oct4 protein. A luciferase reporter assay using the Oct-Sox enhancer region derived from the FGF4 gene was performed in HeLa cells co-transfected with Oct4, SOX2 or Pin1. Although the sole expression of Pin1 had no significant effects, the co-expression of Oct4 and Pin1 produced a significant increase in reporter activity in a dose-dependent fashion (Fig. 6I). This indicated that Pin1 promotes Oct4-mediated transcriptional activation. We performed a parallel experiment using the W34A and K63A Pin1 mutants. Neither of these mutants up-regulated the transcriptional activity of Oct4 to the levels seen with wild-type Pin1 (Fig. 6J), indicating that both the WW and PPIase domains are required for this function.
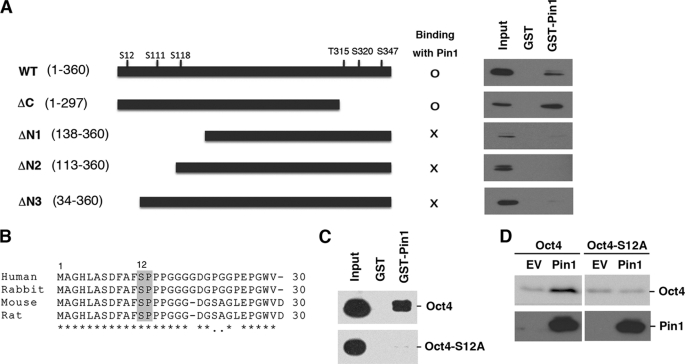
Pin1 Interacts with Ser12-Pro motif of Oct4
To identify the specific Pin1 binding site within the Oct4 protein, we generated several Oct4 deletion mutants and performed GST-pulldown analysis. These experiments revealed that a C-terminal Oct4 deletion mutant (representing amino acids 1–297) could still bind Pin1, but that three extended N-terminal deletion mutants (amino acids 138–360, 113–360, or 34–360) failed to do so (Fig. 7A). These data indicate that Pin1 binds to Oct4 in the region between amino acids 1 and 34. Previous reports have indicated that Pin1 can bind only phosphorylated Ser/Thr-Pro motifs (17, 27) of which only one (Ser12-Pro) exists between residues 1 and 34 in the Oct4 protein. Interestingly, this motif is conserved between various species including human, mouse, rat, and rabbit (Fig. 7B). We generated an Oct4 site-directed mutant at this site by substituting serine 12 for alanine (S12A). GST pulldown analysis subsequently revealed that Pin1 binds wild-type Oct4, but not its S12A mutant (Fig. 7C). These results confirm that Pin1 indeed bind the phosphorylated Ser12-Pro motif of Oct4.
FIGURE 7.
Pin1 interacts with the Ser12-Pro motif of Oct4. A, schematic representation of the Oct4 deletion mutants generated in this study (left panel). HeLa cells were transfected with the indicated Oct4 deletion mutants for 24 h. Cell lysates were then prepared and subjected to GST pulldown analysis with either GST or GST-Pin1 followed by immunoblotting analysis with Oct4 antibodies (right panel). B, amino acid sequence alignment of the human, rabbit, mouse, and rat Oct4 proteins. The conserved Ser12-Pro motifs are boxed. C, HeLa cells were transfected with the Oct4 site-directed mutant Oct4-S12A and subjected to GST pulldown analysis. D, HeLa cells were transfected with wild-type Oct4 or its S12A mutant with or without Pin1. After 24 h, the cells were subjected to immunoblotting analysis with an anti-Oct4 antibody.
To further examine the functional interactions between Pin1 and Oct4 on this site, we next investigated the nature of the S12A mutant in terms of its protein expression in the presence of Pin1. HeLa cells were transfected with either wild-type Oct4 or its S12A mutant and co-transfected with Pin1. This was followed by immunoblotting analysis. We found that Pin1 increased the expression levels of wild-type Oct4, but not the S12A mutant (Fig. 7D).
DISCUSSION
In our present study, we report that Pin1 is an essential regulator of the self-renewal and maintenance of pluripotent stem cells. We further found the following: 1) Pin1 is induced upon the induction of human iPS cells; 2) the co-expression of Pin1 with defined reprogramming factors significantly enhances the frequency of iPS cell induction; 3) the blockade of Pin1 significantly inhibits the colony formation of dissociated human iPS cells and murine ES cells; 4) Pin1 inhibition leads to the aberrant cell differentiation in human iPS cells and murine ES cells after forming colonies; 5) Oct4 is a putative Pin1 substrate in human iPS cells; and 6) Pin1 interacts with Oct4 at its Ser12-Pro motif and facilitates its stability and enhanced transcriptional activity. Our findings thus uncover a novel role of Pin1 as a putative regulator of the self-renewal and survival of pluripotent stem cells via Oct4 function.
Our current results add to previous findings indicating that Pin1 is a multifunctional protein that mediates various phosphorylated proteins involved in divergent cellular processes (17). This implicates Pin1 as a modulator of multiple signaling pathways depending on the cell type and biological context. Indeed, we demonstrate in our present study that Pin1 is a crucial regulator of the phosphorylation-dependent intracellular signaling network that controls cellular stemness and pluripotency. Moreover, iPS cells induced by the expression of four Yamanaka factors (Oct4, SOX2, Klf4, and c-Myc) led to a high expression level of Pin1, and these cells were found to be dependent on Pin1 function. This suggests that Pin1 could be one of the crucial factors in the induction of iPS cells from somatic cells that functions by cooperating with reprogramming transcription factors.
The molecular mechanisms underlying the regulation of Pin1 in the induction and maintenance of pluripotency are likely to be highly complex given that Pin1 interacts with multiple substrates in pluripotent stem cells, as revealed by our proteomics analysis. However, our current findings also indicate that Pin1 is involved in the growth and maintenance of pluripotency in stem cells through its phosphorylation-dependent prolyl isomerization of substrates such as Oct4. In this regard, a recent report by Moretto-Zita et al. (30) has demonstrated that Pin1 can also associate with another pluripotent transcription factor, Nanog, in murine ES cells and sustain the self-renewal and teratoma formation of these cells in immunodeficient mice. These results indicate that Pin1 is a crucial modulator of the transcription factor network governing cellular stemness. It is possible also that Pin1 could regulate this process by modulating the function of other substrates. Further studies of Pin1 function in stem cells at various stages might shed new light on the underlying molecular pathways and factors that control self-renewal and multipotency.
It has been demonstrated that Pin1 knock-out mice develop normally but display some proliferation abnormalities, including a decreased body weight, retinal degeneration, and impaired mammary gland development (31, 32). Pin1 knock-out mice also exhibit testicular atrophy with a significantly impaired proliferation of primordial germ cells and the progressive loss of spermatogenic cells (33). These phenotypes can now be attributed to the impaired maintenance and proliferation of germ-related stem cells due to the loss of Pin1 function.
In many circumstances, Pin1 acts as either a repressor or an enhancer of the degradation of substrate proteins (15–17, 34). Our current data now additionally demonstrate that Pin1 can also prolong the protein half-life of Oct4, thereby enhancing its transcriptional activity. Oct4 has been shown to be regulated by post-translational modifications such as SUMOlylation (35). Our current findings reveal that Oct4 is also regulated by phosphorylation and subsequent prolyl isomerization. Identification of the kinase(s) responsible for the association of Pin1 and Oct4 will enhance our understanding of the regulatory pathways that operate during and after the induction of pluripotency.
It is desirable to utilize pluripotent stem cells such as iPS cells for future regenerative medicine applications. However, there are already concerns surrounding the use of iPS cells in a clinical setting because prior studies have suggested that they are likely to develop cancers (4, 36). Our current findings suggest, however, that the Pin1 inhibition could effectively block the proliferation of iPS cells in an undifferentiated state. Pin1 could therefore act as a molecular switch that can reversibly control the proliferation and survival of iPS cells, thereby reducing the risk of cell transformation and tumor formation.
Acknowledgments
We are grateful to the Riken Bioresource Center for providing human iPS cells and MRC5 fibroblasts. We also thank M. Machida, A. Hosoda, and S. Yoshizaki for comments on the manuscript and critical discussions and S. Baba and T. Taniguchi for technical assistance.
This work was supported in part by grants from the Takeda Science Foundation, Uehara Memorial Foundation, and Kanagawa Nanbyo Foundation (to A. R.).
- iPS
- induced pluripotent stem
- AP
- alkaline phosphatase
- dnPin1
- dominant-negative Pin1
- 4F
- four reprogramming factors
- DMSO
- dimethyl sulfoxide
- SUMO
- small ubiquitin-like modifier
- Oct4
- Octamer 4.
REFERENCES
- 1. Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998) Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 2. Watt F. M., Hogan B. L. (2000) Science 287, 1427–1430 [DOI] [PubMed] [Google Scholar]
- 3. Lewitzky M., Yamanaka S. (2007) Curr. Opin. Biotechnol. 18, 467–473 [DOI] [PubMed] [Google Scholar]
- 4. Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 5. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eiselleova L., Matulka K., Kriz V., Kunova M., Schmidtova Z., Neradil J., Tichy B., Dvorakova D., Pospisilova S., Hampl A., Dvorak P. (2009) Stem Cells 27, 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dvorak P., Dvorakova D., Koskova S., Vodinska M., Najvirtova M., Krekac D., Hampl A. (2005) Stem Cells 23, 1200–1211 [DOI] [PubMed] [Google Scholar]
- 8. Li J., Wang G., Wang C., Zhao Y., Zhang H., Tan Z., Song Z., Ding M., Deng H. (2007) Differentiation 75, 299–307 [DOI] [PubMed] [Google Scholar]
- 9. Sun H., Tonks N. K. (1994) Trends Biochem. Sci. 19, 480–485 [DOI] [PubMed] [Google Scholar]
- 10. Brill L. M., Xiong W., Lee K. B., Ficarro S. B., Crain A., Xu Y., Terskikh A., Snyder E. Y., Ding S. (2009) Cell Stem Cell 5, 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prudhomme W., Daley G. Q., Zandstra P., Lauffenburger D. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2900–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hunter T. (2009) Curr. Opin. Cell Biol. 21, 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu K. P., Hanes S. D., Hunter T. (1996) Nature 380, 544–547 [DOI] [PubMed] [Google Scholar]
- 14. Ryo A., Liou Y. C., Lu K. P., Wulf G. (2003) J. Cell Sci. 116, 773–783 [DOI] [PubMed] [Google Scholar]
- 15. Ryo A., Suizu F., Yoshida Y., Perrem K., Liou Y. C., Wulf G., Rottapel R., Yamaoka S., Lu K. P. (2003) Mol. Cell 12, 1413–1426 [DOI] [PubMed] [Google Scholar]
- 16. Ryo A., Nakamura M., Wulf G., Liou Y. C., Lu K. P. (2001) Nat. Cell Biol. 3, 793–801 [DOI] [PubMed] [Google Scholar]
- 17. Lu K. P., Zhou X. Z. (2007) Nat. Rev. Mol. Cell Biol. 8, 904–916 [DOI] [PubMed] [Google Scholar]
- 18. Esnault S., Shen Z. J., Malter J. S. (2008) Crit. Rev. Immunol. 28, 45–60 [DOI] [PubMed] [Google Scholar]
- 19. Takahashi K., Okita K., Nakagawa M., Yamanaka S. (2007) Nat. Protoc. 2, 3081–3089 [DOI] [PubMed] [Google Scholar]
- 20. Yamada M., Hamatani T., Akutsu H., Chikazawa N., Kuji N., Yoshimura Y., Umezawa A. (2010) Hum. Mol. Genet. 19, 480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Y., Labosky P. A. (2008) Stem Cells 26, 2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A. A., Ko M. S., Niwa H. (2007) Nat. Cell Biol. 9, 625–635 [DOI] [PubMed] [Google Scholar]
- 23. Yang H. M., Do H. J., Oh J. H., Kim J. H., Choi S. Y., Cha K. Y., Chung H. M., Kim J. H. (2005) J. Cell. Biochem. 96, 821–830 [DOI] [PubMed] [Google Scholar]
- 24. Takahashi K., Ichisaka T., Yamanaka S. (2006) Methods Mol. Biol. 329, 449–458 [DOI] [PubMed] [Google Scholar]
- 25. Hennig L., Christner C., Kipping M., Schelbert B., Rücknagel K. P., Grabley S., Küllertz G., Fischer G. (1998) Biochemistry 37, 5953–5960 [DOI] [PubMed] [Google Scholar]
- 26. Shen Z. J., Esnault S., Schinzel A., Borner C., Malter J. S. (2009) Nat. Immunol. 10, 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yaffe M. B., Schutkowski M., Shen M., Zhou X. Z., Stukenberg P. T., Rahfeld J. U., Xu J., Kuang J., Kirschner M. W., Fischer G., Cantley L. C., Lu K. P. (1997) Science 278, 1957–1960 [DOI] [PubMed] [Google Scholar]
- 28. Lu P. J., Zhou X. Z., Liou Y. C., Noel J. P., Lu K. P. (2002) J. Biol. Chem. 277, 2381–2384 [DOI] [PubMed] [Google Scholar]
- 29. Niwa H., Miyazaki J., Smith A. G. (2000) Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 30. Moretto-Zita M., Jin H., Shen Z., Zhao T., Briggs S. P., Xu Y. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13312–13317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fujimori F., Takahashi K., Uchida C., Uchida T. (1999) Biochem. Biophys. Res. Commun. 265, 658–663 [DOI] [PubMed] [Google Scholar]
- 32. Liou Y. C., Ryo A., Huang H. K., Lu P. J., Bronson R., Fujimori F., Uchida T., Hunter T., Lu K. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Atchison F. W., Means A. R. (2003) Biol. Reprod. 69, 1989–1997 [DOI] [PubMed] [Google Scholar]
- 34. Ryo A., Hirai A., Nishi M., Liou Y. C., Perrem K., Lin S. C., Hirano H., Lee S. W., Aoki I. (2007) J. Biol. Chem. 282, 36671–36681 [DOI] [PubMed] [Google Scholar]
- 35. Zhang Z., Liao B., Xu M., Jin Y. (2007) FASEB J. 21, 3042–3051 [DOI] [PubMed] [Google Scholar]
- 36. Knoepfler P. S. (2009) Stem Cells 27, 1050–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]