Abstract
Cystic Fibrosis (CF) is characterized by a massive proinflammatory phenotype in the lung arising from profound expression of inflammatory genes, including interleukin-8 (IL-8). We have previously reported that IL-8 mRNA is stabilized in CF lung epithelial cells, resulting in concomitant hyperexpression of IL-8 protein. However, the mechanistic link between mutations in CFTR and acquisition of the proinflammatory phenotype in the CF airway has remained elusive. We hypothesized that specific microRNAs (miRNAs) might mediate this linkage. To identify the potential link, we screened an miRNA library for differential expression in ΔF508-CFTR and wild type CFTR lung epithelial cell lines. Of 22 differentially and significantly expressed miRNAs, we found that expression of miR-155 was more than 5-fold elevated in CF IB3-1 lung epithelial cells in culture, compared with control IB3-1/S9 cells. Clinically, miR-155 was also highly expressed in CF lung epithelial cells and circulating CF neutrophils biopsied from CF patients. We report here that high levels of miR-155 specifically reduced levels of SHIP1, thereby promoting PI3K/Akt activation. However, overexpressing SHIP1 or inhibition of PI3K in CF cells suppressed IL-8 expression. Finally, we found that phospho-Akt levels were elevated in CF lung epithelial cells and were specifically lowered by either antagomir-155 or elevated expression of SHIP1. We therefore suggest that elevated miR-155 contributes to the proinflammatory expression of IL-8 in CF lung epithelial cells by lowering SHIP1 expression and thereby activating the PI3K/Akt signaling pathway. These data suggest that miR-155 may play an important role in the activation of IL-8-dependent inflammation in CF.
Keywords: Chemokines, Cystic Fibrosis, Inflammation, MicroRNA, RNA Turnover
Introduction
Cystic fibrosis (CF),2 the most common autosomal recessive disease in the United States and Europe, is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene (1–4). CFTR mutations, of which the most common is ΔF508-CFTR, cause a massive proinflammatory phenotype in the lung, which manifests in the airway by high levels of IL-8 and other proinflammatory cytokines and chemokines (5–7). IL-8 is the most potent known chemotactic agent for neutrophils (8) and is constitutively secreted from CF lung epithelial cells (9). The enhanced secretion of IL-8 seems to be an intrinsic property of the CF epithelium because fetal CF lung epithelium also spontaneously secretes high IL-8 levels into the airway as early as the 15th week of gestation (10, 11). However, the mechanism by which a mutation in CFTR causes up-regulated levels of IL-8 expression remains poorly understood.
The expression of proinflammatory genes, such as IL-8, is known to be regulated by post-transcriptional mechanisms. For example, the stability of mRNAs encoding many inflammatory genes, including IL-8, is regulated by interactions between AU-rich elements (AREs) in the 3′-untranslated region (3′-UTR) and specific ARE-binding proteins. In the case of CF lung epithelial cells in culture, which constitutively secrete high levels of IL-8, we have recently reported that high levels of IL-8 mRNA are sustained by mutation-dependent reduction in the ARE-binding protein tristetraprolin (TTP) (12). We also found a similarly low level of TTP in primary CF lung cells, which had been obtained acutely by brush biopsy of CF patients. Finally, we found experimentally that elevation in TTP directly reduced the stability of IL-8 mRNA, and, concomitantly, reduced the level of secreted IL-8. Thus hypersecretion of IL-8 from the CF lung epithelium seems to involve a loss of post-transcriptional regulation by TTP. However, it also became clear that low levels of TTP might not be the only mechanism underpinning CF lung inflammation. For example, as mentioned above, the CF airway is characterized not only by high levels of IL-8 but also by high levels of TNFα, IL-6, and many other potent proinflammatory analytes (5–7). Furthermore, many of the CF-specific proinflammatory processes have been associated with up-regulation of both TNFα/NFκB signaling (13, 14) and TGFβ-1 signaling (15). Thus, the CFTR mutation appears to target a proinflammatory regulatory mechanism with simultaneous deleterious effects on many proinflammatory genes.
MicroRNAs (miRNAs) may provide such a pleiotropic mechanism. miRNAs mediate mRNA instability by action in the 3′-UTRs of target genes (16–19). There are nearly 1000 unique miRNAs in the human genome, each of which individually targets ∼200 different mRNAs (20–25). Furthermore, it has been reported that, in response to certain kinds of stress, miRNAs can switch from a normally translational repressor mode to that of a translational activator (26, 27). Taken together, it appears that a relatively few miRNAs can regulate as much as 20–30% of the human genome. Recently, specific miRNAs have been reported to be associated with diabetes (28, 29), cancer (30–33), heart disease (34, 35), cell cycle (36), and development (17). Importantly, functional suppression of miRNAs can be achieved, both in vitro and in vivo, by antagomirs, which are chemically engineered oligonucleotides that are antisense to miRNAs (37). However, the possibility of an association between specific miRNAs and the pathophysiology of cystic fibrosis has not yet been described. Thus, if aberrant elevation of a CF-specific miRNA could be identified, then it might lead to a candidate therapeutic agent.
We report here the results of a hypothesis-based discovery study for miRNAs associated with the proinflammatory phenotype for cystic fibrosis. We specifically hypothesized that uniquely expressed miRNAs might aid in identifying the mechanism by which mutant CFTR induces the characteristic proinflammatory phenotype in the CF lung. To test this hypothesis, we have performed a comprehensive and systematic analysis of all miRNAs in CF IB3-1 lung epithelial cells that are differentially affected by the presence of natural abundance ΔF508-CFTR. Among 22 miRNAs that are aberrantly expressed in CF cells, we have identified miR-155 as the most abundantly elevated species, both in vitro and in vivo. Furthermore, we found that a reduction in miR-155, mediated by either antisense or antagomir constructs, resulted in suppression of IL-8 mRNA and concomitantly reduced expression of IL-8 protein. The mechanism of miR-155 action in CF cells is to inhibit translation of SHIP1, an inositol phosphate phosphatase. Loss of SHIP1 results in elevated signaling of the PI3K/Akt pathway, with downstream effects on the IL-8 system. We suggest that miR-155 may play an important role in the regulation of inflammation in CF lung epithelial cells.
EXPERIMENTAL PROCEDURES
Reagents
LHC-8 media, trypsin-EDTA (0.05%), and Lipofectamine transfection reagent were purchased from Invitrogen. Bronchial epithelial growth medium and the normal human bronchial epithelial cells were purchased from Lonza. The miRVana kit and RiboPure kit for isolation of total RNA from CF cells were purchased from Ambion Inc. (Austin, TX). Taqman low density V1 arrays, miRNA primer pools, and pMIR-Report luciferase vector were purchased from Applied Biosystems (Foster City, CA). Wortmannin was purchased from EMD Chemicals (Gibbstown, NJ), and CFTRinh-172 was obtained from Sigma.
Cell Culture
IB3-1 CF lung epithelial cells and the control CFTR-repaired IB3-1/S9 cells were maintained in LHC-8 serum-free medium in humidified 5% CO2 as described previously (13).
RNA Isolation
Total RNA was isolated from the IB3-1 and IB3-1/S9 cells using the miRVana isolation kit (Ambion). The primary bronchial epithelial cells were obtained from lung brush biopsies of CF patients as described earlier (12), and the blood was collected from CF patients and controls under a Uniformed Services University of the Health Sciences Institutional Review Board-approved protocol. The cells were stored in RNA later or TRIzol, and total RNA was isolated using the miRVana kit.
Real-time Quantification of miRNAs by Stem-Loop RT-PCR
Multiplex Reverse Transcription was performed with the TaqMan microRNA reverse transcription kit (Applied Biosystems). Following reverse transcription, each reverse transcription reaction was diluted and mixed with TaqMan gene expression Master Mix (2×). 100 μl of the reverse transcription reaction-specific PCR mix was loaded into the corresponding fill ports of the TaqMan low density human microRNA panel version 1.0 (Early Access). The CF brush biopsy samples were similarly analyzed using the Taqman version 2.0 low density arrays. Individual microRNA assays were performed using specific TaqMan MicroRNA assay kit (ABI).
miRNA Expression Arrays
miRNA expression arrays were probed essentially as described (Ambion). Five micrograms of total RNA from IB3-1 or IB3-1/S9 cells were end-labeled with 30 μCi of [γ-33P]dATP (3000 Ci/mmol) by T4 polynucleotide kinase and purified using the QIAgen nucleotide removal kit. Membranes were first prehybridized in MicroHyb hybridization buffer (ResGen) at 37 °C for at least 30 min, followed by an overnight hybridization in the same solution containing RNA probe. Following hybridization membranes were washed twice with 2× SSC, 0.5% SDS at 37 °C. The second wash was performed in 1× SSC, 0.5% SDS at 37 °C. Membranes were exposed to a phosphor storage screen and scanned using a PhosphorImager, and hybridization signals were quantified using ImageQuant software (Amersham Biosciences).
Statistical Data Analyses
Real-time PCR data were analyzed using the R and the Bioconductor package as well as with STATMINER (a statistical analyses package from ABI). Data were filtered for Ct values of <35, and the data were normalized to the endogenous control gene RNU44. If an assay measurement was not detected in both experimental subgroups, the assay was not included in the pairwise statistical analysis. Furthermore, if an assay measurement was not detected in more than 50% of samples in an experimental subgroup, it was deemed undetected for that subgroup. For all detectable assays, an unpaired Student's t test was performed on the ΔCt values. Adjusted p values (false discovery rates) were calculated using the Benjamini Hochberg procedure. -Fold changes were calculated using the comparative Ct method. Hierarchical clustering based upon Euclidean distances was performed on differentially expressed samples with p values of <0.05.
Illumina mRNA Expression Processing and Analysis
Beadarray data were obtained from whole-genome expression HumanRef-8 version 2.0 as well as human HT-12 BeadChips using the iScan system and BeadScan software (Illumina, San Diego, CA). Non-background, non-normalized array data were generated using BeadStudio 3.2.7 software. Preprocessing of array data by model-based or offset background correction and robust spline normalization was performed using MATLAB or the Lumi 1.8.3 package from Bioconductor 2.3 on the R 2.8 programming language platform. The processed Illumina array data are MIAME-compliant and have been submitted to the NCBI Gene Expression Omnibus (GEO) data base. Processed array data were analyzed using ingenuity pathway analysis (IPA). MicroRNA and mRNA relationship analysis was generated using TargetScan release 5.1 (Whitehead Institute) for miRNA biological target prediction and IPA.
RESULTS
CF Lung Epithelial Cells Express Mutation-specific miRNAs
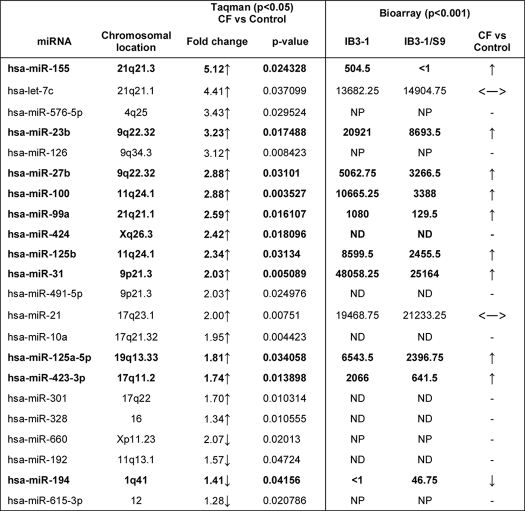
As shown in Table 1, we used two independent methods to identify CF-specific microRNAs in CF lung epithelial cells. Using the new technology, quantitative Taqman® qPCR miRNA array platform, we find that of 365 miRNAs tested, only 22 significantly distinguish between the natural abundance IB3-1 CF cell and the wild type CFTR-repaired daughter cell, IB3-1/S9 (see Table 1, left). For this analysis, we analyzed three independent cultures of both IB3-1 and IB3-1/S9 cells and identified any miRNAs for which the -fold difference was at least ∼50%, and the p value for the difference was <0.05. Of the 22 differentially expressed miRNAs, 18 were elevated in the CF cells, and four were reduced. The data in Table 1, ordered by -fold change, indicate that the miRNAs with the highest differential expression (>4-fold) are miR-155 and let-7c. We also validated the miR-155 and miR-let7c data with independent Taqman® miRNA assays (see Fig. 1B). Of the remaining 16 elevated miRNAs, all were changed by 1.3–3.4-fold. Of the four down-regulated miRNAs, only miR-615 has a >2-fold reduction, whereas miR-660, miR-194, and miR-192 exhibit reductions only in the 1.2–1.6-fold range.
TABLE 1.
miRNA expression levels in IB3-1 CF cells relative to control cells (p < 0.05)
The miRNAs that exhibit altered expression, analyzed by Taqman qPCR miRNA arrays, are listed with respective -fold changes in expression in the IB3-1 CF cells compared with IB3-1/S9 control cells; the 18 miRNAs up-regulated (↑) and the four that are down-regulated (↓) in CF cells are indicated. Additionally, the miRNA expression profile obtained by miRNA microarrays (Bioarray, Ambion) is also depicted. The miRNAs that indicate similar expression profile by these two independent methods are indicated in boldface type. NP, not present; ND, not detectable; ↔, saturated expression.
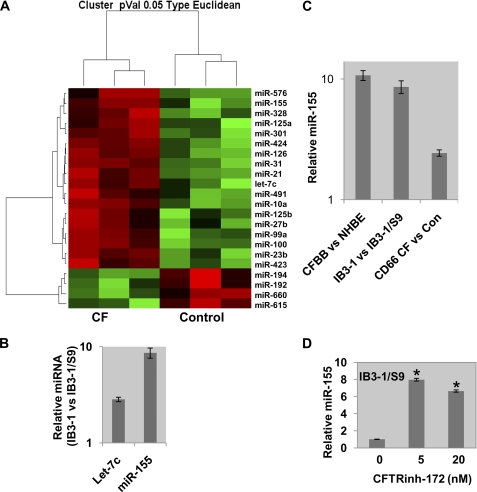
FIGURE 1.
miRNA expression profile in CF cells. A, the miRNA expression profile in IB3-1 CF cells compared with that in control CFTR-repaired IB3-1/S9 cells shows significant differences (p ≤ 0.05, n = 3) in the expression of 22 miRNAs (listed on the right). B, individual miRNA assays were performed for the two miRNA, miR-155 and let-7c, which exhibit maximum up-regulation, to validate the expression profiling data. C, the miR-155 up-regulation was further determined by individual Taqman assays specific for miR-155 in IB3-1 CF cells versus IB3-1/S9 control cells, CF bronchial brushings (CFBB) versus normal human bronchial epithelial cells (NHBE), and CD66+ CF versus controls. D, treatment of the IB3-1/S9 CFTR-repaired control cells with CFTRinh-172 for 1 h with the indicated doses induced miR-155 expression in the WT CFTR cells (p < 0.05). Error bars, S.E.
However, these differently expressed miRNAs all appear to contribute to a composite CF microRNA signature. For example, Fig. 1A shows that when all 22 microRNAs are compared using a hierarchical cluster algorithm, the dendrogram clearly distinguishes between three independent experiments with CF IB3-1 and three independent experiments with CFTR-repaired IB3-1/S9 daughter cells. Thus, despite quantitative differences in expression, all of the significantly aberrant miRNAs, from the most aberrant miR-155 to the least, appear to make a concerted contribution to the CF phenotype in the CF IB3-1 lung epithelial cell system.
To further validate the Taqman® data, we also tested the CF lung epithelial cell samples using a conventional, semiquantitative Ambion Bioarray® platform (see Table 1, right). The data with this different type of system consistently replicated 12 of the 18 miRNAs predicted by the Taqman® platform. The common directions of change are indicated by upward or downward arrows. Horizontal double-headed arrows indicate saturated expression. The validated miRNAs included miR-155 and many of the other miRs with predicted high -fold differences. Several of the low -fold changes are also confirmed. However, consistent saturation problems were encountered with two of the high -fold different miRs, let7c and miR-21, and were not pursued further. Inasmuch as we had independently validated the let7c by PCR (Fig. 1B), we did not pursue this technical saturation problem further. Of the microRNAs with a ≤2-fold difference from control, 6 were not detectable at all on the microarray platform (marked ND). Based on low levels in either platform, we also chose not to pursue this technical sensitivity problem any further. We therefore conclude that both the quantitative Taqman® and the semiquantitative Ambion Bioarray® platforms agree with similar identifications for most of the miRNAs with high -fold differences between CF IB3-1 and wild type CFTR-repaired IB3-1/S9 cells, including miR-155.
Ex Vivo Clinical CF Cells Also Exhibit Elevated miR-155 Levels
To test the extent to which microRNA data from the IB3-1 cell system might parallel microRNA expression in cells from CF patients, we used the Taqman® qPCR miRNA array platform to measure microRNA expression in bronchial brushings and neutrophils obtained from CF patients homozygous for the ΔF508 mutation. As a control for the CF lung epithelial cells obtained by brush biopsy, we measured miR-155 in primary non-CF normal human bronchial epithelial cells. As a control for CF neutrophils, we used neutrophils from normal individuals. Of the 22 differentially expressed microRNAs found to be differentially expressed in the CF IB3-1 cell system, only massively elevated miR-155 expression was noted. We therefore validated this finding using independent Taqman® miRNA assays. As shown in Fig. 1C, we found that miR-155 was significantly elevated in CF bronchial brushings (versus normal human bronchial epithelial cells, 10.8-fold elevated) and in CF neutrophils (CD66+ CF versus control neutrophils, 2.4-fold elevated). We conclude that these differences therefore appear to closely parallel the differences in the CF lung epithelial IB3-1 cell system (IB3-1 versus IB3-1/S9, 8.6-fold elevated). Additionally, the inhibition of WT CFTR function with CFTR-172 inhibitor in CFBE41o-wtCFTR cells, a human bronchial epithelial cell line, has been shown to produce a similar effect on inflammatory signaling as is observed in CF cells (38). As depicted in Fig. 1D, treatment of the IB3-1/S9 CFTR-repaired control cells with CFTR-172 inhibitor induced an 8-fold up-regulation of miR-155. These data indicate that elevation in miR-155 expression is closely tied to failure of CFTR channel activity, either by chemical inhibition or by mutation. The data thus consistently suggest that further specific interest in miR-155 is warranted.
Aberrantly Expressed miRNAs Have Predicted Effects on mRNA Expression in CF Cells
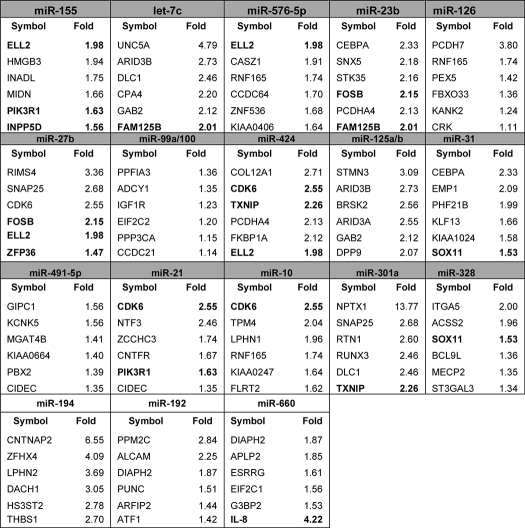
To further understand the mechanism by which miR-155 and the other CF-specific microRNAs might affect the CF phenotype, we used the ILLUMINA® bead chip system to analyze levels of ∼24,000 transcripts, in both CF IB3-1 and wild type CFTR-repaired IB3-1/S9 cells. Table 2 is ordered by starting with top-ranked miR-155 and then proceeding in the order of declining values of -fold elevation, followed by -fold reduction in CF cells. For each of the miRNAs, the changes are listed for the top six specific mRNAs, chosen from among the predicted target mRNAs for the respective miRNA. In each box, mRNAs that are targeted by more than one of the CF-specific microRNAs are shown in boldface type.
TABLE 2.
TargetScan-predicted miRNA target genes detected on Illumina arrays
Shown are the mRNA levels corresponding to the 20 miRNA expressions for CF IB3-1 and CFTR-repaired IB3-1/S9 cells, using ILLUMINA bead chip HumanRef-8 v2.0 arrays. These arrays can analyze ∼24,000 transcripts/sample. The target mRNAs are listed: -fold increases in IB3-1/S9 control cells compared with IB3-1CF cells for the miRNAs up-regulated (gray) and -fold increases in the IB3-1 cells compared with IB3-1/S9 for the miRNAs down-regulated (white) in CF cells. The miRNAs miR-423-3p and miR-615-3p were not present in the TargetScan database and were excluded from this analysis. Genes targeted by multiple miRNAs are indicated in boldface type.
For example, in the case of miR-155, the top six reduced mRNAs are shown. Two mRNAs, ELL2 (elongation factor, RNA polymerase II; 1.98-fold reduced) and PIK3R1 (phosphoinositol 3-kinase regulatory subunit 1 (p85α, GRB1); 1.63-fold reduced), are in boldface type, meaning that they are predicted to also be targets of other miRs in this data set. In this instance, they are also targeted by miR-576–5p and miR-21, respectively. The remaining top four miR-155-dependent mRNAs include HMGB3 (high mobility box 3; 1.94-fold reduced), INADL (channel-interacting PDZ domain protein; 1.75-fold reduced), MIDN (midbrain nucleolar protein; 1.66-fold reduced), and INPP5D (phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 1 (SHIP1); 1.56-fold reduced).
Each of the subtables in Table 2 can be analyzed in the same way. However, because we wish to establish a basis for experimental priority among these 22 microRNAs, we are faced with a complex problem. The hierarchical cluster analysis in Fig. 1A suggests that all 22 miRNAs may cooperate to affect the CF phenotype (supplemental Fig. S1). However, as suggested by Table 2, the boldface target mRNAs suggest that the confluence of effects could be due to multiple microRNAs affecting expression of more than one mRNA. Although this could be a basis for cooperation, it does not help with the priority problem. Alternatively, one mRNA, which is affected by a specific miRNA, could have an indirect, downstream effect on an entirely separate miRNA and thus its mRNA targets. Finally, there might be multiple effects of the CFTR mutation on multiple miRNAs. To bypass these complexities for the moment, we have decided to focus on miR-155. The practical reasons are that (i) miR-155 exhibits the highest level of CF-specific difference; (ii) miR-155 is the only known microRNA in the human genome to target INPP5D/SHIP1, a well known effector for regulation of inflammation; and (iii) ex vivo CF lung epithelial cells and CF neutrophils also express high levels of miR-155.
Antagomir-155 Reduces miR-155 and IL-8 Expression in CF Cells
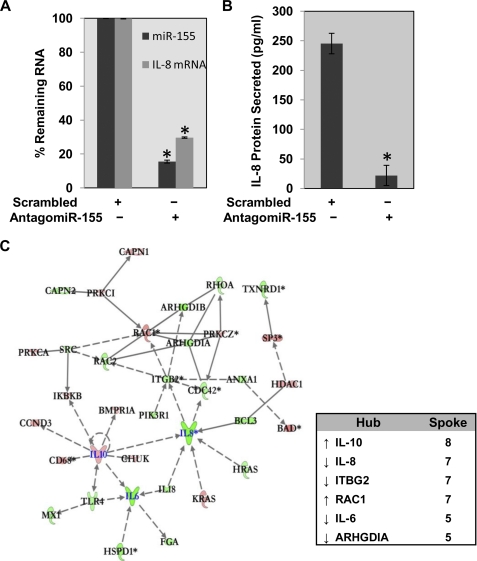
Because miR-155 expression has been consistently associated in the literature with suppression of inflammation and cancer (39–43), we hypothesized that down-regulating miR-155 might also suppress the IL-8-associated proinflammatory phenotype in CF cells. To reduce the levels of miR-155 in CF cells, we decided to deploy an antagomir against miR-155. Antagomirs are metabolically stable antisense constructs against specific microRNAs, which are further modified with cholesterol or other moiety to permit efficient entry into cells. We therefore synthesized antagomir-155 and a scrambled control and tested each for potency and efficiency as a suppressor of both miR-155 and IL-8 expression in our CF cell system. As shown in Fig. 2A, incubation of IB3-1 CF cells with antagomir-155 (2.5 μm) effectively down-regulates miR-155 expression by ∼85%. Concomitantly, we observed a corresponding decrease in IL-8 mRNA levels of ∼70% (Fig. 2A) as well as an ∼11-fold decrease in IL-8 protein levels (see Fig. 2B). The scrambled antagomir control has no effect on miR-155 expression (see Fig. 2A). These data thus firmly establish the specificity of antagomir-155 as a negative effector for IL-8 expression in CF lung epithelial cells, both ex vivo and in cell culture.
FIGURE 2.
Effect of antagomir-155 on IL-8 expression in CF lung epithelial cells. Treatment of IB3-1 CF cells with antagomir-155 (2.5 μm) or with mock treatment for 24 h leads to down-regulation of miR-155 and IL-8 mRNA levels (A) and IL-8 protein levels (B). The mRNA levels were analyzed by quantitative RT-PCR, and IL-8 protein levels were analyzed by ELISA. Data (p < 0.01, denoted by an asterisk) reflect average of three or more independent experiments, and error bars on graphs represent S.E. C, the corresponding mRNA levels were determined with Illumina HT-12 arrays, and subsequent IPA analyses of the data indicate the diminution of the inflammatory phenotype. Whereas both proinflammatory genes, IL-8 and IL-6, were down-regulated in antagomir-155-treated cells, the anti-inflammatory gene IL-10 was up-regulated.
Inflammation-related mRNAs Are Targeted by Antagomir-155 in CF Cells
We next used the ILLUMINA bead chip HT-12 array system to identify those proinflammatory or anti-inflammatory mRNAs, in addition to IL-8, that were significantly targeted by antagomir-155 in CF lung epithelial cells. We found 34 mRNAs in this category whose expression was significantly affected by treatment with antagomir-155. Using a conventional hub-and-spoke connectivity analysis, Fig. 2C uses a color code to depict when the change is elevated (red) or reduced (green). The spokes identify literature-based relationships among the individual mRNAs. As expected, the major down-regulated hubs included IL-8 (seven spokes) but also IL-6 (five spokes), and ITGB2 (integrin β2, seven spokes). Major up-regulated hubs included IL-10 (eight spokes) and RAC1 (seven spokes). However, all of the hubs are connected, either directly or indirectly, and many of the low spoke number singlets and doublets have important inflammatory functions. We conclude that the collective and concurrent changes in expression of the cytokine and chemokine genes in response to antagomir-155 treatment can contribute to attenuation of the inflammatory phenotype in CF cells. Importantly, the reduced mRNAs (green), such as IL-8 and IL-6, are only indirectly connected to the antagomir-155 because a direct effect on the mRNAs would have caused the signals to increase, not decrease. The problem to be solved is therefore what that indirect connection might be in the CF cell.
miR-155 Targets SHIP1 in CF Cells to Promote mRNA Stability of the IL-8
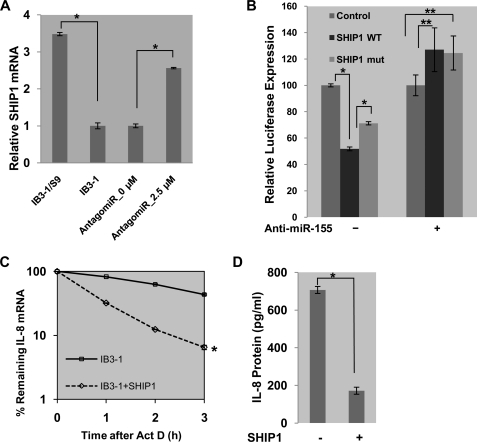
Although INPP5D/SHIP1 is not a classical inflammation-associated gene and is not included as such by the IPA data base in Fig. 2C, it is predicted to be targeted only by miR-155 (44). Furthermore, low levels of SHIP1 have recently been directly linked to proinflammatory processes involving granulocyte/monocyte populations, through regulation by elevated miR-155 (44, 45). We therefore hypothesized that high levels of miR-155 in CF cells might also directly affect the CF proinflammatory phenotype by suppressing SHIP1 expression. We further anticipated that antagomir-155 might reverse the process and the phenotype. Using conventional qPCR, we found that SHIP1 mRNA is expressed at a relatively high level in control IB3-1/S9 cells (∼3.5-fold) compared with CF IB3-1 cells (see Fig. 3A). As further shown in Fig. 3A, we found that suppression of miR-155 in CF cells with antagomir-155 also up-regulated SHIP1 mRNA expression by ∼3-fold. Consistently, we had observed similar suppression of SHIP1 mRNA expression in IB3-1 cells, compared with IB3-1/S9 control cells, when measured by the Illumina bead chip array platform (see Table 2). In this case, we had observed a ∼1.6-fold reduction in INPP5D/SHIP1 in CF IB3-1 cells, when miR-155 levels were elevated by ∼5-fold (see Table 1). Thus, both conventional qPCR and Illumina-based methods suggest that SHIP1 expression is systematically and significantly reduced in CF cells but can be elevated to near control levels when CF cells are treated with antagomir-155.
FIGURE 3.
SHIP1 regulates the expression of IL-8 gene in CF cells. SHIP1 expression was examined as a potential mediator of the miR-155-dependent proinflammatory state in CF. A, SHIP1 expression, examined by qPCR, was significantly diminished in IB3-1 CF cells compared with control IB3-1/S9 cells (∼3.5-fold). This relates to an inverse miR-155 expression profile in these cells. Additionally, suppression of miR-155 in CF cells with antagomir-155 also up-regulated SHIP1 expression as expected. B, luciferase reporter assays were performed in IB3-1 CF cells transfected with pMIR-Report vectors, either controls or those containing SHIP1 3′-UTR target sequences of miR-155 (both WT and mutant), in the presence or absence of anti-miR-155. The data reflect averages of at least three independent experiments (*, p < 0.05; **, p > 0.05). C, IB3-1 cells were transiently transfected with expression plasmid encoding SHIP1 (0.5 μg/2 × 106 cells). RNA was isolated from IB3-1/SHIP1 and IB3-1 cells after actinomycin D treatment for the indicated time intervals, and the remaining mRNA was analyzed by quantitative real-time PCR. Increased expression of SHIP1 promoted rapid degradation of IL-8 mRNA (*, p < 0.001). D, the level of IL-8 protein secreted by IB3-1/SHIP1 cells was analyzed by ELISA 16 h after transfection and also exhibited a diminution in IL-8 protein (*, p < 0.01). Error bars, S.E.
To gain more direct evidence that SHIP1 might be the target of miR-155 in CF cells, we developed a luciferase reporter assay in which the pMIR-Report vector was constructed with wild type or mutant SHIP1 3′-UTR sequences. These reporter plasmids were then transfected into CF IB3-1 cells. As an alternative to treatment with the antagomir-155 construct, we further transfected the CF cells with a conventional anti-miR-155. A demonstration that anti-miR 155 knocks down levels of miR-155 in a dose-dependent manner in CF IB3-1 cells is shown in supplemental Fig. S2. As shown in Fig. 3B, we found that wild type SHIP1 3′-UTR diminished luciferase expression, whereas luciferase expression was returned toward control levels by a partially inactivating mutation engineered into the SHIP1 3′-UTR. However, when these cells were additionally transfected with anti-miR-155, to knock down miR-155 expression, the luciferase reporter activity was only slightly elevated over control and was independent of whether or not wild type or mutant SHIP1 3′-UTR sequences in the luciferase reporter were also present.
In previous work, we have shown that IL-8 mRNA is greatly stabilized in CF cells, both in vitro and in vivo, and that IL-8 protein expression rates were also elevated as a consequence (12, 46). We therefore hypothesized that if SHIP1 levels were exogenously elevated in CF cells, then the half-life of IL-8 mRNA would be reduced, and the expression levels of IL-8 protein would also be reduced. To test this hypothesis, we overexpressed SHIP1 in CF IB3-1 cells and used qPCR to measure the levels of IL-8 mRNA following actinomycin D treatment. As shown in Fig. 3C, 16 h after transfection with an expression plasmid for SHIP1, the half-life of IL-8 in the CF IB3-1 cells was reduced nearly 3-fold when SHIP1 was overexpressed. Corresponding to the enhanced degradation of IL-8 mRNA, we also concomitantly observed a significant reduction in IL-8 protein secretion into the incubation medium at the 16 h post-transfection time point (see Fig. 3D). These data clearly show a close relationship between miR-155-dependent low SHIP1 levels in the CF cell and the proinflammatory CF phenotype manifest by stabilized IL-8 mRNA and elevated IL-8 protein expression.
To further test the potentially countervailing effects of miR-155 and SHIP1, we compared the influence of antagomir-155- and siRNA-mediated knockdown of SHIP1 in CF cells. As shown in Fig. 4A (effect on IL-8 mRNA) and Fig. 4B (effect on SHIP1 mRNA), knockdown of SHIP1 in CF cells reduced the already low levels of SHIP1 (see Fig. 4B) by a further 40%. As anticipated, there was no measurable effect on the already high levels of IL-8 mRNA (see Fig. 4A). As also anticipated, the treatment of the CF cells with antagomir-155 doubled the level of SHIP1 mRNA (see Fig. 4B) while reducing IL-8 mRNA levels by ∼70% (see Fig. 4A). However, when antagomir-155 and siSHIP1 were simultaneously added to the CF cells, SHIP1 mRNA levels were significantly reduced toward SHIP1 knockdown levels, whereas IL-8 levels were slightly but significantly elevated toward SHIP1 knockdown levels. Thus, miR-155 and SHIP1 have mutually countervailing effects on SHIP1 and IL-8 mRNA levels in CF lung epithelial cells. The next critical question is how the information from the miR-155/SHIP1 system is translated to effects on IL-8 mRNA stability in CF cells.
FIGURE 4.
miR-155 and SHIP1 regulate IL-8 expression in CF cells. IB3-1 cells were incubated with siSHIP1 (30 nm) or antagomir-155 alone or with both for 24 h. The isolated RNA was analyzed by quantitative RT-PCR for IL-8 mRNA (A) or SHIP1 mRNA (B) expression. The data reflect averages of at least three independent experiments (*, p < 0.05). Error bars, S.E.
SHIP1 Destabilizes IL-8 mRNA by Suppressing PI3K/Akt Signaling
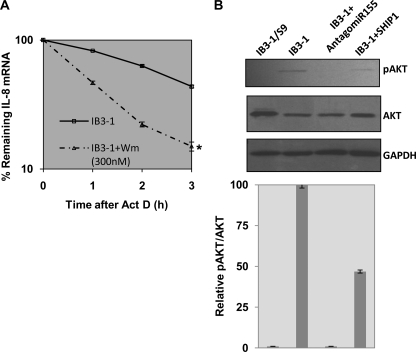
The default function of SHIP1, an inositol 5 phosphatase, is to interfere with PI3K signaling to Akt. The enzymatic function of PI3K is to convert PIP2 to PIP3, and the PIP3 product functions to activate Akt (protein kinase B). However, SHIP1 hydrolyzes PIP3 back to PIP2, thereby blocking PI3K signaling to Akt. Therefore, to test whether this PI3K activity per se might be relevant to how low levels of SHIP might lead to stabilization of IL-8 mRNA in CF cells, we treated CF IB3-1 cells with the PI3K inhibitor wortmannin and tested for effects on IL-8 mRNA stability. As shown in Fig. 5A, 300 nm wortmannin caused a reduction in IL-8 mRNA stability of ∼50% (∼2-fold change in half-life). Thus, either raising the levels of SHIP1 to hydrolyze the PI3K product PIP3 to PIP2 (Fig. 3C) or blocking primary production of PIP3 itself, as evidenced by blocking PI3K with wortmannin (Fig. 5A), has the same destabilizing consequences for IL-8 mRNA in CF cells.
FIGURE 5.
PI3K/Akt activity regulates IL-8 mRNA stability in CF cells. A, IB3-1 cells treated with the pharmacological inhibitor wortmannin (300 nm) had significantly potentiated rapid decay of IL-8 mRNA (*, p < 0.001). B, both phospho-Akt and total Akt protein levels were analyzed by Western blot in IB3-1/S9 CFTR-repaired control cells as well as in the IB3-1 cells mock-treated or treated with antagomir-155 or transfected with SHIP1 expression vector. GAPDH protein levels were also analyzed as endogenous control. The relative quantitation of phospho-Akt/total Akt is also depicted in the bar graph below. Error bars, S.E.
This result suggest that SHIP1-dependent failure of Akt activation by the PIP3 product of PI3K activity may be a crucial signaling intermediate in stabilizing IL-8 mRNA in CF lung epithelial cells. To test this hypothesis, we examined the influence of antagomir-155 and SHIP1 overexpression on the activation state of Akt. Fig. 5B shows that the parental CF IB3-1 cell has high resting levels of phospho-Akt compared with levels in the wild type CFTR-repaired IB3-1/S9 cells. This is consistent with the low levels of SHIP1, which we have shown are typical of CF cells. However, to further test whether Akt activation in CF cells was specifically and reciprocally dependent upon miR-155 and SHIP1, we treated IB3-1 cells with either antagomir-155 or the SHIP1 expression plasmid. As shown in Fig. 5B, antagomir-155, by lowering miR-155 expression, reduced activation of Akt to virtually undetectable levels. On the other hand, expression of SHIP1 in the CF cells raised phospho-Akt levels to ∼50% of the levels seen in the wild type CFTR-repaired IB3-1/S9 cells. We conclude that the mechanism by which miR-155 induces stabilization of IL-8 mRNA depends directly on suppression of Akt signaling by SHIP1.
miR-155 Stabilizes IL-8 mRNA by Activation of MAPKs
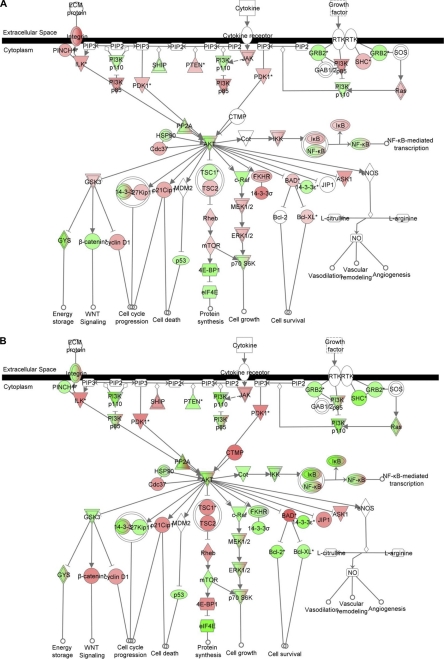
In CF cells, both in vitro and in vivo, we have previously identified intrinsically reduced levels of the ARE-interacting protein TTP (12) and increased levels of MAPK signaling (46) as being independently responsible for IL-8 mRNA stabilization. Nonetheless, in these previous studies, the CF-specific drivers for either of these specific activities had yet to be identified. As shown in Fig. 6A, the base-line transcriptome for CF IB3-1 versus repaired IB3-1/S9 cells shows that in the CF cells, there are intrinsic elevations (color-coded in red) in mRNAs for PI3K/p85 and PDK1. Elevated mRNAs are also found along the downstream signaling pathway toward mTOR, including TSC2, Rheb, and mTOR itself. The downstream PI3K/AKT effector c-Raf is reduced in expression, but MEK1/2 and ERK1/2 are, as anticipated, significantly elevated. However, as shown in Fig. 6B, when these cells are treated with an anti-miR-155, the mRNA for PI3K/p85 is reduced in CF cells relative to control, as is mTOR and the entire signaling pathway comprising c-RAF, MEK1/2, and ERK1/2. Thus, elevated levels of miR-155 differentially control downstream levels and activation of MAPK activities. These data therefore suggest that the mechanism by which elevated miR-155 causes stabilization of IL-8 in CF lung epithelial cells is through loss of SHIP1, activation of PI3K/Akt signaling, and downstream activation of MAPK signaling.
FIGURE 6.
miR-155 stabilizes IL-8 mRNA by activation of MAPKs. The mRNA expression profile in IB3-1 Cf cells, IB3-1/S9 control cells, and IB3-1 cells treated with anti-miR-155 was analyzed by Illumina bead arrays. Subsequently, IPA of the data were performed. A, IB3-1 cells compared with IB3-1/S9 cells; B, IB3-1 cells treated with anti-miR-155 compared with IB3-1 cells.
DISCUSSION
In this paper, we have shown that CF lung epithelial cells, both in vitro and in vivo, hyperexpress miR-155. The downstream consequences of elevated miR-155 include activation of PI3K/Akt signaling through specific reduction in SHIP1. Furthermore, we have shown here that in CF cells, intrinsic activation of PI3K signaling occurs through intrinsic miR-155-dependent down-regulation of SHIP1. This leads to activation of downstream MAPKs with consequent stabilization of IL-8 mRNA and hyperexpression of IL-8 protein (46). SHIP1, an inositol 5-phosphatase, controls PI3K/Akt signaling by hydrolyzing the PIP3 product of PI3K enzymatic activity to PIP2. Thus, the low levels of SHIP1 found in CF cells result in elevated levels of PIP3 and consequent elevated signaling of the PI3K/Akt pathway. We have validated this conclusion by raising or lowering miR-155 using antagomir-155 or using anti-miR-155. We have also used expression plasmids and siRNA for SHIP1 to verify that raising or lowering levels of miR-155 is mirrored by reciprocal effects of SHIP1. Thus, as summarized in Fig. 7, the effect of raising miR-155, lowering SHIP1, and activating PI3K/Akt is to activate MAPKs, thereby stabilizing IL-8 mRNA and increasing the levels of IL-8 protein expression. These data thus lend further support to the concept that the proinflammatory phenotype of the CF airway is due to increased stability of the IL-8 mRNA and subsequent increased expression of IL-8 protein (12, 46).
FIGURE 7.
Regulation of IL-8 gene expression in CF cells. A model appropriately summarizes the proinflammatory status of CF cells due to increased expression of miR-155. The aberrant up-regulation of miR-155 leads to increased IL-8 levels via down-regulation of the expression of intermediate target gene of miR-155, SHIP1 (INPP5D).
SHIP1 and PI3K/Akt Signaling
The only predicted and validated microRNA to target SHIP1 is miR-155 (44), and the connection of SHIP1 to activation of PI3K/AKT signaling has been validated using both pharmacologic and molecular methods. For example, in both hematologic and other cell types, repression of endogenous SHIP1, caused by overexpression of miR-155, leads to an increase in Akt signaling (47). This increase in Akt activity is secondary to the loss of SHIP1 and thus failure to hydrolyze PIP3, the active product of PI3K enzyme activity (48, 49). Consistently, metabolically stabilized analogues of PIP3, utilizing methylene phosphonates or phosphorothioates in place of phosphate, have been shown to inhibit SHIP1, thereby ensuring long lived PI3K/Akt activation (50). A different type of chemical inhibitor for SHIP1, 3α-aminocholestane, has a similar biological effect on blood cell production and numbers of immune regulatory cells in the circulation (51). The opposite experiment, with small molecule agonists of SHIP1, results in inhibition of PI3K/AKT signaling (52). It has also been suggested that SHIP1 can interact directly with the PI3K complex by means of an adaptor protein, immunoreceptor tyrosine-based activation motif-containing receptor (53).
At the level of intact animals, knock-out mice for SHIP1 have been created and studied. It has been reported, for example, that in SHIP1(−/−) knock-out mice, there is an increased number of neutrophils and macrophages, due to enhanced progenitor survival. Consistently, it has been demonstrated in many systems that PI3K activity is necessary for the survival of different types of progenitor cells (54, 55). The SHIP1(−/−) mice also exhibit osteoporosis and significant neutrophil infiltration into the lungs (48). It is possibly not just by coincidence alone that both osteoporosis and neutrophil lung infiltrates are characteristic of the CF phenotype in humans.
miR-155-dependent Activation of PI3K/Akt Signaling and Downstream Activation of MAPKs
In CF cells, both in vitro and in vivo, we have previously shown that increased levels of MAPK signaling contribute to IL-8 mRNA stabilization (46). Nonetheless, in these previous studies, the CF-specific driver for MAPK activation had yet to be identified. In the case of IL-8 mRNA stabilization by MAPK signaling, the experiments showed destabilization of IL-8 mRNA and reduced expression of IL-8 protein by both pharmacological and molecular inhibitors of ERK1/2 (viz. U0126, or Mnk-1) and p38 (viz. SB203580, or p38-AGF); the mediator of p38 action, MK2 (viz. MK2-KR); or JNK-1/2 (JNK-APF and MEK7). Thus, the chemical biology of MAPK-dependent stabilization of IL-8 mRNA is well understood at the pharmacological and molecular levels. However, the puzzle had remained as to how mutations in CFTR might cause MAPK activities to be intrinsically elevated in CF cells, both in vitro and in vivo.
The new data in Fig. 6 suggest that elevated miR-155 contributes to the mechanism of intrinsic MAPK activation and dependent IL-8 mRNA stabilization in CF cells. The data in this paper clearly show that elevated levels of miR-155 cause decreased levels of SHIP1, with enhanced PI3K/Akt signaling as a principal consequence. Mechanistically, activation of PI3K converts PIP2 to PIP3, which binds to and activates PDK1 (PI3K-dependent protein kinase 1). With SHIP1 levels low in CF cells, there is more PIP3 available. Activated PDK1 then binds to and phosphorylates Akt (protein kinase B). This activated complex has multiple signaling possibilities, including interaction with mTOR and activation of cRAF and Rheb. cRAF then activates MEK1/2, ERK1/2, and other MAPK elements. The data in Fig. 6 clearly substantiate this hypothesis in terms of reciprocal miR-155 changes in mRNA expression for the individual signaling proteins. However, exactly how activated MAPKs stabilize IL-8 mRNA remains a subject for future study.
Intrinsic Elevation of miR-155 in Cystic Fibrosis Cells
The specific basis for the proinflammatory phenotype in the CF airway remains an enigma and may be the central pathophysiologic problem confronting our understanding of this disease. Most disease-inducing mutations in the CFTR gene, including ΔF508-CFTR, are associated with intrinsic activation of the TNFα/NFκB signaling pathway (13, 14), leading to production of IL-8 and other proinflammatory mediators. However, how the mutations in CFTR induce TNFα/NFκB signaling remains a mystery. Nonetheless, studies in diffuse large B cell lymphomas have shown that TNFα causes elevated levels of miR-155 (43). Therefore, it is possible that the same mechanism by which mutant CFTR is responsible for activation of TNFα/NFκB signaling may also be responsible for activating miR-155 in CF lung epithelial cells (see Fig. 7).
Potential Translational Impact of Antagomir-155 for Cystic Fibrosis
Our study clearly indicates that antagonizing miR-155 is a potent method of suppressing IL-8 and other proinflammatory genes in CF cells and therefore might have potential as a candidate CF therapeutic. As indicated in Fig. 2C, antagomir-155, while lowering the level of IL-8 mRNA, has the opposite effect on the anti-inflammatory cytokine IL-10. Low levels of IL-10 have recently been suggested as a mechanism for up-regulation of miR-155 (57), possibly via effects on Toll-like receptors (58). Such an approach has recent precedents. Intravenous administration of antagomirs against miR-122, miR-92a, miR-196, and miR-221/222 were among the first examples shown to suppress functions of endogenous miRs in vivo. In the case of antagomir-122, increases in a set of genes associated with cholesterol biosynthesis were demonstrated in mouse liver (37). An antagomir to miR-92a (“antagomir-92a”) has been shown to aid in angiogenesis and functional recovery of ischemic tissues in intact mice (56). Administration of antagomir-196 into intact chicken eggs has been shown to regulate hox gene expression and vertebral development in the enclosed developing chick (57). Finally, antagomir −221/−222 has been shown to regulate the progression of human melanoma Me665/1 cells in athymic nude mice (58). What is most striking is the fact that in all cases cited, the antagomirs were not only functional but also apparently non-toxic. We therefore conclude that the potential translational value of antagomir-155 for cystic fibrosis therapeutics needs to be fully investigated.
Supplementary Material
Acknowledgments
We thank Venkat Gopalan (Ohio State University) for critical reading of the manuscript and Mike Flora (Bio Instrumentation Core, Uniformed Services University of the Health Sciences) for the synthesis of antagomir-155. We also thank Mark Coggeshall (Oklahoma Medical Research Foundation) for the vector encoding SHIP1, Michael David (University of California San Diego) for the pMIR-Report luciferase clones containing SHIP 3′-UTR target sequences for miR-155, Pam Zeitlin and Michael Boyle (The Cystic Fibrosis Center at Johns Hopkins School of Medicine) for patient samples, and Wei Huang (Uniformed Services University of the Health Sciences) for processing of the miRNA bioarrays.
This study was supported by USU-Intramural Funds (to R. B.), National Institutes of Health (RO1-DK053051, to H. B. P.) and Cystic Fibrosis Foundation (to R. B. and H. B. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- CF
- cystic fibrosis
- CFTR
- cystic fibrosis transmembrane conductance regulator
- miRNA
- microRNA
- TTP
- tristetraprolin
- ARE
- AU-rich element
- IPA
- ingenuity pathway analysis
- qPCR
- quantitative PCR
- PIP3
- phosphatidylinositol 3,4,5-trisphosphate
- PIP2
- phosphatidylinositol 4,5-bisphosphate.
REFERENCES
- 1. Pollard H. B. (2000) Anat. Rec. 259, FMIII–FMIX [DOI] [PubMed] [Google Scholar]
- 2. Frizzell R. A. (1999) Physiol. Rev. 79, S1–2 [DOI] [PubMed] [Google Scholar]
- 3. Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. (1990) Cell 63, 827–834 [DOI] [PubMed] [Google Scholar]
- 4. Kopito R. R. (1999) Physiol. Rev. 79, S167–S173 [DOI] [PubMed] [Google Scholar]
- 5. Dean T. P., Dai Y., Shute J. K., Church M. K., Warner J. O. (1993) Pediatr. Res. 34, 159–161 [DOI] [PubMed] [Google Scholar]
- 6. Richman-Eisenstat J. B., Jorens P. G., Hébert C. A., Ueki I., Nadel J. A. (1993) Am. J. Physiol. 264, L413–L418 [DOI] [PubMed] [Google Scholar]
- 7. Armstrong D. S., Grimwood K., Carlin J. B., Carzino R., Gutièrrez J. P., Hull J., Olinsky A., Phelan E. M., Robertson C. F., Phelan P. D. (1997) Am. J. Respir. Crit. Care Med. 156, 1197–1204 [DOI] [PubMed] [Google Scholar]
- 8. Roebuck K. A. (1999) J. Interferon Cytokine Res. 19, 429–438 [DOI] [PubMed] [Google Scholar]
- 9. Bonfield T. L., Panuska J. R., Konstan M. W., Hilliard K. A., Hilliard J. B., Ghnaim H., Berger M. (1995) Am. J. Respir. Crit. Care Med. 152, 2111–2118 [DOI] [PubMed] [Google Scholar]
- 10. Khan T. Z., Wagener J. S., Bost T., Martinez J., Accurso F. J., Riches D. W. (1995) Am. J. Respir. Crit. Care Med. 151, 1075–1082 [DOI] [PubMed] [Google Scholar]
- 11. Tirouvanziam R., de Bentzmann S., Hubeau C., Hinnrasky J., Jacquot J., Péault B., Puchelle E. (2000) Am. J. Respir. Cell Mol. Biol. 23, 121–127 [DOI] [PubMed] [Google Scholar]
- 12. Balakathiresan N. S., Bhattacharyya S., Gutti U., Long R. P., Jozwik C., Huang W., Srivastava M., Pollard H. B., Biswas R. (2009) Am. J. Physiol. Lung Cell Mol. Physiol. 296, L1012–L1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eidelman O., Srivastava M., Zhang J., Leighton X., Murtie J., Jozwik C., Jacobson K., Weinstein D. L., Metcalf E. L., Pollard H. B. (2001) Mol. Med. 7, 523–534 [PMC free article] [PubMed] [Google Scholar]
- 14. Srivastava M., Eidelman O., Zhang J., Paweletz C., Caohuy H., Yang Q., Jacobson K. A., Heldman E., Huang W., Jozwik C., Pollard B. S., Pollard H. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7693–7698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wojnarowski C., Frischer T., Hofbauer E., Grabner C., Mosgoeller W., Eichler I., Ziesche R. (1999) Eur. Respir. J. 14, 1136–1144 [DOI] [PubMed] [Google Scholar]
- 16. Ambros V. (2001) Cell 107, 823–826 [DOI] [PubMed] [Google Scholar]
- 17. Ambros V. (2003) Cell 113, 673–676 [DOI] [PubMed] [Google Scholar]
- 18. Ambros V. (2004) Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 19. von Roretz C., Gallouzi I. E. (2008) J. Cell Biol. 181, 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim V. N. (2005) Nat. Rev. Mol. Cell Biol. 6, 376–385 [DOI] [PubMed] [Google Scholar]
- 21. Kim V. N. (2005) Mol. Cells 19, 1–15 [PubMed] [Google Scholar]
- 22. Bagga S., Pasquinelli A. E. (2006) Genet. Eng. (NY) 27, 1–20 [DOI] [PubMed] [Google Scholar]
- 23. John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004) PLoS Biol. 2, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. John B., Sander C., Marks D. S. (2006) Methods Mol. Biol. 342, 101–113 [DOI] [PubMed] [Google Scholar]
- 25. Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 26. Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. (2006) Cold Spring Harb. Symp. Quant. Biol. 71, 513–521 [DOI] [PubMed] [Google Scholar]
- 27. Vasudevan S., Tong Y., Steitz J. A. (2007) Science 318, 1931–1934 [DOI] [PubMed] [Google Scholar]
- 28. Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P. E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Stoffel M. (2004) Nature 432, 226–230 [DOI] [PubMed] [Google Scholar]
- 29. Poy M. N., Spranger M., Stoffel M. (2007) Diabetes Obes. Metab. 9, Suppl. 2, 67–73 [DOI] [PubMed] [Google Scholar]
- 30. Blenkiron C., Goldstein L. D., Thorne N. P., Spiteri I., Chin S. F., Dunning M. J., Barbosa-Morais N. L., Teschendorff A. E., Green A. R., Ellis I. O., Tavaré S., Caldas C., Miska E. A. (2007) Genome Biol. 8, R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blenkiron C., Miska E. A. (2007) Hum. Mol. Genet. 16, R106–R113 [DOI] [PubMed] [Google Scholar]
- 32. Hwang H. W., Mendell J. T. (2006) Br. J. Cancer 94, 776–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jovanovic M., Hengartner M. O. (2006) Oncogene 25, 6176–6187 [DOI] [PubMed] [Google Scholar]
- 34. van Rooij E., Olson E. N. (2007) Physiol. Genomics 31, 365–366 [DOI] [PubMed] [Google Scholar]
- 35. Ikeda S., Kong S. W., Lu J., Bisping E., Zhang H., Allen P. D., Golub T. R., Pieske B., Pu W. T. (2007) Physiol. Genomics 31, 367–373 [DOI] [PubMed] [Google Scholar]
- 36. Rane S., Sayed D., Abdellatif M. (2007) Cell Cycle 6, 1850–1855 [DOI] [PubMed] [Google Scholar]
- 37. Krützfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., Stoffel M. (2005) Nature 438, 685–689 [DOI] [PubMed] [Google Scholar]
- 38. Vij N., Mazur S., Zeitlin P. L. (2009) PLoS ONE 4, e4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bolisetty M. T., Dy G., Tam W., Beemon K. L. (2009) J. Virol. 83, 12009–12017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stahl H. F., Fauti T., Ullrich N., Bopp T., Kubach J., Rust W., Labhart P., Alexiadis V., Becker C., Hafner M., Weith A., Lenter M. C., Jonuleit H., Schmitt E., Mennerich D. (2009) PLoS ONE 4, e7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tili E., Croce C. M., Michaille J. J. (2009) Int. Rev. Immunol. 28, 264–284 [DOI] [PubMed] [Google Scholar]
- 42. Banerjee A., Schambach F., DeJong C. S., Hammond S. M., Reiner S. L. (2010) Eur. J. Immunol. 40, 225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pedersen I. M., Otero D., Kao E., Miletic A. V., Hother C., Ralfkiaer E., Rickert R. C., Gronbaek K., David M. (2009) EMBO Mol. Med. 1, 288–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Connell R. M., Chaudhuri A. A., Rao D. S., Baltimore D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7113–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cremer T. J., Ravneberg D. H., Clay C. D., Piper-Hunter M. G., Marsh C. B., Elton T. S., Gunn J. S., Amer A., Kanneganti T. D., Schlesinger L. S., Butchar J. P., Tridandapani S. (2009) PLoS ONE 4, e8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bhattacharyya S., Gutti U., Mercado J., Moore C., Pollard H. B., Biswas R. (2011) Am. J. Physiol. Lung Cell Mol. Physiol. 300, L81–L87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamanaka Y., Tagawa H., Takahashi N., Watanabe A., Guo Y. M., Iwamoto K., Yamashita J., Saitoh H., Kameoka Y., Shimizu N., Ichinohasama R., Sawada K. (2009) Blood 114, 3265–3275 [DOI] [PubMed] [Google Scholar]
- 48. Rauh M. J., Sly L. M., Kalesnikoff J., Hughes M. R., Cao L. P., Lam V., Krystal G. (2004) Biochem. Soc. Trans. 32, 785–788 [DOI] [PubMed] [Google Scholar]
- 49. Ooms L. M., Horan K. A., Rahman P., Seaton G., Gurung R., Kethesparan D. S., Mitchell C. A. (2009) Biochem. J. 419, 29–49 [DOI] [PubMed] [Google Scholar]
- 50. Zhang H., He J., Kutateladze T. G., Sakai T., Sasaki T., Markadieu N., Erneux C., Prestwich G. D. (2010) ChemBioChem 11, 388–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brooks R., Fuhler G. M., Iyer S., Smith M. J., Park M. Y., Paraiso K. H., Engelman R. W., Kerr W. G. (2010) J. Immunol. 184, 3582–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ong C. J., Ming-Lum A., Nodwell M., Ghanipour A., Yang L., Williams D. E., Kim J., Demirjian L., Qasimi P., Ruschmann J., Cao L. P., Ma K., Chung S. W., Duronio V., Andersen R. J., Krystal G., Mui A. L. (2007) Blood 110, 1942–1949 [DOI] [PubMed] [Google Scholar]
- 53. Peng Q., Malhotra S., Torchia J. A., Kerr W. G., Coggeshall K. M., Humphrey M. B. (2010) Sci. Signal 3, ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ebner S., Dunbar M., McKinnon R. D. (2000) J. Neurosci. Res. 62, 336–345 [DOI] [PubMed] [Google Scholar]
- 55. Dubrovska A., Kim S., Salamone R. J., Walker J. R., Maira S. M., García-Echeverria C., Schultz P. G., Reddy V. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 268–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bonauer A., Carmona G., Iwasaki M., Mione M., Koyanagi M., Fischer A., Burchfield J., Fox H., Doebele C., Ohtani K., Chavakis E., Potente M., Tjwa M., Urbich C., Zeiher A. M., Dimmeler S. (2009) Science 324, 1710–1713 [DOI] [PubMed] [Google Scholar]
- 57. McGlinn E., Yekta S., Mansfield J. H., Soutschek J., Bartel D. P., Tabin C. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18610–18615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Felicetti F., Errico M. C., Segnalini P., Mattia G., Carè A. (2008) Expert Rev. Anticancer Ther. 8, 1759–1765 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.