Abstract
Mutations in Parkin, an E3 ubiquitin ligase that regulates protein turnover, represent one of the major causes of familial Parkinson disease, a neurodegenerative disorder characterized by the loss of dopaminergic neurons and impaired mitochondrial functions. The underlying mechanism by which pathogenic Parkin mutations induce mitochondrial abnormality is not fully understood. Here, we demonstrate that Parkin interacts with and subsequently ubiquitinates dynamin-related protein 1 (Drp1), for promoting its proteasome-dependent degradation. Pathogenic mutation or knockdown of Parkin inhibits the ubiquitination and degradation of Drp1, leading to an increased level of Drp1 for mitochondrial fragmentation. These results identify Drp1 as a novel substrate of Parkin and suggest a potential mechanism linking abnormal Parkin expression to mitochondrial dysfunction in the pathogenesis of Parkinson disease.
Keywords: E3 Ubiquitin Ligase, Mitochondria, Parkinson Disease, Protein-Protein Interactions, Ubiquitination, Drp1 (Dynamin-related Protein 1)
Introduction
Parkinson disease (PD)4 is one of the most common neurodegenerative diseases affecting over 2% populations over 65 years of age. It is classically characterized by the loss of dopaminergic neurons that project from the midbrain substantia nigra to the striatum (1, 2). Although the loss of dopaminergic neurons is responsible for the symptom of movement disorder in PD, it is now clear that other types of neurons throughout the brain are also affected in the disease (3, 4). The identification of genes linking to PD has greatly advanced our understanding of the molecular pathogenesis of the disease (5–8). Mutations in Parkin represent one of major causes for early onset of familial PD (9–11). Parkin is an E3 ubiquitin ligase that contains two ring finger domains (12–15). A handful of substrates have been identified, including Parkin itself and CDCrel-1, synphilin-1, Pael-R, glycosylated α-synuclein, FBP1 (far upstream element-binding protein 1), and the RNA-processing protein subunit p38/AIMP2 (16–19). A putative mechanism by which mutations of Parkin cause PD would be abnormal accumulation and aggregation of the above substrates due to insufficient E3 ligase activity for ubiquitin-proteasome-dependent protein turnover (18, 20, 21). Surprisingly, only p38/AIMP2 and FBP1 were found to be accumulated in the brain samples of PD patients or in Parkin knock-out mice (16, 17, 19). Even though a number of the putative substrates have been identified, the causative link between these substrates and the PD pathogenesis remains not fully understood.
Over the past few decades, accumulating evidence has suggested that mitochondrial dysfunction and the resulting oxidative damage are associated with PD. This is supported by a large number of reports demonstrating impaired mitochondrial functions in PD patients (22–26). Mitochondria undergo frequent fission, fusion, and redistribution throughout the cytoplasm in response to the energy needs (27, 28). Either disruption of the fusion process or enhancement of the fission process renders the normal, tubular network of mitochondria to fragment into short rods or spheres (29). Abnormal mitochondrial fission or fusion is closely associated with neuronal cell death and a number of neuromuscular diseases (30).
Drp1 is a cytosolic protein responsible for mitochondrial fission (31). It targets to mitochondria to initiate mitochondrial fragmentation (32–34). Enhanced Drp1 expression or a reduced level of mitofusins induces mitochondrial fragmentation, an early event prior to the release of mitochondrial cytochrome c and programmed cell death (20, 35). Strong evidence has shown that Parkin plays a critical role in regulating mitochondrial fission and fusion (36) and mitochondrial quality control (37). Recent studies suggest that Drosophila Parkin genetically interacts with proteins that regulate mitochondrial fission and fusion, although other reports describe inconsistent phenotypes in Parkin- and PINK1-deficient Drosophila cells (38–44). Knockdown of Parkin results in mitochondrial elongation in flies (40). However, studies in mammalian cells suggest that loss of Parkin/PINK1 function may lead to excess mitochondria fragmentation or enhanced mitochondrial biogenesis (45–48). We thus sought to address the molecular details on how Parkin regulates mitochondrial fission and fusion in mammalian systems. To this end, we have identified Drp1 as a novel substrate of Parkin which effectively promotes the proteasome dependent degradation of Drp1. Our results thus uncover a novel mechanism linking loss of Parkin to mitochondrial dysfunction in the pathogenesis of PD and suggest that Drp1 could be a potential target for fighting against this currently incurable disease.
EXPERIMENTAL PROCEDURES
Plasmids
The mammalian expression plasmids for Parkin and Drp1 were generated by PCR and cloned into pEGFPC1 and pRK5-myc vectors. The mammalian expression plasmid for FLAG-ubiquitin was generated by insertion of ubiquitin cDNA in-frame into the pCMV-tag-2B vector. The pCMV-HA-UB and pCMV-HA-UB-K0 plasmids were kindly provided by Dr. Tomohiko Ohta (St. Marianna University, Japan).
Antibodies and Reagents
DAPI and antibodies against FLAG, HA, Myc, and β-actin were purchased from Sigma-Aldrich. Antibodies against Parkin (Cell Signaling), Drp1 (BD Biosciences), GFP (Roche Applied Science), ubiquitin (Santa Cruz), rhodamine- and fluorescein-conjugated secondary antibodies (Jackson ImmunoResearch) were from the indicated sources. MG132, PS341, PMSF, and cycloheximide were obtained from Sigma-Aldrich. MitoTracker-Red, CM-H2XRos, and chloroquine were from Invitrogen, pepstatin was from BioBasic.
Cells, siRNAs, shRNAs, and Transfections
All the cells used in this study were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS at 37 °C in an atmosphere of humidified air and 5% CO2. siRNA oligonucleotides were synthesized by RiboBio with a 2-base (dTdT) overhang and transfected into cells with the Lipofectamine 2000 reagent (Invitrogen). The sequences of the Parkin siRNAs are 5′-CUUGGCUACUCCCUGCCUU-3′ and 5′-CAGCCAAAUUGCAGAAGAA-3′. Drp1 shRNA sequences are: 5′-GATCCGTGGTGCTAGAATTTGTTATTCAAGAGATAACAAATTCTAGCACCACTTTTTTG-3′ and 5′-AATTCAAAAAAGTGGTGCTAGAATTTGTTATCTCTTGAATAACAAATTCTAGCACCACG-3′. Plasmids were transfected into cells with PEI according to the manufacturer's instructions.
Cell Lysate Preparation and Western Blotting
To prepare cell lysates, cells were washed twice with ice-cold phosphate-buffered saline (PBS) and solubilized in lysis buffer (150 m NaCl, 25 mm HEPES, pH 7.4, 1% Chaps, 0.25% sodium deoxycholate, 1 mm EGTA, 1 mm DTT, 1 mm PMSF, 10% glycerol, and 10 mg/ml aprotinin, leupeptin, and pepstatin). The cells were scraped, and the supernatants were collected after 15 min of centrifugation at 14,000 × g and 4 °C. Protein concentrations were determined by using the BCA protein assay kit. Proteins were resolved by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were blocked in PBS containing 0.1% Tween 20 and 5% fat-free dry milk and incubated first with primary antibodies and then with horseradish peroxidase-conjugated secondary antibodies. Specific proteins were visualized with enhanced chemiluminescence detection reagent (Pierce Biotechnology). The intensity of protein bands was determined by the ImageJ software and corrected by subtracting the measured intensity with the background intensity.
Immunoprecipitation and MBP Pulldown
Cell lysate was incubated with specific antibodies at 4 °C for 2 h, and protein A/G-agarose beads (Pierce Biotechnology) were then added to incubate for another 3 h. The beads were washed extensively and boiled in SDS loading buffer, and the precipitated proteins were detected by SDS-PAGE and Western blotting. For MBP pulldown in vitro, MBP or MBP-Parkin fusion protein immobilized on amylose magnetic beads was incubated with in vitro translated Myc-Drp1 at 4 °C for 2 h. The beads were washed and boiled in the SDS loading buffer, and the precipitated proteins were detected by SDS-PAGE and Western blotting.
Ubiquitination Assays
Cells were transfected with GFP-Parkin, Myc-Drp1, and HA-UB or HA-UB-K0 plasmids and incubated with 20 μm MG132 for 8 h before harvest. Cell lysate was immunoprecipitated with an antibody against Myc. The precipitates were subjected to Western blotting with an antibody against HA. In vitro ubiquitination assay was performed in 50 μl of ubiquitination reaction buffer containing 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 2 mm DTT, 2 mm ATP, 10 μg of ubiquitin, 100 ng of E1, 200 ng of E2 (UbcH7), 2 μg of purified MBP-Parkin, 2 μg of immunoprecipitated MARCH5, and 2 μg of in vitro translated Drp1. The reaction was performed for 2 h at 30 °C and terminated by addition of the SDS loading buffer. The reaction products were then subjected to Western blotting with anti-ubiquitin and anti-Drp1 antibodies.
Fluorescence Microscopy
Cells grown on glass coverslips were transfected with Mito-DsRed together with GFP-Parkin. Cells were fixed with 4% paraformaldehyde for 30 min at room temperature, incubated with primary and secondary antibodies, and then stained with DAPI. Coverslips were mounted with 90% glycerol in PBS and examined with a Zeiss fluorescence microscope.
[35S]Methionine Pulse-Chase Experiments
HeLa cells were incubated with the labeling medium containing 10 μCi of Met/Cys Tran35S-label (PerkinElmer Life Sciences) for 3 h. Cells were washed and then incubated with the chase medium containing 2 mm methionine and cysteine (Sigma). Cells were harvested at different time points, and proteins were then immunoprecipitated with anti-Drp1 and anti-β-actin antibodies, separated by SDS-PAGE, and analyzed by autoradiography.
Statistical Analysis
Statistical analysis between groups was performed by unpaired two-tailed Student's t test. Data are presented as means ± S.E.
RESULTS
Alteration of Parkin Expression in Cells Affects Mitochondrial Morphology
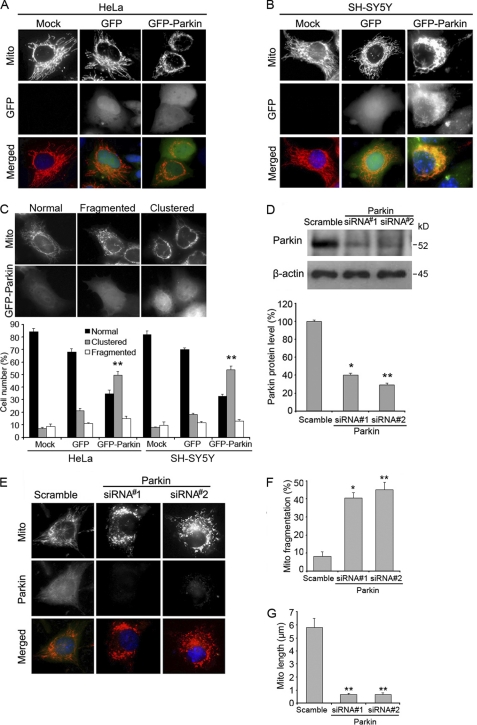
We first examined mitochondrial morphology in cells with altered Parkin expression. Fluorescence microscopy revealed that overexpression of Parkin resulted in perinuclear clustering of mitochondria in both HeLa and SH-SY5Y cells (Fig. 1, A and B), and nearly half of the cells had this phenotype (Fig. 1C). To assess the role of endogenous Parkin in the regulation of mitochondrial morphology, we inhibited the expression of Parkin in SH-SY5Y cells with specific siRNAs (Fig. 1D). Strikingly, although the majority of the cells transfected with control siRNA had a normal reticulum of mitochondria, nearly half of the cells transfected with Parkin siRNAs had fragmented mitochondria (Fig. 1, E and F), which was in accordance with previous reports (48). The length of mitochondria was also significantly decreased in cells transfected with Parkin siRNAs (Fig. 1G). These results indicate that Parkin expression can significantly alter mitochondrial morphology in cells.
FIGURE 1.
Alteration of Parkin expression in cells affects mitochondrial morphology. A, HeLa cells were transfected with Mito-DsRed together with GFP-Parkin or GFP vector for 24 h, and cells were then stained with DAPI and analyzed by fluorescence microscopy. Cells with a dense cluster of mitochondria were classified as perinuclear mitochondrial clustering. B, experiments were performed as in A except that SH-SY5Y cells were used. C, experiments were performed as in A and B, and the percentage of cells with mitochondrial clustering was quantified. Cells with an intact network of tubular mitochondria were defined as normal, cells with disrupted and predominantly spherical mitochondria were defined as fragmented mitochondria, and cells with a dense cluster of mitochondria were classified as perinuclear mitochondrial clustering. D, Western blot analysis of the expression of Parkin and β-actin in SH-SY5Y cells transfected with two different Parkin siRNAs or a scramble siRNA control was performed. Parkin protein level was determined by dividing the intensity of Parkin with the intensity of β-actin on the blot. E, SH-SY5Y cells were transfected with control or Parkin siRNAs, stained with Mito-Tracker Red and an anti-Parkin antibody, and then analyzed by immunofluorescence microscopy. F, experiments were performed as in E, and the percentage of cells with mitochondrial truncation or fragmentation was quantified. G, experiments were performed as in E, and the length of mitochondria was measured with ImageJ. All immunofluorescence data shown in the bar graphs represent means ± S.E. (error bars) from three independent experiments, with at least 100 cells counted in a blinded manner. *, p < 0.01 and **, p < 0.001.
Parkin Induces the Degradation of Drp1 through the Proteasome-dependent Pathway
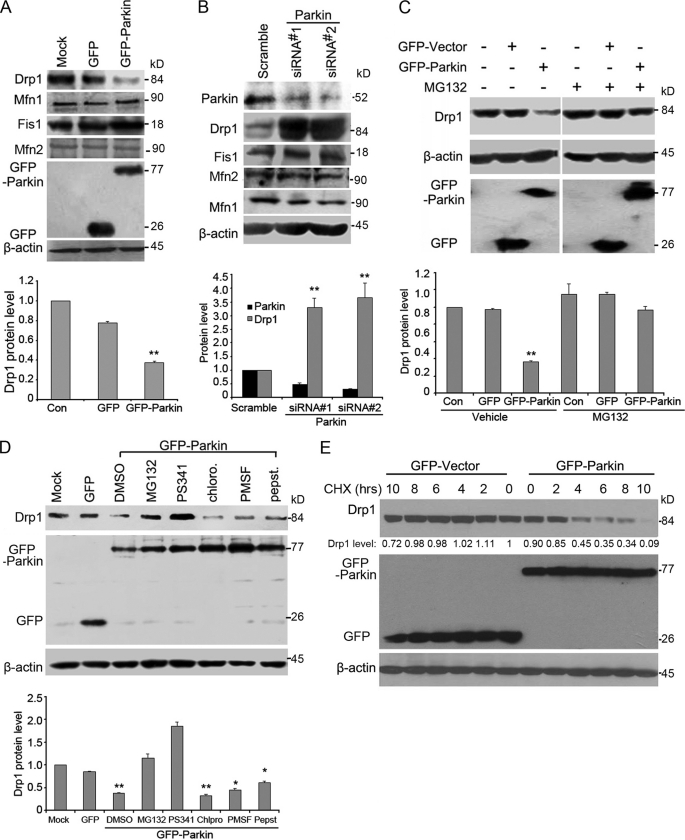
The remarkable effect of Parkin on mitochondrial morphology suggests that it may affect the level of either the pro-fission proteins or the pro-fusion protein, key regulators of mitochondrial dynamics. To test this possibility, we chose 293T cells which have high transfection efficiency for the overexpression experiments. We examined the expression of Drp1, Mfn1/2 (mitofusion 1/2) and Fis1 (another pro-fission molecule) in cells transfected with GFP-Parkin by Western blotting. As shown in Fig. 2A, the level of Drp1 was significantly decreased by GFP-Parkin, whereas the levels of Mfn1/2 and Fis1 were not affected. siRNA-mediated knockdown of Parkin expression was performed in SH-SY5Y cells that have endogenous expression of Parkin. We found that Drp1 expression was significantly increased, but the expression of Mfn1/2 and Fis1 was not affected (Fig. 2B).
FIGURE 2.
Parkin induces the degradation of Drp1 through the proteasome. A, 293T cells were untransfected or transfected with GFP-Parkin or GFP vector, and Western blotting was performed to examine the expression of Drp1, Mfn1/2, Fis1, β-actin, and GFP proteins. Drp1 protein level was quantified according to the results of three independent blots. B, Western blot analysis of the expression of Parkin, Drp1, Mfn1/2, Fis1 and β-actin in SH-SY5Y cells transfected with control or Parkin siRNAs was performed. Parkin and Drp1 protein levels were quantified according to the results of three independent blots. C, Western blot analysis of the expression of Drp1, β-actin, and GFP proteins in 293T cells untransfected or transfected with GFP-Parkin or GFP vector, in the presence or absence of the proteasome inhibitor MG132, was performed. Drp1 protein level was quantified according to the results of three independent blots. D, 293T cells were untransfected, transfected with GFP vector, or transfected with GFP-Parkin in the presence of various inhibitors, including two proteasome inhibitors (MG132 and PS341), a lysosome inhibitor (chloroquine), or two protease inhibitors (PMSF and pepstain). Western blotting was then performed to examine the expression of Drp1, β-actin, and GFP proteins. E, CHX chase assay for the half-life of Drp1 is shown. 293T cells were transfected with GFP and GFP-Parkin for 24 h. Cells were then treated with CHX (100 μg/ml) for the indicated hours, and Western blotting was performed. The level of remaining Drp1 at different time points was quantified as the percentage of initial Drp1 level (0 h of CHX treatment). All data are representative of at least three independent experiments. *, p < 0.01 and **, p < 0.001.
We further investigated whether the down-regulatory effect of Parkin on Drp1 was due to an increase in Drp1 degradation through the proteasome-dependent pathway, similar to the effect of Parkin toward other substrates. To test this possibility, cells were transfected with GFP-Parkin in the absence or presence of MG132, a specific proteasome inhibitor. We found that the reduction of Drp1 by GFP-Parkin was remarkably blocked by MG132 (Fig. 2C). A similar result was obtained by treatment of these cells with PS341, another proteasome inhibitor (Fig. 2D). In contrast, neither chloroquine, a lysosome inhibitor, nor PMSF and pepstatin, protease inhibitors, could inhibit the down-regulatory effect of Parkin on Drp1 expression (Fig. 2D). We further examined the effect of Parkin on Drp1 stability by measuring the half-life of Drp1 via the cycloheximide (CHX)-chase assay and [35S] methionine pulse-chase assay (supplemental Fig. S1). Our result revealed a striking decrease of Drp1 half-life in cells overexpressing GFP-Parkin (Fig. 2E and supplemental Fig. S1). Taken together, these data demonstrate that Parkin induces Drp1 degradation through the proteasome-dependent proteolytic machinery.
Parkin Interacts with Drp1 through Its Second Ring Finger Domain
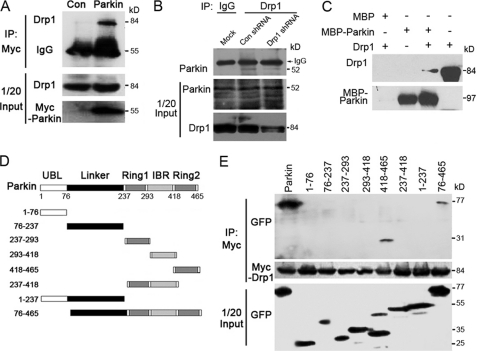
Parkin, as an E3 ubiquitin ligase, can regulate the ubiquitination and proteasome-dependent degradation of its substrates. Our finding that Parkin is capable of regulating Drp1 expression suggests that Drp1 might be a new substrate of Parkin. To test this, we first examined whether these two proteins interact. Cells were transfected with plasmids expressing either Myc-Parkin or Myc-Vector, and cell lysates were then analyzed by immunoprecipitation with anti-Myc antibody. As shown in Fig. 3A, endogenous Drp1 was immunoprecipitated by Myc-Parkin.
FIGURE 3.
Parkin interacts with Drp1 in vivo and in vitro. A, 293T cells were transfected with Myc-Parkin or the empty vector (control), and immunoprecipitation (IP) and Western blotting were performed to examine the interaction between Myc-Parkin and endogenous Drp1. B, endogenous Parkin (kD) interacts with endogenous Drp1. SH-SY5Y cells were transfected with Drp1 or control shRNAs. Cell lysates were subjected to immunoprecipitation with an anti-Drp1 antibody or an IgG control, and the immunoprecipitates were examined by Western blotting using an anti-Parkin antibody. The middle and bottom panels show the expression of Drp1 and Parkin in the cell lysates (1/20 input). C, Parkin and Drp1 interact in vitro. In vitro translated Drp1 was incubated with bacterially purified MBP-Parkin or MBP immobilized on MBP beads. The presence of Drp1 in the pulldown preparation was examined by Western blotting. The last lane shows 1/20 input of Drp1. D, various truncated forms of GFP-Parkin are shown as a schematic. E, 293T cells were transfected with Myc-Drp1 together with a series of truncated forms of GFP-Parkin. Immunoprecipitation and Western blotting were performed to examine the critical domains in Parkin that mediate its interaction with Myc-Drp1. All data are representative of at least three independent experiments.
Next, we investigated whether endogenous Drp1 interacts with endogenous Parkin. Cells were transfected with Drp1 or control shRNAs. Cell lysates were subjected to immunoprecipitation with an anti-Drp1 antibody or an IgG control, and Western blotting was then performed. We found that endogenous Parkin was immunoprecipitated by endogenous Drp1, and the immunoprecipitated Parkin protein decreased when endogenous Drp1 protein level was down-regulated by Drp1 shRNA (Fig. 3B). To study whether Drp1 and Parkin interact directly, in vitro translated Drp1 was incubated with bacterially purified MBP-Parkin or MBP immobilized on amylose beads, and MBP pulldown assay was performed. Drp1 was detected in the pulldown preparation of MBP-Parkin but not in that of MBP (Fig. 3C). Thus, our results demonstrate that Drp1 and Parkin can interact both in cells and in vitro.
Parkin contains two ring finger domains separated by an in-between ring finger domain important for its interaction with most of the substrate proteins. We next determined the domain mediating the interaction of Parkin with Drp1. A series of Parkin truncated constructs were constructed to determine the domain of Parkin that interacted with Drp1 (Fig. 3D). Cells were transfected with the plasmids that express various truncations of GFP-Parkin, together with Myc-Drp1, and immunoprecipitation assay was then performed. Such an experiment revealed that the second ring finger domain of Parkin mediated its interaction with Drp1 (Fig. 3E). CHX assay indicated that the decrease of Drp1 half-life by Parkin was inhibited by deletion of the second ring finger of Parkin (supplemental Fig. S1).
Parkin Promotes the Ubiquitination of Drp1 in Cells
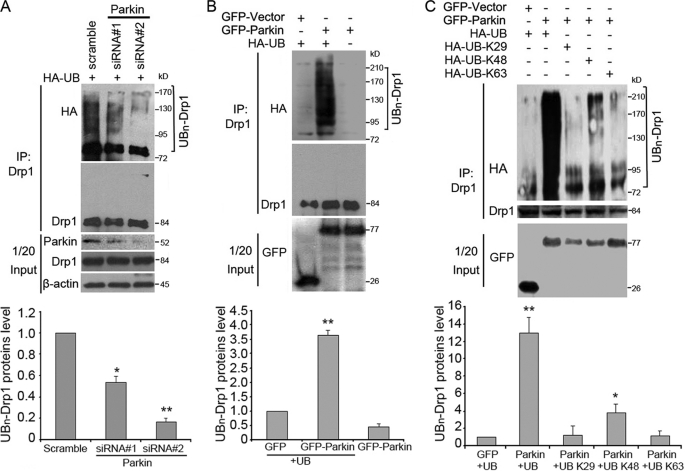
We then examined whether Drp1 is ubiquitinated by Parkin. To test this possibility, SH-SY5Y cells were transfected with HA-UB and control or Parkin siRNAs and treated with MG132 before harvest. Immunoprecipitation analysis indicated that the down-regulation of Parkin expression significantly reduced the ubiquitination of Drp1 (Fig. 4A).
FIGURE 4.
Parkin promotes the ubiquitination of Drp1 in cells. A, SH-SY5Y cells were transfected with HA-UB together with control or Parkin siRNAs as indicated. Cells were treated with MG132 before harvest. Cell lysates were then subjected to immunoprecipitation (IP) and Western blotting to examine the ubiquitination of Drp1. Uniquitinated Drp1 protein level was quantified according to the results of three independent blots. B, 293T cells were transfected with HA-UB together with GFP-Parkin or GFP vector. Cells were treated with MG132 for 8 h before harvest. Cell lysates were then subjected to immunoprecipitation and Western blotting. Uniquitinated Drp1 protein level was quantified according to the results of three independent blots. C, 293T cells were transfected with various HA-UB constructs together with GFP-Parkin or GFP vector. Cells were treated with MG132 for 8 h before harvest. Cell lysates were then subjected to immunoprecipitation and Western blotting. All Western blot data shown in the bar graphs represent means ± S.E. (error bars) from three independent experiments. *, p < 0.01 and **, p < 0.001.
We then examined whether Drp1 ubiquitination was enhanced by Parkin overexpression. 293T cells were transfected with HA-UB together with Myc-Drp1 and GFP-Parkin and treated with MG132 before harvest. By immunoprecipitation and Western blotting, we found that Parkin significantly increased the ubiquitination of Drp1 (Fig. 4B). Lys48-linked polyubiquitination acts as the canonical signal for targeting the substrate to the proteasome for degradation (14). Based on our finding that Parkin could ubiquitinate Drp1 leading to its degradation by the proteasome, we hypothesized that Parkin may mediate the polyubiquitination of Drp1 via Lys48-linked ubiquitin chains. To test this, cells were transfected with Myc-Drp1, GFP-Parkin, and various HA-UB constructs that encode wild-type ubiquitin or ubiquitin mutants containing arginine substitutions of all the lysine residues except the one at position 29, 48, or 63, respectively. Immunoprecipitation analysis revealed that Parkin was able to induce the polyubiquitination of Drp1 in the presence of wild-type or Lys48 ubiquitin, but not Lys29 or Lys63 ubiquitin (Fig. 4C). Thus, our results suggest that Parkin mediates the polyubiquitination of Drp1 mainly via Lys48-linked ubiquitin chains.
Mutations in Parkin Affect Its Ability to Ubiquitinate Drp1 for Degradation
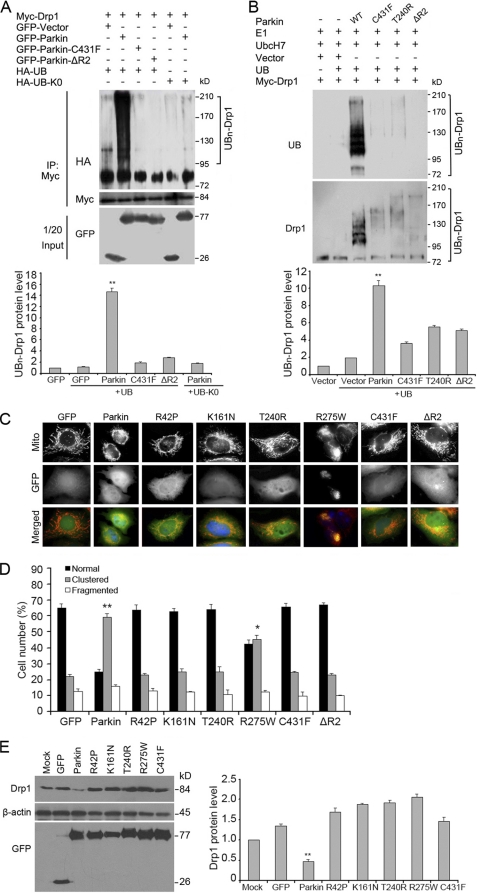
We then investigated whether familial PD-causing mutation (C431F) of Parkin, which is known to have attenuated E3 ligase activity (49), impaired its ability to ubiquitinate Drp1. 293T cells were transfected with HA-UB together with Myc-Drp1 and GFP-Parkin or GFP-Parkin (C431F), and treated with MG132 before harvest. By immunoprecipitation and followed by Western blotting, we found that Parkin mutation significantly decreased the ubiquitination of Drp1 (Fig. 5A). Similar results were achieved when the second ring finger domain of Parkin was deleted (Fig. 5A).
FIGURE 5.
Parkin mutations affect its ability to ubiquitinate Drp1 and to cause Drp1 degradation. A, 293T cells were transfected with HA-UB or HA-UB-K0 together with Myc-Drp1 and various GFP-Parkin constructs as indicated. Cells were treated with MG132 before harvest. Cell lysates were then subjected to immunoprecipitation (IP) and Western blotting to examine the ubiquitination of Drp1. Uniquitinated Drp1 protein level was quantified according to the results of three independent blots. B, in vitro translated Drp1 was incubated with purified ubiquitin, E1, E2 (UbcH7), and various Parkin proteins. The reaction products were analyzed by Western blotting with anti-ubiquitin and anti-Drp1 antibodies. Uniquitinated Drp1 protein level was quantified according to the results of three independent blots. C, cells were transfected with various GFP-Parkin constructs, stained with Mito-Tracker Red and then analyzed by fluorescence microscopy. D, experiments were performed as in C, and the percentage of cells with mitochondrial clustering was quantified. For each experiment, at least 100 cells were counted in three independent experiments. E, cells were transfected with various GFP-Parkin constructs, and Western blotting was performed. Drp1 protein level was quantified according to the results of three independent blots. All immunofluorescence data shown in the bar graphs represent mean ± S.E. (error bars) with at least 100 cells counted in a blinded manner. *, p < 0.01 and **, p < 0.001.
Next, we attempted to determine whether Parkin could directly ubiquitinate Drp1 and whether mutations in Parkin could result in the loss of its activity for Drp1 ubiquitination, by using an in vitro ubiquitination assay. To reconstitute the ubiquitin-conjugation-ligation reaction, in vitro translated Drp1 was incubated with purified ubiquitin, E1, E2 (UbcH7), and various Parkin proteins. MARCH5, an E3 ligase known to ubiquitinate Drp1, was used as a positive control for Parkin. The reaction products were analyzed by Western blotting with an anti-ubiquitin antibody. We found that wild-type Parkin, but not the C431F, T240R, or ΔR2 mutants, induced a significant increase in ubiquitinated protein bands (Fig. 5B). Western blotting with an anti-Drp1 antibody further confirmed that the high molecular weight ubiquitinated proteins represent the ubiquitinated Drp1 (Fig. 5B). We also found that the effect of Parkin on Drp1 ubiquitination was comparable with that of MARCH5 (supplemental Fig. S2). Taken together, these results demonstrate that Parkin is able to ubiquitinate Drp1 directly.
Mutations in Parkin account for 50% of all recessively transmitted early onset PD cases (50). We found that Parkin mutations R42P, K161N, T240R, R275W, and C431F could reduce its activity toward perinuclear mitochondrial clustering (Fig. 5, C and D) and Drp1 degradation (Fig. 5E). These data suggest that Parkin-induced ubiquitination and degradation of Drp1 might directly impact the regulation of mitochondrial morphology by Parkin and mutations of Parkin affecting its E3 ligase activity increase Drp1 expression, resulting in mitochondrial dysfunction.
DISCUSSION
In this study, we have found that Drp1 is a novel substrate of Parkin. Parkin effectively promotes the ubiquitination and proteasome-dependent degradation of Drp1. Our results implicate a potential role for Parkin dysregulation in mitochondrial dynamics and PD pathogenesis. Because the role of the Parkin mutation in the pathogenesis of PD has been well established, it has long been speculated that this event could result in the accumulation of its substrates affecting mitochondrial morphology and functions (18). A number of substrates of Parkin have been identified, and deregulation of these substrates are responsible for the symptoms of neurodegenerative disease such as PD (14, 18). However, the direct connection of these substrates with the mitochondrial dysfunction widely observed in PD is remains elusive. Our results thus suggest a straightforward mechanism for how mutations of Parkin impact mitochondrial integrity and functions, leading to PD.
Our results demonstrate that Parkin interacts with and subsequently ubiquitinates Drp1, a mitochondrial fission factor, resulting in the degradation of Drp1 by the proteasome-dependent pathway. The second ring finger domain of Parkin appears to be important for its interaction with Drp1. Mutations or deletion in this region may result in the loss of the E3 ligase activity of Parkin and/or affect its interaction with Drp1, thereby reducing the level of ubquitination and leading to mitochondrial fragmentation. Strong evidence has shown that, in mammalian cells, increased level of Drp1 causes mitochondrial fragmentation (31–34). Accumulation of mitofusin leads to enhanced mitochondrial aggregation and is protective to the mitochondrial dysfunctions and apoptosis (51). These results are in contrast to the recent reports showing that, in Drosophila, Parkin promotes mitochondrial fission (39, 40), and knockdown of Parkin results in mitochondrial elongation and increase of Mfn levels for mitophagy in Drosophila cells (38). The reasons for this discrepancy may be because Parkin regulates mitochondrial fission and fusion differently in distinct systems, and this warrants further clarification. Nevertheless, our results are consistent with the recent report that knockdown of Parkin leads to mitochondrial fragmentation (48). Our data suggest that one of the important functions of Parkin is to keep the cellular Drp1 level under control for proper mitochondrial functions.
Drp1 is the major factor responsible for mitochondrial fission (29, 33), which is one of the early events for neuronal cell death and the development of PD and other types of neurodegenerative diseases (20). Our results uncover a novel regulatory pathway of Drp1 by ubiquitination and proteasome-dependent degradation involving a cytosolic E3 ligase. E3 ubiquitin ligases localized in mitochondria, such as MARCH5/MITOL and Mulan, have been shown to regulate mitochondrial fission by interaction with Drp1 and another mitochondrial fission factor Fis1 (52, 53). To our knowledge, Parkin is the first cytosolic E3 ligase identified to regulate Drp1 turnover. Parkin is known to interact with PINK1/DJ1 (54) to regulate protein ubiquitination and degradation. It would be interesting to examine whether this type of interaction is also important for monitoring the Drp1 level in mitochondria. Recently, strong evidence has shown that Parkin is recruited onto damaged mitochondria to mediate mitophagy, a process for eliminating damaged or unwanted mitochondria (55, 56). Loss of PINK1 function promotes mitophagy in a Parkin- and Drp1-dependent manner (46). It is possible that Parkin has dual roles for mitochondrial homeostasis and quality control. On one hand, it is able to keep cellular Drp1 levels in check to prevent mitochondrial fragmentation. Once it is recruited toward depolarized mitochondria, Parkin may help remove damaged mitochondria. It is of interest to note that ubiquitination of proteins promotes both protein degradation and selective removal of unwanted protein aggregates or damaged organelles. Dysregulation of these processes may cause mitochondrial fragmentation and accumulation of damaged mitochondria for subsequent cell death and neural diseases.
It is possible that dysregulation of the Parkin/Drp1 axis represents one of the early events predisposing susceptibility of neuronal cells to intracellular or environmental changes because mitochondrial fragmentation is tightly associated with the loss of mitochondrial functions and enhanced mitochondrial oxidative stress. Investigation of how the Parkin/Drp1 axis is regulated may help elucidate the molecular pathogenesis of PD and design new treatment strategies, in addition to a better understanding of mitochondrial homeostasis.
Supplementary Material
Acknowledgments
We are grateful to Drs. Ted Dawson and Jian Feng for generously providing the plasmids. We are also grateful to Dr. Aimin Zhou from Cleveland State University for a critical reading of the manuscript.
This work was supported by Key Project Grant 30630038 and 973 Program Grants 2006CB910102 and 2010CB912200 from the National Natural Science Foundation of China (to Q. C.) and Grant 30825022 from the National Natural Science Foundation of China (to J. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PD
- Parkinson disease
- CHX
- cycloheximide
- Drp1
- dynamin-related protein 1
- UB
- ubiquitin
- MBP
- maltose-binding protein.
REFERENCES
- 1. Ishikawa A., Tsuji S. (1996) Neurology 47, 160–166 [DOI] [PubMed] [Google Scholar]
- 2. Thomas B., Beal M. F. (2007) Hum. Mol. Genet. 16, R183–194 [DOI] [PubMed] [Google Scholar]
- 3. Braak H., Ghebremedhin E., Rüb U., Bratzke H., Del Tredici K. (2004) Cell Tissue Res. 318, 121–134 [DOI] [PubMed] [Google Scholar]
- 4. Ferrer I. (2009) Prog. Neurobiol. 88, 89–103 [DOI] [PubMed] [Google Scholar]
- 5. Farrer M. J. (2006) Nat. Rev. Genet. 7, 306–318 [DOI] [PubMed] [Google Scholar]
- 6. Dawson T. M. (2007) Parkinsonism Relat. Disord. 13, S248–249 [DOI] [PubMed] [Google Scholar]
- 7. Dawson T. M., Dawson V. L. (2003) Science 302, 819–822 [DOI] [PubMed] [Google Scholar]
- 8. Bossy-Wetzel E., Schwarzenbacher R., Lipton S. A. (2004) Nat. Med. 10, S2–9 [DOI] [PubMed] [Google Scholar]
- 9. Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. (1998) Nature 392, 605–608 [DOI] [PubMed] [Google Scholar]
- 10. Lesage S., Brice A. (2009) Hum. Mol. Genet. 18, R48–59 [DOI] [PubMed] [Google Scholar]
- 11. Biskup S., Gerlach M., Kupsch A., Reichmann H., Riederer P., Vieregge P., Wüllner U., Gasser T. (2008) J. Neurol. 255, 8–17 [DOI] [PubMed] [Google Scholar]
- 12. Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., Suzuki T. (2000) Nat. Genet. 25, 302–305 [DOI] [PubMed] [Google Scholar]
- 13. Marín I., Lucas J. I., Gradilla A. C., Ferrús A. (2004) Physiol. Genomics 17, 253–263 [DOI] [PubMed] [Google Scholar]
- 14. Tanaka K., Suzuki T., Hattori N., Mizuno Y. (2004) Biochim. Biophys. Acta 1695, 235–247 [DOI] [PubMed] [Google Scholar]
- 15. Dev K. K., van der Putten H., Sommer B., Rovelli G. (2003) Neuropharmacology 45, 1–13 [DOI] [PubMed] [Google Scholar]
- 16. Corti O., Hampe C., Koutnikova H., Darios F., Jacquier S., Prigent A., Robinson J. C., Pradier L., Ruberg M., Mirande M., Hirsch E., Rooney T., Fournier A., Brice A. (2003) Hum. Mol. Genet. 12, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 17. Ko H. S., von Coelln R., Sriram S. R., Kim S. W., Chung K. K., Pletnikova O., Troncoso J., Johnson B., Saffary R., Goh E. L., Song H., Park B. J., Kim M. J., Kim S., Dawson V. L., Dawson T. M. (2005) J. Neurosci. 25, 7968–7978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kahle P. J., Haass C. (2004) EMBO Rep. 5, 681–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ko H. S., Kim S. W., Sriram S. R., Dawson V. L., Dawson T. M. (2006) J. Biol. Chem. 281, 16193–16196 [DOI] [PubMed] [Google Scholar]
- 20. Cho D. H., Nakamura T., Fang J., Cieplak P., Godzik A., Gu Z., Lipton S. A. (2009) Science 324, 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H., Guo M. (2009) J. Clin. Invest. 119, 442–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin M. T., Beal M. F. (2006) Nature 443, 787–795 [DOI] [PubMed] [Google Scholar]
- 23. Knott A. B., Perkins G., Schwarzenbacher R., Bossy-Wetzel E. (2008) Nat. Rev. 9, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orth M., Schapira A. H. (2001) Am. J. Med. Genet. 106, 27–36 [DOI] [PubMed] [Google Scholar]
- 25. Schon E. A., Manfredi G. (2003) J. Clin. Invest. 111, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schapira A. H. (2008) Lancet Neurol. 7, 97–109 [DOI] [PubMed] [Google Scholar]
- 27. Yaffe M. P. (1999) Nat. Cell Biol. 1, E149–150 [DOI] [PubMed] [Google Scholar]
- 28. Okamoto K., Shaw J. M. (2005) Annu. Rev. Genet. 39, 503–536 [DOI] [PubMed] [Google Scholar]
- 29. Chen H., Chan D. C. (2005) Hum. Mol. Genet. 14, R283–289 [DOI] [PubMed] [Google Scholar]
- 30. Detmer S. A., Chan D. C. (2007) Nat. Rev. Mol. Cell Biol. 8, 870–879 [DOI] [PubMed] [Google Scholar]
- 31. Zhu P. P., Patterson A., Stadler J., Seeburg D. P., Sheng M., Blackstone C. (2004) J. Biol. Chem. 279, 35967–35974 [DOI] [PubMed] [Google Scholar]
- 32. Wells R. C., Picton L. K., Williams S. C., Tan F. J., Hill R. B. (2007) J. Biol. Chem. 282, 33769–33775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoon Y., Krueger E. W., Oswald B. J., McNiven M. A. (2003) Mol. Cell. Biol. 23, 5409–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S. O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y., Taguchi N., Morinaga H., Maeda M., Takayanagi R., Yokota S., Mihara K. (2009) Nat. Cell Biol. 11, 958–966 [DOI] [PubMed] [Google Scholar]
- 35. Frank S., Gaume B., Bergmann-Leitner E. S., Leitner W. W., Robert E. G., Catez F., Smith C. L., Youle R. J. (2001) Dev. Cell 1, 515–525 [DOI] [PubMed] [Google Scholar]
- 36. Van Laar V. S., Berman S. B. (2009) Exp. Neurol. 218, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Whitworth A. J., Pallanck L. J. (2009) J. Bioenerg. Biomembr. 41, 499–503 [DOI] [PubMed] [Google Scholar]
- 38. Ziviani E., Tao R. N., Whitworth A. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poole A. C., Thomas R. E., Andrews L. A., McBride H. M., Whitworth A. J., Pallanck L. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deng H., Dodson M. W., Huang H., Guo M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang Y., Ouyang Y., Yang L., Beal M. F., McQuibban A., Vogel H., Lu B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7070–7075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park J., Lee G., Chung J. (2009) Biochem. Biophys. Res. Commun. 378, 518–523 [DOI] [PubMed] [Google Scholar]
- 43. Greene J. C., Whitworth A. J., Kuo I., Andrews L. A., Feany M. B., Pallanck L. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4078–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. (2006) Nature 441, 1162–1166 [DOI] [PubMed] [Google Scholar]
- 45. Cui M., Tang X., Christian W. V., Yoon Y., Tieu K. (2010) J. Biol. Chem. 285, 11740–11752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dagda R. K., Cherra S. J., 3rd, Kulich S. M., Tandon A., Park D., Chu C. T. (2009) J. Biol. Chem. 284, 13843–13855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kuroda Y., Mitsui T., Kunishige M., Shono M., Akaike M., Azuma H., Matsumoto T. (2006) Hum. Mol. Genet. 15, 883–895 [DOI] [PubMed] [Google Scholar]
- 48. Lutz A. K., Exner N., Fett M. E., Schlehe J. S., Kloos K., Lämmermann K., Brunner B., Kurz-Drexler A., Vogel F., Reichert A. S., Bouman L., Vogt-Weisenhorn D., Wurst W., Tatzelt J., Haass C., Winklhofer K. F. (2009) J. Biol. Chem. 284, 22938–22951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Giasson B. I., Lee V. M. (2001) Neuron 31, 885–888 [DOI] [PubMed] [Google Scholar]
- 50. Mata I. F., Lockhart P. J., Farrer M. J. (2004) Hum. Mol. Genet. 13, R127–133 [DOI] [PubMed] [Google Scholar]
- 51. Sugioka R., Shimizu S., Tsujimoto Y. (2004) J. Biol. Chem. 279, 52726–52734 [DOI] [PubMed] [Google Scholar]
- 52. Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. (2008) PloS One 3, e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakamura N., Kimura Y., Tokuda M., Honda S., Hirose S. (2006) EMBO Rep. 7, 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xiong H., Wang D., Chen L., Choo Y. S., Ma H., Tang C., Xia K., Jiang W., Ronai Z., Zhuang X., Zhang Z. (2009) J. Clin. Invest. 119, 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.