Abstract
Tubulin post-translational modifications generate microtubule heterogeneity and modulate microtubule function, and are catalyzed by tubulin tyrosine ligase-like (TTLL) proteins. Using antibodies specific to monoglycylated, polyglycylated, and glutamylated tubulin in whole mount immunostaining of zebrafish embryos, we observed distinct, tissue-specific patterns of tubulin modifications. Tubulin modification patterns in cilia correlated with the expression of ttll3 and ttll6 in ciliated cells. Expression screening of all zebrafish tubulin tyrosine ligase-like genes revealed additional tissue-specific expression of ttll1 in brain neurons, ttll4 in muscle, and ttll7 in otic placodes. Knockdown of ttll3 eliminated cilia tubulin glycylation but had surprisingly mild effects on cilia structure and motility. Similarly, knockdown of ttll6 strongly reduced cilia tubulin glutamylation but only partially affected cilia structure and motility. Combined loss of function of ttll3 and ttll6 caused near complete loss of cilia motility and induced a variety of axonemal ultrastructural defects similar to defects previously observed in zebrafish fleer mutants, which were shown to lack tubulin glutamylation. Consistently, we find that fleer mutants also lack tubulin glycylation. These results indicate that tubulin glycylation and glutamylation have overlapping functions in maintaining cilia structure and motility and that the fleer/dyf-1 TPR protein is required for both types of tubulin post-translational modification.
Keywords: Development, Microtubules, Post-translational Modification, Tubulin, Zebrafish, Cilia, Tubulin Tyrosine Ligase-like
Introduction
Glycylation and glutamylation are post-translational modifications of tubulin subunits within stable microtubules of cilia, centrioles, neuronal axons, and mitotic spindles (1–3). Variable lengths of added glycyl or glutamyl side chains generate diverse tubulin subtypes and contribute to heterogeneity of tubulin function by modulating microtubule-protein interactions (2, 4, 5). For instance, glutamyl side chain length has been shown to differentially affect recruitment of the microtubule-associated proteins MAP1A and MAP2 (6), and to selectively enhance KIF1A-dependent vesicle transport in hippocampal neurons (7). In vertebrates, glutamylated tubulin is associated with cilia and neuronal axons, whereas glycylated tubulin is primarily localized to cilia (8–11). Both modifications occur on an overlapping set of glutamates near the tubulin C terminus (3, 12, 13). Deletion of the β-tubulin C terminus is lethal in Tetrahymena and generates non-motile, short cilia lacking a portion of the microtubule doublet B-subfiber (14–16). Drosophila mutants lacking the β-tubulin C terminus fail to assemble sperm axonemes, whereas cytoplasmic microtubules are not affected (17, 18). Axonemal B-subfiber defects and reduced cilia beat amplitude are also observed in the zebrafish fleer mutant, which lacks a TPR protein required for cilia polyglutamylation (19), as well as in Tetrahymena overexpressing the tubulin glutamylase, TTLL6Ap (20, 21). Evidence for a role for tubulin glycylation in cilia elongation in zebrafish has recently been reported (22). Thus whereas, the importance of tubulin polymodifications are clear, the roles of glycylation versus glutamylation in regulating cilia function have only recently emerged with the identification of enzymes responsible for tubulin-amino acid ligation.
Tubulin polymodifications are catalyzed by members of the tubulin tyrosine ligase-like (TTLL)2 protein family. The 13 mammalian TTLL proteins are distinct with respect to preference toward α- or β-tubulin as substrates, ability to add either glycine or glutamate residues, and activity as initiating or elongating post-translational amino acid linkages (3, 23, 24). Substrate recognition by TTLLs can also be influenced by interactions between TTLL proteins and non-catalytic components of a tubulin-modifying complex (20, 25). Recent reports indicate that TTLL tubulin glutamylases in Tetrahymena and Chlamydomonas preferentially modify B subfibers of axonemal doublets and are required for normal cilia motility by modulating dynein inner arm-microtubule interactions (26, 27). TTLL1 has been implicated in mouse lung cilia tubulin glutamylation and regulation of asymmetric cilia waveform that is essential for directed fluid flow (28).
Although these studies underscore the essential role of TTLL proteins in generating tubulin diversity, less is known about spatio-temporal regulation of TTLL activity and how multiple TTLL activities may affect cilia functions (19, 20, 22, 23). The zebrafish embryo provides a good model for in vivo functional analysis of ttll genes because cilia in zebrafish exhibit tissue-specific differences in length, number, and motility (19, 29). We find that tubulin modifications are specific to different types of cilia and in many cases both glutamylation and glycylation occur in single axonemes. We present evidence that the ttll3 glycylase and ttll6 glutamylase are the main tubulin modifying enzymes of ciliated cells and whereas, disruption of either tubulin glycylation or glutamylation alone causes mild cilia defects, combined loss of both tubulin modifications generates severe defects in cilia structure and motility. The results suggest that different tubulin modifications can have overlapping or redundant activities in maintaining cilia function.
MATERIALS AND METHODS
Zebrafish Strains and Maintenance
The wild type TuAB and CD-41 GFP zebrafish lines were used for morpholino injections and these were maintained according to standard procedures. For maintenance of optical clarity, 1-phenyl 2-thiourea was added to the E3 water between 20 and 24 h.
Zebrafish ttll EST/cDNA Clones
Zebrafish ttll orthologs were identified by blast search using mammalian proteins as query against the current zebrafish genome sequence (Zv9; tblastn). Best hits for each TTLL gene were identified by reverse blasting against all GenBankTM protein sequence (nr; blastp). A full-length ttll6 cDNA (EU124004) and partial ttll10 cDNA were cloned in pCS 2Gateway. Full or partial EST clones of additional zebrafish ttll genes were purchased from Open Biosystems. Accession numbers of ttll gene sequences used are ttll1 (BC053185), ttll2 (EE324724), ttll3 (BC117656), ttll4 (EG567349), ttll7 (BC129350), ttll9 (BC115266), ttll11 (BC124694), ttll12 (BC048032).
Morpholino Knockdown of ttll3 and ttll6
Antisense morpholino ttll3ex7dMo was designed to target the ttll3 splice donor site at exon 7; ttll6ATGMo morpholino was designed to block the ttll6 initiation codon. Morpholino sequences with the targeted splice and translation initiation sites underlined: ttll3ex7dMo, GATTAAGAGCACTGACCAATAAAAG; ttll6ATGMo, CTGGTGTCCCCATTCTGATCTCTTC; the ttll3ATG sequence was identical to the one used by Wloga et al. (22), GTGTTGGTGCATGTTTGAGTTAACC. Control injections were performed using a heterologous morpholino: Kif17conMo, CTGTTAGGTCCATTGGTCAAATGTT (30).
Morpholino oligonucleotides were dissolved in RNase-free water to a 2 mm stock concentration. Working dilutions were prepared in 100 mm KCl, 10 mm Hepes (pH 7.4), 0.1% phenol red, and 4.6-nl volumes were injected in 2–4 cell wild type TuAB or transgenic CD41-GFP embryos, using a nanoliter 2000 microinjector (WPI instruments, Sarasota, FL). To detect morpholino-induced splicing defects of ttll3ex7dMo, nested RT-PCR was performed on total RNA extracted from individual embryos using the following primers: ttll3 Ex4F1, CCCGACGACATGACCATGAAACT; ttll3 Ex5F2, ATGACCTAATGTCGCGGCTGGTT; ttll3 Ex9R1, CGTGTCTTCCTCAGACCATGGGATT; ttll3 Ex8R2, GCACTTCTCCCATTTTCTCCCCTTC.
For statistical analysis of ttll3/ttll6 knockdown synergy, the equation: (Fttll3Mo × Fttll6Mo) − Fttll3/6Mo = D (31), was evaluated where F equaled the fraction of wild type embryos remaining in each condition and a positive number for D indicated synergy (double knockdown effects greater than combined effects of individual gene knockdowns). To assess statistical significance, the null hypothesis that (Fttll3Mo × Fttll6Mo) − Fttll3/6Mo ≤ 0 was tested using the one-tailed Student's t test for four independent experimental replicates.
RNA in Situ Hybridization
Digoxigenin-labeled antisense riboprobes of ttll genes were synthesized by individual in vitro transcription reactions using the T7 RNA polymerase. Host plasmid/linearizing restriction enzyme/RNA polymerase were: ttll1 (pCMVSport6.1EcoRI/T7), ttll2 (pExpress1/EcoRI/T7), ttll3 (pExpress1/EcoRI/T7), ttll4 (pExpress1/EcoRI/T7), ttll6 (pCS2Gateway/BamHI/T7), ttll7 (pME18S-FL3/PCR generated T7 promoter), ttll9 (pExpress1/EcoRI/T7), ttll10 (cDNA/PCR generated T7 promoter), ttll11 (pME18S-FL3/PCR generated T7 promoter), and ttll12 (PME18S-FL3/PCR generated T7 promoter). Whole mount in situ with individual digoxigenin-labeled probes was performed as described previously (29, 32). Double in situ hybridization analysis was performed using digoxigenin-labeled riboprobe for either ttll3 or ttll6 and the fluorescein-labeled shpp1 probe by light and fluorescent microscopy based color detection methods as described previously (29). Stained embryos were dehydrated and transferred to a 2:1 mixture of benzyl benzoate:benzyl alcohol or alternatively the stained larvae were cleared in dimethylformamide and mounted in 80% glycerol. Images were obtained using a Spot image digital camera mounted on a Leitz MZ12 stereomicroscope or Nikon E 800 microscope or via the Zeiss LSM5 confocal microscope.
Immunofluorescence Microscopy
Whole zebrafish larvae were fixed for immunolabeling in Dent's fixative (80% methanol, 20% dimethyl sulfoxide) at 4 °C overnight. For visualization of pronephric cilia, distension of the pronephric lumen was induced by mechanical obstruction of the cloaca 1 h prior to fixation. Fixed specimens were rehydrated gradually and washed with PBS containing 0.5% Tween 20 (PBST) and blocked for 2 h with 5% normal goat serum, prior to antibody labeling. Larvae were sequentially incubated with primary and secondary antibodies overnight at 4 °C. The dilutions and primary antibodies used were: TAP952 (1:200), R PolyG (1:300), mAb GT335 (1:400), mAb 6-11B-1 (1:800), and mAb Gtu88 (1:800). Immunoreactivity was detected using Alexa 546-conjugated goat anti-rabbit (1:800) and Alexa 546-conjugated goat anti-mouse and Alexa 488-conjugated donkey anti-mouse (1:800). Immunolabeling with two mouse antibodies was performed as described earlier where the first primary and secondary antibodies were briefly fixed with 4% paraformaldehyde and their reactive sites blocked by sequential incubations in 10% unconjugated mouse serum and 5% unconjugated mouse Fab′ fragments (19). Two color confocal Z-series were acquired using sequential laser excitation on a Zeiss Pascal LSM5 laser scanning microscope.
High-speed Video Microscopy Analysis
54 hpf control and morpholino-injected 1-phenyl 2-thiourea-treated embryos of wild type TuAB or CD-41 GFP transgenic zebrafish (to identify pronephric multiciliated cells) were maintained alive in an anesthetic/immobilizing mixture (E3 egg water containing tricaine (1:25 dilution of 4.1% stock) and the heartbeat blocker 2,3-butanedione monoxime (20 mm final concentration). The immobilized larvae were placed on 3% methylcellulose immersed in the anesthetic/immobilizing mixture and observed by Nomarski optics using a ×40/0.55 water immersion lens mounted on the Nikon E-800 microscope. Images of moving cilia were obtained using the Dragonfly2 CCD camera (Point Gray Research, Richmond, BC, Canada) at 240 frames/s. The QuickTime movies were imported into ImageJ software where the cilia beat frequency and amplitude were measured using the line scan function. Beat amplitude was determined from the pixel displacement of the cilium at extreme positions and converted to micromole scale (1 μm = 2.7 pixels in a 324 × 242 tiff image).
Electron Microscopy
Fixation and embryo processing for electron microscopy were as previously described (33). Quantification of cilia defects was performed by identifying orthogonal sections of pronephric cilia with intact peripheral membrane and clear outer doublet structure and counting B-subfiber defects per total number of doublets and other structural defects per total number of cilia.
RESULTS
Distinct Patterns of Glutamyl- and Glycyl-tubulin in Zebrafish Cilia and Neurons
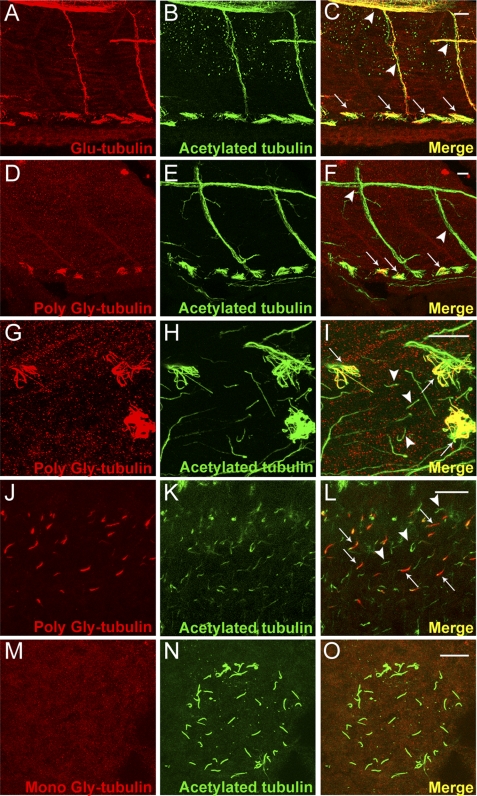
To examine the distribution of glutamylated and glycylated tubulin in microtubules in vivo, we performed whole mount confocal immunofluorescence on wild type zebrafish embryos and larvae with antibodies specific to glutamylated tubulin (mAb GT335), monoglycylated tubulin (mAb TAP952), and polyglycylated tubulin (rabbit polyclonal Ab R PolyG). In 2-day-old zebrafish larvae (Fig. 1, A–F) glutamylated tubulin was detected in pronephric single and multicilia, intersomitic motor neuron axons (Fig. 1, A–C), cilia basal bodies in Kupffer vesicle, the zebrafish organ of laterality (supplemental Fig. S1, A–C) and, as previously reported, in olfactory multicilia (19) (supplemental Fig. S1, D–F). In contrast, polyglycylated tubulin was present in pronephric multicilia (Fig. 1, D–I, arrows in F and I) and olfactory placode cilia (supplemental Fig. S1, G–I) but excluded from intersomitic neurons (Fig. 1, E, and arrowheads in F) and from single cilia in the pronephros (Fig. G–I, arrowheads in I). Polyglycylated tubulin was also detected in a subset of spinal canal cilia within the caudal spinal cord (Fig. 1, J–L). Cilia tubulin monoglycylation (TAP952) has previously been reported to be ubiquitous (22) (supplemental Fig. S1, J–L). However, we found unexpectedly that cilia of Kupffer vesicle were unique in not showing any evidence of tubulin mono- or polyglycylation (TAP952, Fig. 1, M–O; R PolyG, data not shown).
FIGURE 1.
In vivo distribution of glutamyl and glycyl-modified tubulin in zebrafish. A–C, confocal projection of the trunk region of a 2.5-day post-fertilized wild type zebrafish larva, double immunolabeled with Glu-tubulin specific mAb GT335 (red) (A), acetylated tubulin-specific mAb 6-11B-1 (B), and the merged image (C). Glutamylated and acetylated tubulin overlap in motor neurons (arrowheads in C) and all types of pronephric cilia (arrows at bottom in C). D–F, confocal projections of the trunk region of a 2.5-day post-fertilized wild type zebrafish larva double immunolabeled with poly-Gly tubulin-specific R PolyG (red) (D), acetylated tubulin specific mAb 6-11B-1 (green) (E), and the merged image (F). Poly-Gly tubulin was localized specifically in pronephric cilia (arrows) and excluded from the primary motor neurons (arrowheads). G–I, high magnification view of the pronephros showing cilia labeled with poly-Gly tubulin-specific R PolyG (G), acetylated tubulin specific 6-11B-1 (H), and the merged image (I). Polyglycylated tubulin occurred only in the multicilia subtype (arrows in I) and not in adjacent, single pronephric cilia (arrowheads in I). J–L, confocal projection of spinal canal cilia immunolabeled with poly-Gly tubulin-specific R PolyG (J) (red), acetylated tubulin-specific mAb 6-11B-1 (K) (green), and the merged image (L). Polyglycylated tubulin was present in a specific subset of cilia (arrows in L). M–O, confocal projections of the Kupffer vesicle in a 10-somite wild type zebrafish embryo, double immunolabeled with mono-Gly tubulin-specific mAb TAP952 (M); acetylated tubulin specific mAb 6-11B-1 (N), and the merged image (O). Monoglycylated tubulin was absent from all Kupffer vesicle cilia. Scale bars = 10 μm.
Expression of the Tubulin Tyrosine Ligase-like Enzymes ttll3 and ttll6 Is Enriched in Ciliated Epithelia
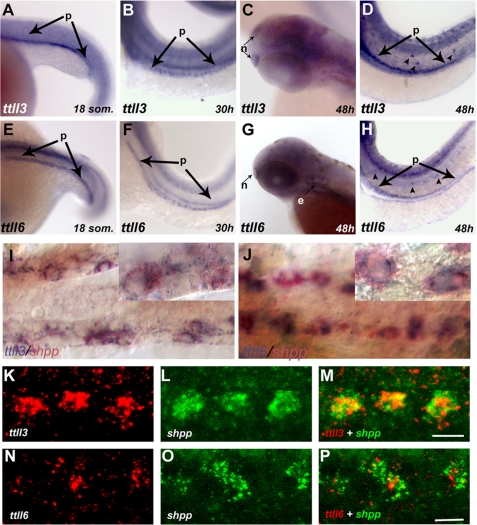
Tubulin modifying enzymes are members of the TTLL protein family (20). To characterize tissue- and cell-type specific patterns of ttll gene expression, we performed a whole mount RNA in situ hybridization screen of all zebrafish ttll genes. We identified zebrafish homologs of mammalian TTLL genes (supplemental Table S1) and analyzed expression of ttll1, -2, -3, -4, -5, -6, -7, -9, -10, -11, and -12 during development. No zebrafish homolog for mammalian TTLL8 or TTLL13 was found in exhaustive searches of the zebrafish genome. No in situ expression of ttll5 was observed. For the remaining ttll genes, ttll1, -2, -3, -4, -6, -7, -9, -10, -11, and -12, we observed abundant maternal transcripts as early as the 8-cell stage (shown representatively for ttll1; supplemental Fig. S2, I1). Later in development, the majority of ttll genes were widely expressed in the central nervous system (supplemental Fig. S2). Unique expression patterns were observed for ttll1 (proximal pronephros, brain neurons; supplemental Fig. S2, A3 and A4), ttll3 (ciliated epithelia; supplemental Fig. S2, C1–C5, and Fig. 2, A–D), ttll4 (somites, heart, and gut; supplemental Fig. S2, D1, D4, and D6), ttll6 (ciliated epithelia; supplemental Fig. S2, E1–E6, and Fig. 3, E–H), and ttll7 (otic placode; supplemental Fig. S2, F2 and F4).
FIGURE 2.
Expression of ttll3 and ttll6 is enriched in ciliated tissues. A–D, RNA in situ hybridization showing ttll3 is expressed during early development in distinct ciliated tissues: A, in the presumptive pronephros (p) at the 18-somite stage; B, at 30 hpf in discrete cells of the pronephros and at 48 hpf in olfactory placode (C) (n); D, lateral line organs (arrowheads) and in the pronephros (p; arrows). E–H, RNA in situ hybridization showing ttll6 is expressed in distinct ciliated tissues: E, the presumptive pronephros of an 18-somite embryo; F, in discrete cells of the pronephros at 30 and 48 hpf (G) in the olfactory placode; H, lateral line organs (arrowheads) and the pronephros (p). I and J, bright field images of two-color RNA in situ hybridization, showing transcripts of both ttll3 (I) (blue) and ttll6 (J) (blue) , respectively, colocalized with shpp (red), a marker of multiciliated cells. Insets within each panel show individual cells at higher magnification. K–P, fluorescent two-color RNA in situ hybridization confocal images showing antisense probes of ttll3 (red) (K); ttll6 (red) (N) and the multiciliated cell marker shpp (green) (L and O) in a single confocal slice of the pronephros (30 hpf). In the merged panels, coexpression of ttll3 (M) with shpp (green) (red) and ttll6 (red) with shpp (green) (P) indicates that transcripts of both ttll3 and ttll6 are enriched in multiciliated cells. Scale bars in M and P = 10 μm.
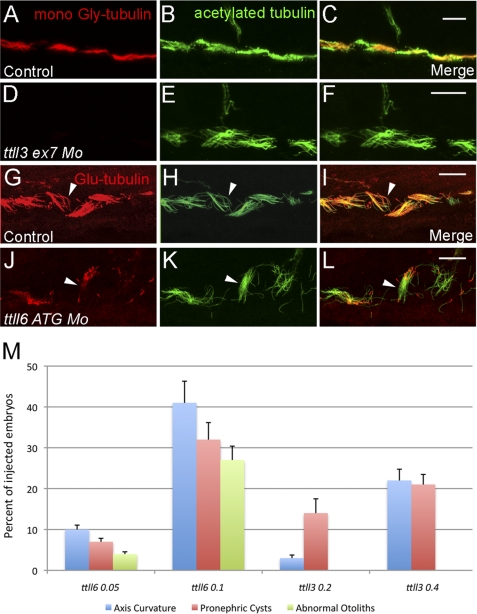
FIGURE 3.
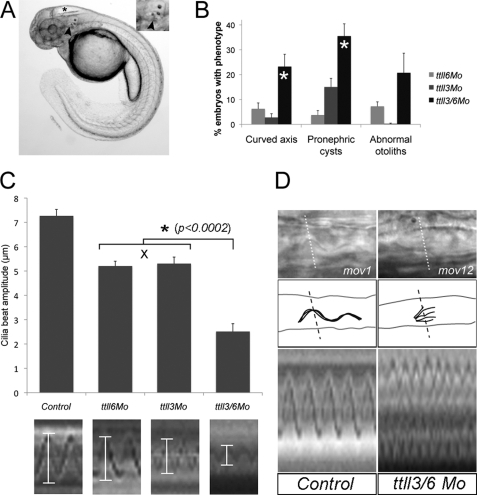
Tubulin glycylation and glutamylation is inhibited in ttll3 and ttll6 morphants. A–C, pronephric cilia from 2.5-day-old embryo injected with the control morpholino and double immunolabeled with mono-Gly tubulin-specific mAb TAP952 (A), acetylated tubulin-specific mAb 6-11B-1 (B), and the merged image (C). D–F, pronephric cilia from 2.5-day-old embryos injected with ttll3ex7dMo (4.6 nl of 0.4 mm stock) and double immunolabeled with mono-Gly tubulin-specific mAb TAP952 (D), acetylated tubulin-specific mAb 6-11B-1 (E), and the merged image (F). In the merged panel (F), multicilia in ttll7 knockdown embryos show absence of TAP952 labeling and appear normal with respect to acetylated tubulin content and length. G–H, pronephric cilia of 2.5-day-old embryos injected with control morpholino and double immunolabeled with mono- and poly-Glu tubulin-specific mAb GT335 (G), acetylated tubulin-specific mAb6-11B-1 (H), and the merged image (I). In the merged panel (I) note that glutamylated tubulin is abundant within the basal bodies and distributed along the axonemes in a decreasing gradient from the base to the tip. J–L, pronephric cilia of 2.5-day-old embryos injected with ttll6ATGMo (4.6 nl of 0.1 mm stock) and double immunolabeled with mono- and poly-Glu tubulin-specific mAbGT335 (J), acetylated tubulin-specific mAb 6-11B-1 (K), and the merged image (L). Both single and multicilia contain Glu-tubulin in basal bodies but show a significant reduction of Glu-tubulin in axonemes (arrowheads). Scale bars = 10 μm. M, graph showing that frequencies of cilia phenotypes (axis curvature, pronephric cysts and abnormal otolith number, shown on the y axis) increase as a function of the morpholino dose in embryos injected with ttll6ATGMo and ttll3ex7dMo. All phenotype frequencies were significantly increased over control embryos that never manifested these defects.
Given the abundant expression of ttll3 and ttll6 in ciliated cells and their importance in axonemal tubulin modification (34), we further characterized their cell type-specific expression. High-resolution RNA in situ images of the tubulin glycylase ttll3 (Fig. 2, A–D) and the tubulin glutamylase ttll6 (Fig. 2, E-H) demonstrated expression of both of these genes in multiple ciliated epithelia including the pronephros (Fig. 2, A, B, E, and F), olfactory placodes (Fig. 2, C and G), and lateral line organs (Fig. 2, D and H). Double label RNA in situ hybridization analysis with the pronephric multiciliated cell marker shpp1 (29) revealed that expression of both ttll3 (Fig. 2, I–M) and ttll6 (Fig. 2, J and N-P) were enriched in the multiciliated cells of the pronephros, similar to the expression of fleer, a gene required for tubulin glutamylation in zebrafish and Caenorhabditis elegans (19).
Functional Significance of Tubulin Post-translational Modifications in Zebrafish
Both tubulin glycylation and glutamylation have been reported to be essential in cilia function and biogenesis (23, 34, 35). The presence of both modifications on a subset of zebrafish cilia and the overlapping expression of ttll3 and ttll6 in ciliated cells raised the possibility that tubulin glycylation and glutamylation could have unique roles in cilia or alternatively, that tubulin modifications could be functionally redundant. To resolve cilia-specific functions of ttll3 and ttll6, we knocked down each gene alone or in combination and assayed cilia-related phenotypes.
We first disrupted ttll3 expression using antisense morpholinos targeting the ttll3 exon7 splice donor (supplemental Fig. S3A). ttll3 knockdown with the ttll3ex7dMo (4.6 nl of 0.4 mm stock) resulted in complete absence of normal ttll3 mRNA and appearance of a misspliced mRNA (exon skipping) predicted to encode a truncated enzyme lacking the Ttll3 catalytic domain (supplemental Fig. S3B). Consistent with complete loss of ttll3 function, tubulin glycylation (TAP952 immunofluorescence) was completely eliminated in pronephric cilia in 100% of the examined embryos (n = 30) (Fig. 3, D–F, and supplemental Fig. S4). Loss of tubulin glycylation in ttll3 knockdown embryos resulted in a significant dose-dependent, but relatively low frequency of cilia-related phenotypes including ventral axis curvature and pronephric kidney cysts (Fig. 3M). Cilia length in the pronephros was not affected (Fig. 3E).
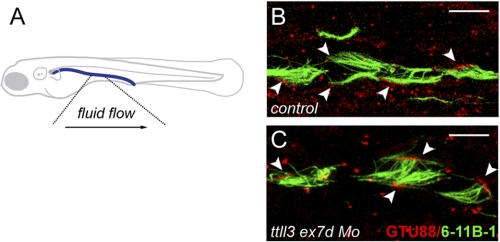
The partial penetrance of pronephric cysts in ttll3-deficient embryos suggested that fluid flow directed by cilia motility in the pronephros could be variably compromised by lack of tubulin glycylation. In wild type embryos, multicilia beat unidirectionally toward the cloaca (supplemental Movie S1), driving fluid flow out of the pronephros (36). In ttll3-deficient embryos that developed kidney cysts, isolated bundles of multicilia were observed to beat in reverse orientation, opposite to the direction of normal fluid flow (supplemental Movies S2–S5). To determine whether these cilia bundles were truly reversed or represented artifacts induced by tubule distension, we assessed cilia orientation by immunostaining basal bodies with anti-γ-tubulin mAb GTU88 and axonemes with anti-acetylated tubulin (Fig. 4). In the pronephric lumen of wild type zebrafish, the basal bodies and axonemes of all multicilia show a posterior polarization toward the cloaca (Fig. 4B), even when luminal distension was experimentally induced (n = 8 embryos). In contrast, in ttll3 morphants, 2 to 3 misoriented cilia bundles in each nephron were observed in 5 of 8 (63%) embryos imaged (Fig. 4C). The results suggest that loss of tubulin polyglycylation contributes to randomization of multicilia orientation in ttll3-deficient embryos.
FIGURE 4.
Randomization of multicilia orientation in ttll3 morphants. A, schematic view of the zebrafish indicating the direction of fluid flow in the highlighted area sampled to determine multicilia orientation. B and C, pronephric cilia in 2.5-day-old larvae injected with control morpholino (B) and ttll3ex7dMo (C) and double immunolabeled to reveal the axonemes (mAb 6-11B-1; green) and basal bodies (γ-tubulin-specific mAb GTU88). Note that axonemes and basal bodies of all wild type multicilia (B) are oriented to the posterior, whereas ttll3 morphants, basal bodies, and axonemes (C) orientation of some cilia (arrowheads) is reversed toward the anterior. Scale bars = 10 μm.
Surprisingly, using the ttll3ex7dMo that eliminated tubulin glycylation, we did not observe significant cilia-related left-right asymmetry defects previously reported with ttll3 loss of function (22) (supplemental Fig. S5). Injection of the ttll3 ATG morpholino previously used to block ttll3 translation initiation did replicate previously reported left-right asymmetry defects (supplemental Fig. S5S) (22) but also caused significant developmental delay and brain necrosis (supplemental Fig. S5B), common nonspecific side effects of morpholino toxicity (37). Mitigation of nonspecific morpholino effects by co-injection of p53 and ttll3 ATG morpholinos together significantly abated laterality defects (supplemental Fig. S5A). Thus ttll3 was required for cilia tubulin glycylation but, consistent with the lack of tubulin glycylation in Kupffer vesicle cilia (Fig. 1M), was not required for establishing normal left-right asymmetry.
We showed previously that inactivation of zebrafish ttll6 by a splice blocking morpholino affected glutamylation and assembly of olfactory cilia (19) but did not generate other strong cilia phenotypes, possibly due to maternally contributed ttll6 mRNA (supplemental Fig. S2). We therefore tested whether ttll6 knockdown using a translation blocking morpholino (ttll6 ATG Mo; 4.6 nl of a 0.1 mm stock) would further reduce cilia tubulin glutamylation. ttll6 knockdown by a translation blocking morpholino significantly reduced tubulin glutamylation in pronephric cilia (Fig. 3, J–L) compared with controls (Fig. 3, G–I). Loss of glutamylation was consistently observed in single cilia whereas, glutamylation in pronephric multicilia was more variable. ttll6 knockdown did not affect tubulin glutamylation in basal bodies or cilia transition zones (Fig. 3, J–L). Knockdown of ttll1, ttll3, or injection of two different control morpholinos had no effect on cilia glutamylation, arguing that the effects we observe are specific to ttll6 deficiency. ttll6 knockdown induced a dose-dependent set of cilia phenotypes including axis curvature, pronephric cysts, and abnormal otolith number (Fig. 3M). Because cilia length was not significantly altered in ttll6-deficient embryos (Fig. 3K), we examined whether altered cilia beat pattern might underlie observed ciliopathy phenotypes. One instance of reversed multicilia polarity, similar to ttll3 knockdown embryos, was seen but this was not common (1 cilia bundle in 4 embryos examined; supplemental Movie S6). More frequently, ttll6 knockdown caused a reduction in cilia beat amplitude in isolated cilia (supplemental Movies S7 and S8, see below). All embryos examined (n = 6) showed reduced beat amplitude but cilia defects varied in different cells (supplemental Movies S7 and S8).
Combined Deficiency in Tubulin Glutamylation and Polyglycylation Results in Severe Cilia Defects
Despite the complete absence of tubulin glycylation in ttll3 morphants and strong reduction in cilia microtubule glutamylation in ttll6 morphants, cilia-related phenotypes were only partially penetrant (Fig. 3M), suggesting that reduction in either tubulin modification alone may not be sufficient to cause severe cilia defects. We therefore tested whether combined knockdown of ttll3 and ttll6 might have synergistic effects on cilia function. To avoid nonspecific morpholino toxicity, we injected half-maximal doses of each morpholino and assayed cilia phenotypes. All frequencies of cilia phenotypes were synergistically increased in combined ttll3/ttll6-deficient embryos compared with single ttll gene knockdowns (Fig. 5B), with axis curvature and pronephric cyst formation showing statistically significant synergism. Functional synergy between ttll3 and ttll6 knockdown most strongly reduced cilia beat amplitude but not average beat frequency (Fig. 5C; control, 40 ± 2 beats/s (bps); ttll3ex7dMo, 34 ± 6 bps; ttll6ATGMo, 35 ± 2 bps; and ttll3/6Mo, 40 ± 7 bps; n ≥ 8 for each condition). In wild type embryos, the majority of pronephric multicilia beat with an amplitude between 6 and 8 μm (Fig. 5C and supplemental Movie S1). Disruption of either ttll3 or ttll6 alone reduced multicilia beat amplitude, with a majority of cilia exhibiting beat amplitudes of 4–6 μm (Fig. 5C and supplemental Movies S2–S8). Strikingly, knockdown of both ttll3 and ttll6 synergistically reduced multicilia beat amplitude (the majority to 0–4 μm; p < 0.0002), approaching paralysis in some multicilia (Fig. 5, C and D, and supplemental Movies S9-S12). Combined ttll3/6 knockdown also markedly disrupted cilia beat coordination in multiciliated cell bundles. Control pronephric cilia bundles emanating from multiciliated cells beat as a coherent group, as revealed in line scans of high speed videos (Fig. 5D and supplemental Movie S1), whereas all combined ttll3/6-deficient embryos (n = 6) exhibited cilia in bundles that beat independently and out of phase (Fig. 5D and supplemental Movie S12).
FIGURE 5.
Cilia defects increase synergistically in ttll3/6 double morphants. A, ciliopathy phenotypes in a 2.5-day-old ttll6 morphants manifest as hydrocephalus (asterisk), abnormal otolith number (arrowhead and inset), and ventrally curved body axis. B, percentages of morphant embryos showing axis curvature defects, pronephric cysts, and abnormal otolith number when injected with half-maximal dose of ttll6ATGMo alone (0.05 mm; light grey box), ttll3ex7dMo alone (0.2 mm; medium grey box), and a combination of both ttll6ATGMo and ttll3ex7dMo (0.05 + 0.2 mm; dark grey box). All frequencies of phenotypes were synergistically increased in ttll3/6 double morphants with statistically significant synergy in curved axis and pronephric cyst phenotypes (marked by asterisk; axis, p = 0.03; cysts, p = 0.02; otoliths, p = 0.08; see “Materials and Methods”). C, average cilia beat amplitude in control and ttll knockdown embryos. Statistical analysis of synergy was performed as described under “Materials and Methods.” Error bars represent mean ± S.E. Representative ImageJ cilia beat amplitude line scans are shown at the bottom for each condition. D, line scan analysis of cilia beat coordination in control (left; see supplemental Movie S1) and combined ttll3/6 knockdown (right; see supplemental Movie S12) embryos. Dashed lines indicate the position of the line scan. Control cilia bundles beat coordinately (bottom), whereas ttll3/6 knockdown embryo cilia bundles beat out of phase and at a reduced amplitude.
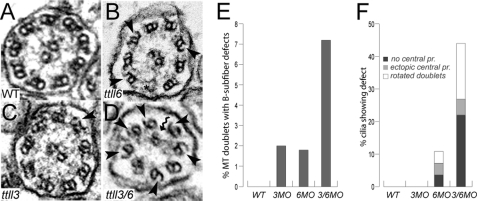
Axonemal microtubule doublets in cilia lacking tubulin modifications often exhibit loss of the outer aspect of B-subfiber microtubules (14, 16, 19). We assessed whether similar ultrastructural defects could be observed upon inactivation of ttll3 and ttll6 alone or in combination. B-tubule defects were rarely observed in pronephric cilia of individual ttll6 (Fig. 6, B and E) or ttll3 (Fig. 6, C and E) single knockdown embryos. However, combined knockdown of ttll3 and ttll6 (Fig. 6, D–F) induced multiple ultrastructural defects including gaps in B-subfiber microtubules (Fig. 6D, arrowheads), misplaced or rotated microtubule doublets (Fig. 6D, kinked arrow, and supplemental Fig. S6, H, J, and K), and ectopic, supernumerary microtubule central pairs (straight arrows in supplemental Fig. S6, I and L). Quantification of B-subfiber defects (Fig. 6E) or various structural defects (Fig. 6F) revealed a synergistic increase when both ttll3 and ttll6 were knocked down compared with single knockdowns of either gene. Cilia structural defects may represent the most severely affected axonemes or cilia degeneration, whereas all cilia exhibited reduced motility in ttll3/6-deficient embryos (Fig. 5, C and D), structural defects were more limited. The results suggest that ttll3 (glycylation) and ttll6 (glutamylation) are likely to have overlapping functions in maintaining axonemal structure and cilia motility.
FIGURE 6.
Synergistic increase in cilia ultrastructural defects in ttll3/6 double morphants. A, axoneme of wild type pronephros multicilia showing the normal 9 + 2 microtubule structure. B, polyglycylation-deficient cilia of ttll3 morphants are normal with rare instances of small B-subfiber breaks (arrowhead) in the outer microtubule doublets. C, glutamylation-deficient cilia of ttll6 morphants show rare instances of B-subfiber breaks (arrowheads). D, cilia of the ttll3/6 double morphants show a range of ultrastructural defects including missing dynein arms, rotated outer doublets (kinked arrows), and frequent loss of B-subfiber outer segments (arrowheads). E, quantification of microtubule doublets showing missing B-subfiber outer segments (% of doublets showing defect; total doublets counted: n = 171 wt; n = 100 ttll3MO, n = 217 ttll6MO, n = 305 ttll3/6MO). F, percent of cilia showing missing central pair microtubules (dark grey), misplaced or ectopic central pairs (light grey), or rotated microtubule doublets (white) (total cilia sections counted: n = 19 wt; n = 14 ttll3MO; n = 28 ttll6MO; n = 41 ttll3/6MO).
fleer Mutants Lack Both Tubulin Glycylation and Glutamylation
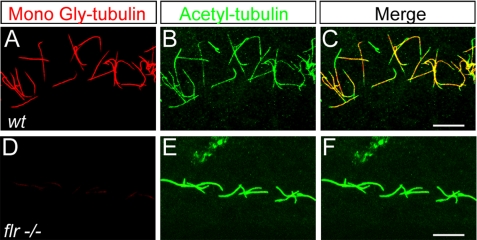
The zebrafish fleer gene encodes an ortholog of the Dyf-1/IFT70 tetratricopeptide repeat protein that regulates tubulin glutamylation in zebrafish and C. elegans (19). fleer mutants have paralyzed cilia and show ultrastructural defects in B-subfiber microtubules (19), similar to embryos deficient in both tubulin glycylation and glutamylation (Figs. 5 and 6). When stained with the TAP952 monoclonal antibody to detect tubulin monoglycylation, pronephric cilia in all fleer mutant embryos examined (n = 5) showed a strong reduction in tubulin glycylation (Fig. 7, D–F) compared to controls (Fig. 7, A–C), indicating that the Fleer/Dyf-1/IFT70 protein regulates both tubulin glutamylation and glycylation.
FIGURE 7.
fleer mutant cilia lack tubulin glycylation. A–C, confocal projections of pronephric cilia double immunolabeled with mono-Gly tubulin-specific mAbTAP952 (A), acetylated-tubulin specific mAb 6-11B-1 (B), and the merged image (C). D–F, 2.5-day-old fleer larva immunolabeled with (D) mono-Gly tubulin-specific mAbTAP952 (D), 6-11B-1 (E), and the merged image (F). Mono-Gly tubulin images (C and F) were acquired under identical imaging parameters indicate that mono-Gly tubulin levels are drastically reduced in fleer mutant cilia. Scale bars = 10 μm.
DISCUSSION
Polymodifications of the tubulin C terminus are now known to be critical for a variety of microtubule-based cellular processes (1, 34, 38). Focusing on the tubulin tyrosine ligase like (ttll) gene family in zebrafish, we found that 1) tubulin modifications occur in specific cilia subtypes, with pronephric and olfactory multicilia associated with both tubulin glycylation and glutamylation, 2) loss of tubulin glycylation or glutamylation separately had only mild effects on cilia whereas, combined loss of both modifications dramatically affected cilia ultrastructure and motility, and 3) in addition to its role in cilia tubulin glutamylation, the Fleer/Dyf-1 protein also regulates cilia tubulin glycylation.
Tubulin Modifications and Zebrafish Cilia
Recent studies have reported that most if not all cilia in zebrafish embryos contain monoglycylated tubulin whereas, a subset of cilia are polyglycylated (22). This finding would be consistent with the wide distribution of monoglycylated tubulin in protozoan and mammalian cilia (8, 11, 39–41). Our results showing complete elimination of tubulin glycylation by ttll3 knockdown are consistent with previous reports establishing an essential role for ttll3 as a tubulin glycylase (22). The loss of tubulin glycylation in ttll3-deficient embryos argues that other ttll genes, for instance ttll8 (24, 25, 42), are not sufficient to initiate tubulin glycylation in zebrafish and may have been lost in the course of zebrafish evolution. Given the essential role for tubulin glycylation in Tetrahymena cilia assembly and similar claims put forward for zebrafish (22), we were surprised to find that loss of ttll3-mediated tubulin glycylation did not have a strong effect on cilia assembly, ultrastructure, or beat rate. Our results are in contrast Wloga et al. (22) who reported that knockdown of the ttll3 tubulin glycylase in zebrafish caused shortening of KV cilia with subsequent defects in establishing left-right asymmetry, concluding that the main effect of ttll3 depletion was shortening and loss of axonemes. This discrepancy may be due to the reliance by Wloga et al. (22) on a morpholino that in our hands produced significant off-target effects. When off-target effects of this morpholino were prevented by co-knockdown of p53 (preventing apoptosis), cilia functional deficits (randomized left-right asymmetry) were also prevented. Our results showing mild effects of ttll3 loss of function are unlikely to be due to incomplete knockdown because RTPCR verification of ttll3 knockdown demonstrated complete absence of normal ttll3 mRNA, and immunostaining for monoglycylated tubulin (TAP952 antibody) showed a complete absence of glycyl-tubulin in cilia. We conclude that whereas Ttll3 is an essential tubulin glycylase, loss of glycylation by itself is not sufficient to significantly disrupt cilia function in zebrafish, perhaps due to the presence of additional, possibly redundant, tubulin modifications on axonemal microtubules (mono- and polyglutamylation; see below). Although we could not confirm a specific role for tubulin glycylation in cilia assembly, we did observe a randomization of multicilia orientation in the pronephros of ttll3-deficient embryos. Tubulin polyglycylation in both fish and mammalian cilia is largely restricted to multiciliated cells (8, 43) that support bundles of synchronously beating cilia, suggesting a role for polyglycylation in contexts where cilia dynamics must be coordinated. Recent studies of Ttll1 function in mouse lung cilia have shown that tubulin glutamylation is required to generate a curved cilia waveform and effective fluid propulsion (28), indicating that tubulin modifications play a role in generating asymmetric properties in cilia. Our finding that disrupting tubulin glycylation leads to cilia misorientation further supports this idea that polyglycylation could be important, either directly or indirectly, for proper cilia or basal body polarity in the zebrafish kidney. Planar cell polarity components such as Dishevelled and Van Gogh-like protein have been implicated in apical basal body docking and orientation (44, 45). Randomization of multicilia orientation in ttll3 morphants could arise from defects in delivery or assembly of cell polarity determinants in the apical cytoplasm (46). Our results suggest that deficiency in tubulin glycylation does not affect the initial docking of basal bodies but may interfere with subsequent basal body alignment.
ttll6 knockdown caused loss of axonemal tubulin glutamylation but not basal body tubulin glutamylation, suggesting that other ttll family genes maintain tubulin glutamylation in basal bodies (19). For example, in addition to ttll6, we also detected transient expression of ttll1, ttll4, ttll7, and ttll9 glutamylases in ciliated cell types (supplemental Fig. S1). ttll1 would perhaps be the best candidate for a redundant glutamylase in cilia because it is expressed in the presumptive pronephros up to 30 hpf and functions in mouse lung cilia (28). However, ttll1 knockdown did not affect pronephric cilia glutamylation (data not shown), suggesting that other ttll are responsible for basal body tubulin glutamylation in the absence of ttll6.
fleer and ttll Function
The fleer gene is homologous to the C. elegans dyf-1 gene and encodes an essential regulator of cilia tubulin glutamylation (19) and glycylation (this work). fleer mutants exhibit cilia immotility and delayed multiciliated cell differentiation with misoriented cilia bundles (19). These defects in cilia function, along with specific ultrastructural gaps in axonemal microtubule B-subfibers, resemble defects seen in cilia lacking tubulin glycyl or glutamyl addition sites (14, 16) and in embryos described here deficient in both ttll-mediated tubulin glycylation and glutamylation. These observations suggest that a combined lack of tubulin post-translational modifications may partially account for the fleer mutant phenotype, specifically with respect to axonemal structure and cilia beat amplitude. The requirement for Fleer in mediating tubulin post-translational modifications and the known axonemal transport of the Fleer homolog Dyf-1 by intraflagellar transport (47) might suggest that Fleer/Dyf-1 could act as a cargo adaptor for Ttll protein delivery to cilia via IFT (19). However, protein interaction studies have not identified Ttll proteins as direct Fleer interactors,3 suggesting that the role of Fleer may be regulatory or indirect. Also, because cilia length was not affected by ttll3/6 knockdown, whereas fleer mutants show shorter axonemes and defects in multiciliated cell development, it is likely that fleer has additional functions in ciliogenesis. Strong ciliogenesis defects in Tetrahymena and Trypanosome Dyf-1 mutants also suggest additional roles for Fleer/Dyf-1 in IFT processes underlying ciliogenesis (48, 49).
Tubulin Post-translational Modifications: Unique Activities or Functionally Redundant?
Although all cilia microtubules are polymodified, the patterns of glycylation versus glutamylation vary widely in different cilia compartments and between species (14, 50–52). For instance, axonemal microtubules in Paramecia are polyglycylated; glutamylation is a quantitatively minor modification that is not detectable by mass spectrometry (14). The reverse is true in Trypanosomes and Plasmodia where tubulin glycylation is not detected (51, 52). These observations suggest that one or the other modification may be sufficient for cilia function and that tubulin glycylation and glutamylation may be functionally redundant (38). On the other hand, specific depletion of glycylated tubulin in Drosophila sperm by TTLL3B RNAi knockdown results in male infertility, despite the continued presence of glutamylated tubulin in sperm axonemes (24). Similarly, a dominant-negative Tetrahymena TTLL3Ap inhibits tubulin glycylation and ciliogenesis, whereas tubulin glutamylation is unaffected or increased (22). In addition, both glycylation and glutamylation occur on the same C-terminal glutamate residues in the β-tubulin primary sequence suggesting that different TTLL modifying enzymes may compete for substrate availability (14, 53). Although attractive in its simplicity, a competitive model does not explain why only glutamylated tubulin is glycylated in Drosophila sperm (50) or why increasing monoglycylation of Tetrahymena tubulin by Ttll3Ap overexpression does not significantly affect tubulin polyglutamylation (22). Further studies using antibodies with well defined specificity will be required to fully assess the relationship of tubulin glutamylation and glycylation.
Our results show that in zebrafish, strong cilia defects are only observed when both tubulin glycylation and glutamylation are reduced by combined ttll3 and ttll6 knockdown, arguing that in this context, tubulin glycylation and glutamylation may be functionally redundant. At first view, functional redundancy might be unexpected because the chemical properties of glycine (neutral) and glutamate (acidic) are quite different. However, a common feature of both glycyl and glutamyl (or any amino acid) side chains is the presence of an acidic C-terminal carboxylate group that would extend out from the tubulin primary sequence (54) forming a flexible extension of tubulin surface negative charge. Tubulin glycyl side chains, whereas not expected to increase the tubulin C-terminal net negative charge, have in fact been shown to interact strongly with monovalent cations, most likely due to the acidic properties of their C-terminal carboxylate groups (54). Flexible negative charges tethered to microtubule surfaces may provide more degrees of freedom for microtubule-protein interactions and enhance dynamic interactions of positively charged motor proteins with microtubules (1, 54). Although specific functions for glutamyl and glycyl tubulin modifications exist (26, 27), functional redundancy of these modifications appears to be the simplest explanation for why profound structural defects in cilia are seen only when 1) tubulin polymodification sites for both glycylation and glutamylation are mutated (14, 16, 55), 2) both tubulin glutamylation and glycylation are prevented in fleer mutants (19), and 3) when both tubulin glycylation and glutamylation are reduced by combined ttll3/ttll6 knockdown (this work). Now that the TTLL enzymes responsible for tubulin modification have been characterized, it should be possible to determine in which contexts tubulin modifications are interchangeable or functionally unique.
Supplementary Material
Acknowledgments
We gratefully acknowledge input from members of the Drummond lab during preparation of this manuscript. We thank Dr. Martin Gorovsky for the gift of R PolyG antibody, Dr. Carsten Janke for the GT335 antibody, and Dr. Nicolette Levilliers for the TAP952 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants K01DK078741 (to N. P.) and RO1DK053093 (to I. A. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, Figs. S1–S7, and Movies S1–S12.
N. Pathak, unpublished observation.
- TTLL
- tubulin tyrosine ligase-like
- hpf
- hours post-fertilization.
REFERENCES
- 1. Gaertig J., Wloga D. (2008) Curr. Top. Dev. Biol. 85, 83–113 [DOI] [PubMed] [Google Scholar]
- 2. Hammond J. W., Cai D., Verhey K. J. (2008) Curr. Opin. Cell Biol. 20, 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janke C., Rogowski K., van Dijk J. (2008) EMBO Rep. 9, 636–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Verhey K. J., Gaertig J. (2007) Cell Cycle 6, 2152–2160 [DOI] [PubMed] [Google Scholar]
- 5. Maas C., Belgardt D., Lee H. K., Heisler F. F., Lappe-Siefke C., Magiera M. M., van Dijk J., Hausrat T. J., Janke C., Kneussel M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8731–8736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonnet C., Boucher D., Lazereg S., Pedrotti B., Islam K., Denoulet P., Larcher J. C. (2001) J. Biol. Chem. 276, 12839–12848 [DOI] [PubMed] [Google Scholar]
- 7. Ikegami K., Heier R. L., Taruishi M., Takagi H., Mukai M., Shimma S., Taira S., Hatanaka K., Morone N., Yao I., Campbell P. K., Yuasa S., Janke C., Macgregor G. R., Setou M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3213–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dossou S. J., Bré M. H., Hallworth R. (2007) Cell Motil. Cytoskeleton 64, 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rüdiger M., Plessmann U., Rüdiger A. H., Weber K. (1995) FEBS Lett. 364, 147–151 [DOI] [PubMed] [Google Scholar]
- 10. Davenport J. R., Watts A. J., Roper V. C., Croyle M. J., van Groen T., Wyss J. M., Nagy T. R., Kesterson R. A., Yoder B. K. (2007) Curr. Biol. 17, 1586–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Adoutte A., Delgado P., Fleury A., Levilliers N., Lainé M. C., Marty M. C., Boisvieux-Ulrich E., Sandoz D. (1991) Biol. Cell 71, 227–245 [DOI] [PubMed] [Google Scholar]
- 12. Redeker V., Levilliers N., Schmitter J. M., Le Caer J. P., Rossier J., Adoutte A., Bré M. H. (1994) Science 266, 1688–1691 [DOI] [PubMed] [Google Scholar]
- 13. Eddé B., Rossier J., Le Caer J. P., Desbruyères E., Gros F., Denoulet P. (1990) Science 247, 83–85 [DOI] [PubMed] [Google Scholar]
- 14. Redeker V., Levilliers N., Vinolo E., Rossier J., Jaillard D., Burnette D., Gaertig J., Bré M. H. (2005) J. Biol. Chem. 280, 596–606 [DOI] [PubMed] [Google Scholar]
- 15. Xia L., Hai B., Gao Y., Burnette D., Thazhath R., Duan J., Bré M. H., Levilliers N., Gorovsky M. A., Gaertig J. (2000) J. Cell Biol. 149, 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thazhath R., Liu C., Gaertig J. (2002) Nat. Cell. Biol. 4, 256–259 [DOI] [PubMed] [Google Scholar]
- 17. Popodi E. M., Hoyle H. D., Turner F. R., Raff E. C. (2005) Cell Motil. Cytoskeleton 62, 48–64 [DOI] [PubMed] [Google Scholar]
- 18. Fackenthal J. D., Turner F. R., Raff E. C. (1993) Dev. Biol. 158, 213–227 [DOI] [PubMed] [Google Scholar]
- 19. Pathak N., Obara T., Mangos S., Liu Y., Drummond I. A. (2007) Mol. Biol. Cell 18, 4353–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Janke C., Rogowski K., Wloga D., Regnard C., Kajava A. V., Strub J. M., Temurak N., van Dijk J., Boucher D., van Dorsselaer A., Suryavanshi S., Gaertig J., Eddé B. (2005) Science 308, 1758–1762 [DOI] [PubMed] [Google Scholar]
- 21. Wloga D., Dave D., Meagley J., Rogowski K., Jerka-Dziadosz M., Gaertig J. (2009) Eukaryot. Cell 9, 184–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wloga D., Webster D. M., Rogowski K., Bré M. H., Levilliers N., Jerka-Dziadosz M., Janke C., Dougan S. T., Gaertig J. (2009) Dev. Cell 16, 867–876 [DOI] [PubMed] [Google Scholar]
- 23. van Dijk J., Rogowski K., Miro J., Lacroix B., Eddé B., Janke C. (2007) Mol. Cell 26, 437–448 [DOI] [PubMed] [Google Scholar]
- 24. Rogowski K., Juge F., van Dijk J., Wloga D., Strub J. M., Levilliers N., Thomas D., Bré M. H., Van Dorsselaer A., Gaertig J., Janke C. (2009) Cell 137, 1076–1087 [DOI] [PubMed] [Google Scholar]
- 25. Ikegami K., Horigome D., Mukai M., Livnat I., MacGregor G. R., Setou M. (2008) FEBS Lett. 582, 1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kubo T., Yanagisawa H. A., Yagi T., Hirono M., Kamiya R. (2010) Curr. Biol. 20, 441–445 [DOI] [PubMed] [Google Scholar]
- 27. Suryavanshi S., Eddé B., Fox L. A., Guerrero S., Hard R., Hennessey T., Kabi A., Malison D., Pennock D., Sale W. S., Wloga D., Gaertig J. (2010) Curr. Biol. 20, 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ikegami K., Sato S., Nakamura K., Ostrowski L. E., Setou M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 10490–10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Y., Pathak N., Kramer-Zucker A., Drummond I. A. (2007) Development 134, 1111–1122 [DOI] [PubMed] [Google Scholar]
- 30. Insinna C., Pathak N., Perkins B., Drummond I., Besharse J. C. (2008) Dev. Biol. 316, 160–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bloomfield P., Eastwood B. (1987) Toxicology 46, 237–245 [DOI] [PubMed] [Google Scholar]
- 32. Thisse C., Thisse B. (1998) in The Zebrafish Science Monitor, Vol. 5, pp. 8–9, University of Oregon, Eugene, OR [Google Scholar]
- 33. Drummond I. A., Majumdar A., Hentschel H., Elger M., Solnica-Krezel L., Schier A. F., Neuhauss S. C., Stemple D. L., Zwartkruis F., Rangini Z., Driever W., Fishman M. C. (1998) Development 125, 4655–4667 [DOI] [PubMed] [Google Scholar]
- 34. Wloga D., Gaertig J. (2010) J. Cell Sci. 123, 3447–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wloga D., Rogowski K., Sharma N., Van Dijk J., Janke C., Eddé B., Bré M. H., Levilliers N., Redeker V., Duan J., Gorovsky M. A., Jerka-Dziadosz M., Gaertig J. (2008) Eukaryot. Cell 7, 1362–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kramer-Zucker A. G., Olale F., Haycraft C. J., Yoder B. K., Schier A. F., Drummond I. A. (2005) Development 132, 1907–1921 [DOI] [PubMed] [Google Scholar]
- 37. Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., Ekker S. C. (2007) PLoS Genet. 3, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Westermann S., Weber K. (2003) Nat. Rev. Mol. Cell Biol. 4, 938–947 [DOI] [PubMed] [Google Scholar]
- 39. Bré M. H., Redeker V., Quibell M., Darmanaden-Delorme J., Bressac C., Cosson J., Huitorel P., Schmitter J. M., Rossler J., Johnson T., Adoutte A., Levilliers N. (1996) J. Cell Sci. 109, 727–738 [DOI] [PubMed] [Google Scholar]
- 40. Iftode F., Clérot J. C., Levilliers N., Bré M. H. (2000) Biol. Cell 92, 615–628 [DOI] [PubMed] [Google Scholar]
- 41. Brown J. M., Marsala C., Kosoy R., Gaertig J. (1999) Mol. Biol. Cell 10, 3081–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ikegami K., Setou M. (2009) FEBS Lett. 583, 1957–1963 [DOI] [PubMed] [Google Scholar]
- 43. Million K., Larcher J., Laoukili J., Bourguignon D., Marano F., Tournier F. (1999) J. Cell Sci. 112, 4357–4366 [DOI] [PubMed] [Google Scholar]
- 44. Park T. J., Mitchell B. J., Abitua P. B., Kintner C., Wallingford J. B. (2008) Nat. Genet. 40, 871–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ross A. J., May-Simera H., Eichers E. R., Kai M., Hill J., Jagger D. J., Leitch C. C., Chapple J. P., Munro P. M., Fisher S., Tan P. L., Phillips H. M., Leroux M. R., Henderson D. J., Murdoch J. N., Copp A. J., Eliot M. M., Lupski J. R., Kemp D. T., Dollfus H., Tada M., Katsanis N., Forge A., Beales P. L. (2005) Nat. Genet. 37, 1135–1140 [DOI] [PubMed] [Google Scholar]
- 46. Vladar E. K., Axelrod J. D. (2008) Trends Cell Biol. 18, 517–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ou G., Blacque O. E., Snow J. J., Leroux M. R., Scholey J. M. (2005) Nature 436, 583–587 [DOI] [PubMed] [Google Scholar]
- 48. Absalon S., Blisnick T., Kohl L., Toutirais G., Doré G., Julkowska D., Tavenet A., Bastin P. (2008) Mol. Biol. Cell 19, 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dave D., Wloga D., Sharma N., Gaertig J. (2009) Eukaryot. Cell 8, 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoyle H. D., Turner F. R., Raff E. C. (2008) Cell Motil. Cytoskeleton 65, 295–313 [DOI] [PubMed] [Google Scholar]
- 51. Fennell B. J., Al-shatr Z. A., Bell A. (2008) Int. J. Parasitol. 38, 527–539 [DOI] [PubMed] [Google Scholar]
- 52. Schneider A., Plessmann U., Weber K. (1997) J. Cell Sci. 110, 431–437 [DOI] [PubMed] [Google Scholar]
- 53. Bulinski J. C. (2009) Dev. Cell 16, 773–774 [DOI] [PubMed] [Google Scholar]
- 54. Vinh J., Langridge J. I., Bré M. H., Levilliers N., Redeker V., Loyaux D., Rossier J. (1999) Biochemistry 38, 3133–3139 [DOI] [PubMed] [Google Scholar]
- 55. Thazhath R., Jerka-Dziadosz M., Duan J., Wloga D., Gorovsky M. A., Frankel J., Gaertig J. (2004) Mol. Biol. Cell 15, 4136–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.