Abstract
Myosin VI is unique in its directionality among myosin superfamily members and also displays a slow and strain-dependent rate of ATP binding that allows for gating between its heads. In this study we demonstrate that leucine 310 is positioned by a class VI-specific insert, insert-1, so as to account for the selective hindrance of ATP versus ADP binding. Mutation of leucine 310 to glycine removes all influence of insert-1 on ATP binding. Furthermore, by analyzing myosin VI structures with either leucine 310 substituted to a glycine or complete removal of insert-1, we conclude that nucleotides may initially bind to myosin by their purine rings before docking their phosphate moieties. Otherwise, insert-1 could not exert a differential influence on ATP versus ADP binding.
Keywords: Kinetics, Molecular Motors, Myosin, Nucleotide, X-ray Crystallography
Introduction
Myosin VI is the only myosin superfamily member that has been shown to traffic toward the minus (pointed) end of actin filaments (1). This unique directionality is made possible due to a myosin VI class-specific structural element that is known as insert-2 and enables myosin VI to perform unique cellular functions (2). This insert wraps around the myosin VI converter and binds a calmodulin via an unusual calmodulin binding motif (3). This redirects the lever arm of myosin VI ∼120° as compared with plus-end-directed myosin motors.
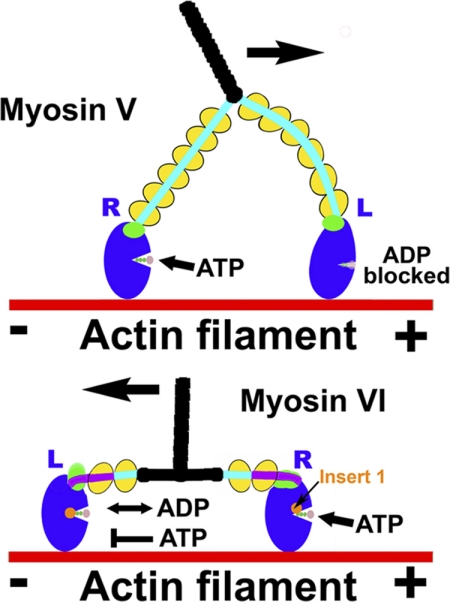
Like myosin V, dimeric myosin VI can function as a processive motor on actin (4–7). That is to say that the dimer can move along actin in a hand-over-hand fashion with steps of 30–36 nm. As in the case of the plus-end directed processive dimer of myosin Va (8–11), the processive movement of myosin VI is optimized by “gating” between the heads. Intramolecular strain stalls the lead head until the rear head has detached from actin as the dimer moves along the filament (8–12). However, the mechanism of how this is accomplished differs between myosin V and VI, necessitated by their opposite directionalities. In the case of myosin V, intramolecular strain greatly decreases the rate of dissociation of MgADP from the lead head (8–11), whereas in the case of myosin VI, ADP dissociation is unaffected, but ATP binding is greatly slowed on the lead head, preventing its detachment (12). This is illustrated in Fig. 1.
FIGURE 1.
Gating in processive myosin dimers. A schematic representation of double-headed myosin V and myosin VI bound to actin illustrates the different gating mechanisms these motors use to optimize their processive movement on actin. Because of their opposite directionality, the lead head (L) experiences the opposite intramolecular strain in myosin VI as compared with myosin V. In myosin V, ADP release is slowed, and the lead head is stalled in a strongly bound ADP state until the rear head (R) detaches after ATP binding. In myosin VI, strain from the rear head does not affect ADP release and binding from the lead head but instead greatly slows ATP binding, preventing dissociation of the lead head from actin until the rear head has detached.
Based on the structure of myosin VI (3), it appeared that another myosin VI class-specific insert, insert-1, is responsible for impeding ATP binding. Indeed, even in the absence of strain, binding of ATP to monomeric myosin VI is at least an order of magnitude slower than binding to myosin V (3), and removal of the 26-residue insert increased the rate of ATP binding (3, 12). The mechanism of how this is achieved was not entirely clear, although we postulated that insert-1 positioned the side chain of leucine 310 physically in the way so that it impeded ATP but not ADP entry into the pocket.
In this study we examine if positioning of leucine 310 does indeed provide steric hindrance for ATP binding and is in fact the mechanism by which myosin VI gating is achieved. Although we have examined the effects of insert-1 on nucleotide binding to myosin VI in the presence of actin, we had not previously examined the impact of insert-1 in the absence of actin. We do so in this study to enable interpretation of myosin VI structures with and without either insert-1 or mutation of leucine 310 to glycine.
EXPERIMENTAL PROCEDURES
Protein Constructs and Expression
A series of truncations of porcine myosin VI cDNA were generated. C-terminal truncations were made corresponding to amino acids 816 (wild type MDins2, Δ-insert-1 MDins2 (in which residues Cys-278 to Ala-303 have been deleted), and L310G mutant of MDins2) and 839 (S1). Each of these had a FLAG tag (encoding DYKDDDDK) appended via a glycine to either the N terminus (MDins2) or the C terminus (S1) to facilitate purification. As previously described (13), a “zippered” dimer myosin VI construct (HMM) was created by truncation at Arg-994 followed by a leucine zipper (GCN4 (14) to ensure dimerization. This sequence was followed by myc and FLAG tags for motility assays and purification, respectively, as described previously (15). These constructs were used to create a baculovirus for expression in SF9 cells, as previously described (16).
ATPase Assays and Transient Kinetics
Actin-activated ATPase assays and transient kinetic measurements shown in Tables 1 and 2 were performed as described (13, 16). Briefly, steady state ATPase activities were measured at 25 °C in KMg50 buffer (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 1 mm DTT, 10 mm imidazole, pH 7.0, at 25 °C) to which 1 mm MgATP was added and then supplemented with the NADH-coupled assay components (17). Transient kinetic measurements were made in KMg50 buffer at 25 °C with an Applied Photophysics SX.18MV stopped-flow having a 1.2-ms dead time. A 400-nm colored glass emission filter was used to monitor pyrene (λex = 365 nm), mantADP3 (λex = 365 nm), mantATP (λex = 365 nm), or NADH (λex = 340 nm) fluorescence. Nonlinear least-squares fitting of the data was done with software provided with the instrument or with KaleidaGraph (Synergy Software, Reading, PA). Uncertainties reported are S.E. in the fits of 3–4 independent protein preparations and sets of measurements.
TABLE 1.
Steady state and transient kinetic parameters of myosin (M) VI-S1 constructs in the presence of actin at 25 °C
| Kinetic parameters | MVI MDins2IQ WT | MVI MDins2IQ L310G | MVI MDins2IQ (Δ-Insert-1) |
|---|---|---|---|
| Steady state parameters | |||
| Vmax (s−1)a | 6.8 ± 0.2 | 10.3 ± 0.4 | 17.7 ± 1.8 |
| KATPase (μm)a | 8.3 ± 0.2 | 8.5 ± 0.3 | 20.4 ± 3.4 |
| ADP binding to actomyosin VI (k−5′) (μm−1s−1)b | 0.27 ± 0.06 | 0.37 ± 0.08 | 0.43 ± 0.05 |
| ADP release from actomyosin VI (k+5′) (s−1)b | 6.3 ± 0.50 | 10.2 ± 0.10 | 18.0 ± 0.7 |
| ATP-induced binding and dissociation from pyrene-actin | |||
| K1′k+2′ (μm−1·s−1)c | 0.023 ± 0.01 | 0.28 ± 0.06 | 0.32 ± 0.04 |
a Measured in the NADH coupled assay.
b Measured with mantADP.
c Measured with pyrene actin.
TABLE 2.
Steady state and transient kinetic parameters of myosin (M) VI-S1 constructs in the absence of actin at 25 °C
| Kinetic parameters | MVI MDins2IQ WT | MVI MDins2IQ L310G | MVI MDins2IQ (Δ-Insert-1) |
|---|---|---|---|
| Steady state | |||
| V0 (-actin) (s−1)a | 0.03 ± 0.01 | 0.06 ± 0.02 | 0.18 ± 0.05 |
| ADP binding to myosin VI (k−5) (μm−1s−1)b | 0.68 | 0.64 | 0.66 |
| ADP release from myosin VI (k+5) (s−1)b | 6.3 ± 0.20 | 10.5 ± 0.30 | 2.2 ± 0.01 |
| ATP binding to myosin VI (K1k+2) (μm−1s−1)b | 0.15 ± 0.02 | 1.15 ± 0.12 | 1.09 ± 0.08 |
a Measured in the NADH-coupled assay.
b Measured with mantATP or mantADP.
Crystallization, X-ray Data Collection, and Structure Determination
The myosin VI L310G MDins2 mutant was crystallized using the hanging drop vapor diffusion method at 4 °C by 1 μl of 19 mg/ml protein solution mixed with 1 μl of reservoir solution (4% PEG 8000, 50 mm glycine, pH 9, 3% isopropyl alcohol, 3% tert-butyl alcohol, 1 mm Tris(2-carboxyethyl)phosphine). The crystals were cryoprotected in the reservoir solution complemented with 20% of ethylene glycol and flash-frozen in liquid nitrogen.
Crystals of myosin VI Δ-insert-1 MDins2 were grown using reservoir solution (containing 10.8% PEG 8000, 50 mm MES, pH 6.75, 175 mm ammonium sulfate, 3% isopropyl alcohol, 3% tert-butyl alcohol, and 4 mm EGTA) and a stock solution of Δ-insert-1 myosin VI at 18 mg/ml. The protein/precipitant drops were microseeded the next day by streak-seeding from previous crystallizations, and crystals appeared overnight. The crystals were cryo-protected by soaking in 15% PEG 8000, 50 mm MES, pH 6.75, 175 mm ammonium sulfate, 4 mm EGTA, and 30% glycerol and then flash-frozen in liquid nitrogen.
X-ray diffraction data were collected at 100 K at the Soleil synchrotron radiation source, Beamline Proxima 1, or the European Synchrotron Radiation Facility Beamline ID-29. The data were processed using the XDS program suite (18, 19).
The mutant MVI crystal structures were determined by the molecular replacement method with Molrep (20) using the wild type myosin VI structure (PDB code 2BKH) as a search model. For the myosin VI Δ-insert-1 mutant, the solution was improved using rigid body and simulated annealing refinements with CNS (21). Several iterative cycles of crystallographic refinement with Refmac5 and manual model rebuilding with Coot were performed to obtain the final structures deposited to the Protein Data Bank (3L9I for L310G MDins2, 2X51 for Δ-insert-1 MDins2). Table 3 lists the data collection and refinement statistics.
TABLE 3.
Data collection and refinement statistics
| M6 L310G | M6 Δ Insert1 | |
|---|---|---|
| Data collection | ||
| Space group | P2(1) | C2 |
| Cell dimensions a, b, c (Å); β (°) | 68.541 | 163.88 |
| 104.403 | 61.017 | |
| 90.335 | 133.27 | |
| 91.087° | 116.19° | |
| X-ray source | Soleil synchrotron, Proxima 1 | ESRF ID29 |
| Resolution (Å) | 19.7-2.2 (2.25-2.2) | 30-2.2 (2.3-2.2) |
| Rsym | 9.1 (45) | 6.8 (43.7) |
| I/sI | 14.4 (3.7) | 14.44 (3.92) |
| Completeness (%) | 99.5 (99.9) | 99.7 (99.9) |
| Redundancy | 3.9 (3.9) | 4.6 (3.9) |
| Refinement | ||
| Resolution (Å) | 19.7-2.2 | 30-2.2 |
| No. reflections | 60,958 | 57,330 |
| Rwork/Rfree(%) | 18.5/23.5 | 19.4/24.26 |
| No. atoms (protein/ligand/waters) | 7,700/37/829 | 7,118/37/324 |
| Average B-factor (Å2) | 40.9 | 56.18 |
| Root mean square deviations | ||
| Bond lengths (Å)/bond angles (°) | 0.014/1.35 | 0.007/1.001 |
RESULTS
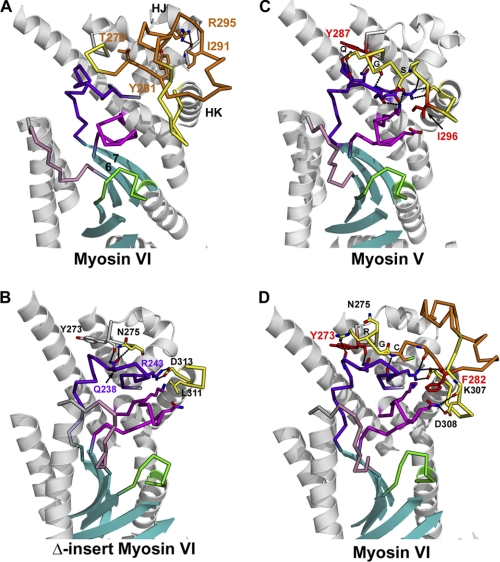
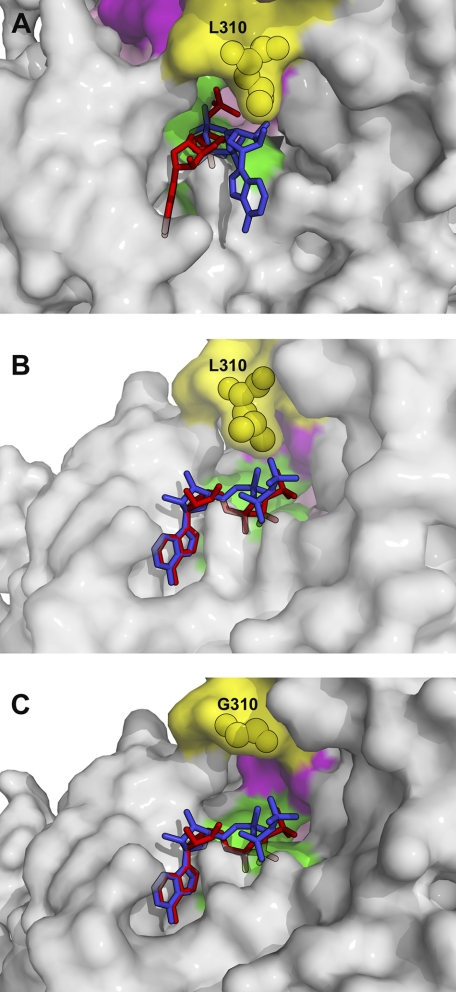
We first substituted leucine 310 with a glycine to test if this leucine provided steric hindrance necessary for gating. The structural basis for this hypothesis comes from the nucleotide-free structure of the wild type myosin VI MDins2 fragment truncated at residue 816 (PDB code 2BKH) (3). As shown in Fig. 2A, leucine 310 can be seen to protrude into the nucleotide binding pocket of myosin VI as a result of the insertion of insert-1. To ascertain the impact of substitution of Leu-310 to glycine or the complete removal of insert-1 on the conformation of the rigor state of myosin VI (state in which ATP must enter to dissociate myosin from actin), we solved two new structures in the rigor-like state, one with insert-1 deleted and the other with leucine 310 substituted to glycine. Note, as shown in Fig. 2B, wherein the structure of the motor domain with insert-1 deleted is depicted, this leucine is not in fact a part of insert-1 but is repositioned by the presence of insert-1. For the L310G mutation, the structure of the myosin VI motor (Fig. 2C) reveals that insert-1 is in the identical conformation as in the wild type motor. The only difference is that there is no longer a leucine side chain protruding into the nucleotide pocket.
FIGURE 2.
Nucleotide binding pocket in the nucleotide-free structures of wild type myosin VI (A), Δ-insert-1 mutant (B), and L310G mutant (C). Three nucleotide binding elements compose the active site of myosin: P-loop (green) from the N-terminal subdomain that binds the phosphate moiety of the nucleotide, Switch I (purple) from the U50 subdomain involved in the binding of Mg2+ and the γ-phosphate when nucleotide binds strongly, and Switch II (pink) that plays an important role for hydrolysis of the nucleotide. The ATP molecule (modeled in these structures) enters in the active site of these nucleotide-free molecules and binds via interactions with the P-loop (phosphate moiety) and the N-terminal subdomain (adenine ring). On the right in these panels, part of the U50 subdomain close to the active site is represented. Insert-1 (residues Cys-278–Ala-303) is represented in orange and is deleted in the Δ-insert-1 mutant. A, in wild type myosin VI, its presence repositions the loop (yellow) that precedes helix HK and notably positions Leu-310 close to the ATP molecule. B, in the Δ-insert-1 mutant, deletion of the 26-residue insert repositions Leu-310 (yellow) far from the nucleotide binding pocket, and the active site is much wider in this mutant. The length of the remaining loop that contains Leu-310 is identical to that found in other myosin such as myosin V. The main chain conformation of the loop differs, however, from that found in myosin V and six residues in the middle of the loop (Gly-277 linked to Gly-304–Ser-305–Leu-306–Lys-307) are too disordered in the structure to be modeled. C, in the L310G mutant, substitution of the leucine by glycine does not introduce any significant changes in the conformation of this loop or in insert-1. The only difference in this mutant is the absence of the side chain of Leu-310, which protrudes within the nucleotide binding pocket in the wild type structure, shown in white in this panel. To illustrate how the leucine side chain may interfere with different conformations explored by the nucleotide upon binding, an ATP molecule in the conformation found in a scallop structure (PDB 1S5C), in which nucleotide is partially bound via the adenine ring only, is represented in blue.
Comparisons of the structures of myosin VI with that of other myosins such as myosin V show that the presence of insert-1 does not influence the conformation of any of the elements of the upper 50 kDa (U50) subdomain. In the Δ-insert-1 structure (residues Cys-278 to Ala-303 deleted), removal of the insert also has no consequence in the conformation of the rest of the U50 subdomain, although several interactions that this insert makes with the rest of the subdomain are lost in the mutant (Fig. 3). Interestingly, the interactions that the beginning of the loop in which insert-1 is found makes with an important linker of the U50 subdomain (residues Leu-229—Asn-244) are lost in the Δ-insert-1 structure, suggesting that the dynamics of the linker may differ in this mutant compared with either wild type myosin VI or the L310G mutant (Fig. 3). This U50 linker may influence the rearrangements of the so-called transducer region (22) upon nucleotide binding and dissociation. It is connected to the last strand of the seven-stranded central β-sheet of the molecule, which undergoes twisting upon nucleotide binding and dissociation (see Fig. 3 and “Discussion”).
FIGURE 3.
Insert-1 interactions with the U50 subdomain and their possible influence on the dynamics necessary for transition between myosin states. Insert-1 (orange) is located in a loop (yellow) of the U50 subdomain that makes interactions with the U50 linker (dark purple) and Switch I (magenta). Note that the U50 linker and switch I are connected to the last two strands of the central β-sheet (cyan), which is part of the transducer in myosin (19) that undergoes major rearrangements upon nucleotide binding and dissociation. A, in myosin VI, insert-1 corresponds to a 26-residues insertion in the loop that connects helix HJ to helix HK. In all wild type myosin VI structural states solved to date, several conserved interactions are observed between insert-1 and the rest of the U50 subdomain and stabilize its conformation. In particular, insert-1 residues Thr-279, Tyr-281, Ile-291, Arg-295 play an anchoring role for the insert. Of note, arginine 295 plays an important role in anchoring insert-1 to the linker found between helices HI and HJ by interacting with the main chain carbonyl of Ala-255. B, in all myosin structures solved to date, including the Δ-insert-1 structure, the first residue of helix HK that follows the insert-1 loop is an aspartate (Asp-313 in myosin VI), which makes a conserved salt bridge interaction with the arginine Arg-243 of the U50 linker (dark purple). At the beginning of the loop, another conserved interaction involves the main chain carbonyl of Tyr-273 that interacts with Q238 of the U50 linker. C, in myosin V the loop (Lys-289—Asp-301; yellow) in which insert-1 is found in myosin VI makes several interactions with the U50 linker and Switch I, in particular via myosin V residues Q290 (Q), G291 (G), S293 (S), Ile-296. B, in the Δ-insert-1 structure, the main chain conformation of the loop differs from that of myosin V, although it also contains 13 residues. This results in a weakening of the interactions mediated by insert-1 residues with the U50 linker and Switch I compared with either myosin V or wild type myosin VI. D, in wild type myosin VI, the conformation of the beginning of the loop is similar to that found in myosin V, and myosin VI residues Arg-276 (R), Gly-277 (G), Cys-278 (C) interact similarly with the U50 linker as do Gln-290, Gly-291, and Ser-293 in the myosin V structure. Residue Phe-282 of insert-1 is found in similar position as Ile-296 in myosin V and also interacts with Switch I. The nature of the contacts is different in the two myosins, however, which likely influences the U50 linker dynamics and the rates of rearrangements within the transducer.
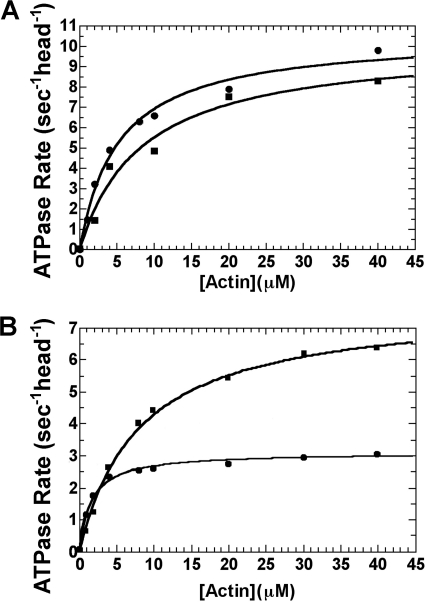
As shown in Fig. 4A, substitution of leucine 310 with a glycine resulted in increased actin-activated ATPase activity for the dimeric HMM construct (zippered double-headed molecule truncated at arginine 994 (12)). As also shown in Fig. 4, the single-headed S1 construct had a slight increase in steady state activity when leucine 310 was mutated to glycine. The net result was that the activity per head of the S1 and HMM L310G mutant constructs was nearly identical. This is in sharp contrast to the wild type constructs (Fig. 4B) in which the HMM has approximately half the activity per head as compared with the S1, indicative of gating.
FIGURE 4.
Actin-activated ATPase activity of myosin VI with or without mutation of leucine 310 to glycine (L310G). The ATPase activity was determined at 25 °C as described under “Experimental Procedures.” A, the actin concentration dependence of the steady state ATPase activities (s−1head−1) of single-headed (S1) myosin VI-L310G (■) and dimeric (HMM) myosin V-L310G (●) is shown for a single, representative preparation. For multiple preparations (four), the average Vmax and KATPase values for the single- and double-headed myosin VI L310G mutants were similar, indicating a lack of gating between the heads. For the single-headed myosin VI-L310G, the average Vmax was 10.3 ± 0.4 versus 10.5 ± 0.7 s−1head−1 for the HMM, with KATPase values of 8.5 ± 0.3 and 4.8 ± 0.8 μm, respectively. B, the actin concentration dependence of the steady state ATPase activities (s−1head−1) of single-headed (S1) wild type myosin VI (■) and dimeric (HMM) wild type myosin VI (●) is shown for a single, representative preparation. For multiple preparations (four), the average Vmax and KATPase values for the single- and double-headed wild type myosin VI differ by ∼2-fold due to the presence of gating in the dimeric construct. For the single-headed myosin VI, the average Vmax was 6.8 ± 0.2 versus 3.1 ± 0.5 s−1head−1 for the dimer, with average KATPase values of 8.3 ± 0.2 and 1.8 ± 0.6 μm, respectively.
Multiple preparations (four) gave average Vmax and KATPase values for the single- and double-headed myosin VI L310G mutants that were similar (10.3 ± 0.4 versus 10.5 ± 0.7 s−1head−1, respectively), confirming a lack of gating between the heads. The average KATPase values differed for the single- and double-headed constructs (8.5 ± 0.3 and 4.8 ± 0.8 μm, respectively), with the higher apparent actin affinity (KATPase) indicative of cooperative actin binding in the two-headed molecule. In contrast, the average Vmax for multiple preparations (four) of the single- and double-headed wild type myosin VI differed by ∼2-fold, indicating the presence of gating in the dimeric construct. For the single-headed myosin VI, the average Vmax was 6.8 ± 0.2 versus 3.1 ± 0.5 s−1head−1 for the dimer, with average KATPase values of 8.3 ± 0.2 and 1.8 ± 0.6 μm, respectively. Thus, leucine 310 is sufficient to confer gating in myosin VI if positioned properly by the remainder of insert-1.
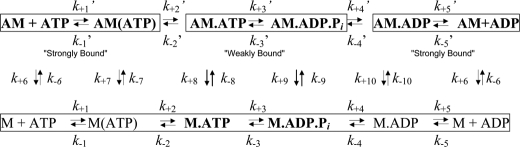
To avoid the complications of the influence of intramolecular strain on rate constants (12) and to facilitate interpretation of the unstrained structures, we focused the transient kinetic analyses in this study on single-headed (S1) constructs. These constructs contained all of the myosin VI motor and terminated immediately after the short myosin VI lever arm containing two bound calmodulin molecules. Transient kinetic properties of the wild type, L310G, and Δ-insert-1 single-headed constructs were determined using the kinetic scheme (Scheme 1) below. The resulting data are summarized in Tables 1 and 2.
SCHEME 1.
Kinetic and equilibrium constants for myosin VI-S1. M indicates myosin, and AM indicates the actomyosin complex.
As summarized in Table 1, the Vmax of the steady state actin-activated ATPase activity is higher for the insert-1 deletion than for the L310G mutant, which in turn is slightly higher than wild type. As shown in Table 1, the increase in the steady state actin-activated ATPase of the L310G and Δ-insert-1 mutants is due to an increase in the rate of ADP dissociation from the actomyosin VI complex (k+5′), which is the rate-limiting step in the actin-activated ATPase cycle for myosin VI (13). Also note that the KATPase is similar for the single-headed wild type and L310G mutant constructs, but it is much lower for the single-headed Δ-insert-1 mutant. This likely indicates that a kinetic step(s) other than nucleotide binding and release are altered by complete removal of insert-1 (likely k+4′).
Because gating is achieved by preventing ATP-induced dissociation of the lead head from actin, we examined the impact of leucine 310 on ATP-induced actin dissociation. This involved determining the rate of transition from strong to weak actin binding (K1′k+2′), as monitored by pyrene actin fluorescence (23). As shown in Table 1, removal of insert-1 increases the rate of ATP-induced dissociation by 10-fold, consistent with our earlier reports (3, 12). Also shown in Table 1 is that mutation of leucine 310 to glycine results in the same 10-fold increase in this rate. This reveals that leucine 310 was indeed the primary modulator of the rate of ATP binding and complex dissociation.
We next characterized the impact of the leucine 310 and insert-1 on nucleotide binding in the absence of actin. We wanted to establish whether or not either leucine 310 or insert-1 confer nucleotide binding selectivity in the absence of actin. This is necessary to interpret the high resolution structures of wild type and mutant myosin VI motors as representative of the state that myosin VI populates on actin. Binding and release of ADP and binding of ATP were, thus, determined, and the resulting data are summarized in Table 2.
The examination of ATP binding kinetics determined by mantATP binding to myosin VI without actin (K1′k+2′) revealed that leucine 310, even in the absence of actin, is the primary modulator of the rate of ATP binding. The rates of ATP binding were similar for the L310G mutant or the insert-1 deletion in the absence of actin and ∼10-fold higher than the wild type construct.
In contrast, ADP binding to myosin VI in the absence of actin (k−5′) was unaffected by deletion of insert-1 or the L310G mutation. ADP dissociation (k+5′) was slightly affected (∼2-fold faster than wild type) with the L310G mutation, and the opposite effect (nearly 3-fold slowing) was observed upon removal of insert-1. In the presence of actin (Table 1), insert-1 has a small influence on the ADP binding kinetics. Binding of MgADP to the actomyosin VI complex (k−5′) was about 1.5-fold faster with either the L310G or deletion of insert-1 mutants. In contrast to the situation in the absence of actin in which removal of insert-1 slows ADP dissociation, this rate is faster for both mutants bound to actin. It was increased ∼1.5-fold in the L310G mutant and ∼2.5-fold with the deletion of insert-1 (Table 1).
DISCUSSION
What is clear is that when myosin VI is either bound or not bound to actin, leucine 310 is positioned by insert-1 to selectively interfere with ATP binding, while having little or no effect on ADP binding. As we demonstrated previously (12), intramolecular strain experienced by the heads of the HMM bound to actin causes the lead head to have a much slower rate of ATP-induced detachment than we see in the absence of strain. Also shown by Sweeney et al. (12), the rear head of the processive dimer has a rate of ATP-induced detachment from actin that is similar to that seen in the absence of strain (i.e. in the S1 construct (12)). Based on the data of this study, it is likely that rearward strain on the lead head positions leucine 310 of insert-1 to provide increased steric hindrance for ATP entry as compared with its position in our single-headed constructs, as replacing leucine 310 with glycine totally abolishes gating (Fig. 4).
Examination of the effects of ADP binding and dissociation revealed that both leucine 310 and the rest of insert-1 have distinct effects. ADP binding in the absence of actin is unaffected by either. ADP dissociation is slightly slowed by the presence of Leu-310 either in the presence or absence of actin, whereas the removal of all of insert-1 increases dissociation to a greater extent in the presence of actin but slows release in the absence of actin. This last result in particular suggests that beyond the steric effects of leucine 310, insert-1 also influences transitions between the states that bind and release MgADP in the presence and absence of actin. The states that bind MgADP are not equivalent depending on whether or not actin is present, which can explain how insert-1 can exert the opposite effect on ADP release.
In the absence of actin, myosin binds ADP strongly in a state that is known as the post-rigor state (24). It is not entirely clear what is the structural transition that releases MgADP from this state, but for myosin VI it is likely that it is a transition to the rigor-like state, which is the state in which myosin VI crystallizes in the absence of nucleotide (3). A major difference in these two states is the twist of the β-sheet associated with the nucleotide binding pocket. The rigor position of the β-sheet represents an unstrained conformation, whereas the MgADP (or MgATP) post-rigor conformation is strained. A possible interpretation of these data is that insert-1 favors the rigor-like conformation of the β-sheet and disfavors the post-rigor conformation. Thus, removal of insert-1 stabilizes post-rigor, slowing ADP dissociation in the absence of actin.
When bound to actin, ADP release occurs during an isomerization between a state that binds MgADP strongly and one that can bind ADP weakly (22, 25). The state that binds ADP weakly is close to the rigor-like state with an unstrained conformation for the β-sheet. Although the nature of the β-sheet conformation in the strong MgADP, strong actin binding conformation is unknown, it must be different from either the rigor-like or post-rigor conformations for both the ADP and actin affinity to be strong. The rigor-like state has a low ADP affinity, whereas the post-rigor state binds actin weakly. Sequential distortion of this β-sheet may resemble the conformational changes visualized in the F1-ATPase in different biochemical states upon release of its products (26). The simplest interpretation of the ADP dissociation data in Table 1 is that insert-1 favors the novel β-sheet conformation found in the strong ADP binding state on actin as compared with the rigor conformation, and thus, insert-1 slows ADP release from actomyosin VI. One cannot, however, simply postulate that insert-1 affects the rate of β-sheet transitions, as in the absence of actin it promotes ADP release, and in the presence of actin it slows ADP release.
Thus, a second consequence of insert-1 on myosin VI kinetics is that it stabilizes the strong binding ADP state on actin, decreasing the rate of ADP release and increasing the duty ratio. Note that these observations mirror those from studies of the alternatively spliced exon 7 of Drosophila melanogaster myosin II (27). Those investigators demonstrated a similar influence of substitutions within the upper 50-kDa subdomain on nucleotide binding and release kinetics. The region coding for this alternative exon encompasses the region where insert-1 is found in myosin VI.
The structures of the myosin VI mutants solved in this study suggest that insert-1 may influence the transitions necessary for binding and release of nucleotide in different ways. First, insert-1 may influence nucleotide binding and release via the direct interactions it makes with an important linker of the U50 subdomain (residues Leu-229–Asn–244) (see Fig. 3). Variability in the dynamics of this U50 linker could influence the transitions between states, as this U50 linker is found to play multiple roles in myosin. Its surface comprises part of the U50 side of the cleft that closes in rigor in the molecule. It also interacts with the Switch I element necessary for strong binding of Mg2+ and nucleotide, and it directly follows the last strand of the central β-sheet, which adopts different conformations upon nucleotide binding and release from myosin. It is also possible that to allow a novel β-sheet conformation, there must be movements within the upper 50-kDa subdomain that are also necessary for the formation of strong binding to actin with ADP bound (following phosphate release). In this regard, the interactions that insert-1 makes with the rest of the U50 subdomain could greatly influence the rearrangements necessary to populate the strong-ADP state. These considerations highlight a critical role for this insert-1 region in being part of the pathway of communication between the nucleotide binding site, the transducer, and the actin binding interface via rearrangements within the U50 subdomain.
Consistent with this hypothesis, it is noteworthy that hypertrophy cardiomyopathy can be caused by β-cardiac myosin heavy chain mutations F312C (28) and V320M (29). These residues are equivalent to residues Tyr-273 and Phe-282 in myosin VI and Tyr-287 and Ile-296 in myosin V (see the residues in red in Fig. 3, C and D). Although Phe-282 interacts with Switch I, Tyr-273 is involved in internal interactions within the U50 subdomain and interacts with the U50 linker.
To explain the effect of Leu-310 on ATP binding in the absence of actin, we studied the structures shown in Fig. 2 in more detail. As noted in the results, leucine 310 is the closest insert-1 residue to the nucleotide binding pocket. Removal of its side chain in the L310G mutant leads to a myosin that binds MgATP as fast as if all of insert-1 is removed. The structure of this mutant myosin in the nucleotide-free state shows that the insert-1 main chain conformation is not affected by the mutation (Fig. 2). Thus, one must conclude that it is the side chain of Leu-310 of insert-1 that is responsible for the large difference in MgATP binding kinetics. Without actin, the nucleotide-free myosin VI structure (rigor-like) shows that it has a more open front door than in the post-rigor conformation. It is clear, however, that the insert-1, in particular Leu-310, restricts the size of this front door in myosin VI compared with other myosins that do not have this insert.
The selectivity of leucine 310 for ATP versus ADP binding has implications for how nucleotides can bind to myosin. It is generally assumed that the phosphates enter the phosphate tube and bind first; however, the narrowed phosphate tube of myosin VI may prevent that from occurring rapidly. The selective influence of leucine 310 on ATP suggests a second possible mechanism. The difference in the influence of insert-1 for the MgATP and MgADP kinetics indicates that nucleotide binding is likely to initially occur via the docking of the purine base to the N-terminal subdomain of myosin. This has previously been suggested by a scallop-striated muscle myosin II structure with ADP bound (30), which describes a conformation of the nucleotide that has different torsional angles between the purine base and the ribose and between the ribose and the C5′ of ADP as shown in Figs. 2C and 5. This structure suggests that the nucleotide can bind to myosin via the purine base interactions only and likely undergoes a number of conformations when partially bound to myosin. Note that this structure (30) had a sulfate ion bound to the P-loop, however, so whether the nucleotide binds first via the purine base interactions could not be deduced from these data.
FIGURE 5.
Nucleotide binding pocket accessibility in wild type and L310G mutant myosin VI. A surface representation of the rigor-like state of the myosin VI molecule is shown, focused on the region where ATP binds. The P-loop (green), Switch I (purple), and the loop that follows insert-1 (yellow) are colored as in Fig. 1. A and B, all atoms of Leu-310, which belong to the loop that follows insert-1, are represented in yellow spheres. In C, the glycine 310 of the L310G mutant structure is represented in yellow spheres. An ATP molecule fully bound via interactions of the phosphate moiety with the P-loop and the adenine ring with residues of the N-terminal subdomain is depicted in red. Note the role of the Leu-310 side chain in narrowing the entrance of the phosphate tube. A, to illustrate how this restricts the conformations that an ATP molecule can undergo to bind via first entering the phosphate tube via its phosphate moieties, an ATP molecule (blue) that starts to enter this tube is represented. B, binding of the nucleotide by first docking the adenine ring in the N-terminal subdomain is represented in this figure in which an ATP molecule (blue) adopts a similar conformation for the nucleotide as that observed for ADP in the scallop striated myosin II structure (PDB 1S5G). Note that after docking of the ring, conformational changes in the nucleotide are necessary to slide the phosphate moieties into the phosphate tube, as presented for the red nucleotide. Note that this is easily done by the diphosphate moiety of an ADP molecule but would lead to collisions with the Leu-310 side chain in the case of a triphosphate moiety of an ATP molecule. C, the absence of the leucine side chain in the L310G mutant is sufficient to remove such collisions. This can explain why ATP binding is 10-fold faster in this mutant and highlights the role of Leu-310 in the selectivity of the myosin VI binding for ADP versus ATP.
What is clear is that the side chain of Leu-310 greatly reduces the opening of the nucleotide binding site and restricts the number of conformations the nucleotide may adopt to enter the site if it would first bind via the phosphate moiety (Fig. 5A). Note, however, that this should affect both ADP and ATP binding kinetics similarly. The phosphate moieties bind to the P-loop, which is found at the bottom of the pocket opening. The γ-phosphate of ATP occupies the deepest site, whereas the position for the β- and α-phosphates are identical in ADP and ATP once they are docked into the pocket and associated with the P-loop of myosin. The rest of the nucleotide would be positioned similarly for ADP and ATP, and thus, one should not expect any difference on the influence of insert-1 in the binding kinetics of these two nucleotides in a scheme in which the phosphates dock first.
This study, thus, supports that nucleotide binding occurs by first docking the purine moiety of the nucleotide (Fig. 5B). The nucleotide-bound myosin structures and, in particular the ADP bound scallop structure mentioned above, show that this purine moiety is likely to interact with a pocket in the N-terminal subdomain composed of residues Asn-98—Tyr-107 and Gly-156—Phe-163. Most of these interactions are hydrophobic and hold the purine moiety of the nucleotide. Of note, a hydrogen bond involving Tyr-107 is conserved in all myosins and helps correctly position the purine moiety.
Docking nucleotide in the nucleotide-free (rigor-like) structure via the purine moiety shows that the diphosphate moiety of ADP can rotate and explore different conformations with minimal hindrance from leucine 310 of insert-1, in agreement with the observation that insert-1 only minimally influences the binding of ADP in myosin VI. In contrast, the triphosphate moiety of ATP is longer and rigidified by the binding of Mg2+ between the β- and γ-phosphate. Thus, collision with the side chain of leucine 310 of insert-1 upon rotation of the triphosphate moiety will occur, whereas the purine ring is docked, accounting for the reduction in the rate of binding of MgATP to myosin VI (Fig. 5, B and C).
Although the structures obtained in this study do provide a mechanism to explain the differential ability of ATP and ADP to bind in the absence of strain, they do not reveal the position of insert-1 and, importantly, of Leu-310, in the presence of intramolecular strain, as would be experienced by the lead head of a processive myosin VI dimer. It is also true that we have no idea of how strain is communicated to the nucleotide binding pocket of a plus-end directed motor, such as myosin V, in a manner that greatly slows nucleotide release (8–11).
Our hypothesis is that strain in the myosin VI motor must be interpreted in a different manner than for myosin V, as the strain on the lead head of myosin VI is applied in the reverse direction as compared with myosin V. Based on single molecule experiments, we recently proposed a model in which intramolecular strain prevents the converter subdomain of the lead head of a myosin VI dimer from undergoing its rearrangement to its rigor conformation until the trailing head detaches from actin (31). The implication of this model is that preventing the converter rearrangement somehow positions the subdomains of the motor such that Leu-310 creates an even greater barrier to ATP binding (but not to ADP binding) than we see in our current structures.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grant DC009100 (to H. L. S.). This work was also supported by CNRS (to A. H.), ANR Blanche BLAN07-3_193368 (to A. H.), and ACI BCMS (to A. H.).
The atomic coordinates and structure factors (codes 3L9I and 2X51) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Mant nucleotides are 2′(3′)-O-(N-methylanthraniloyl)-labeled nucleotides.
REFERENCES
- 1. Wells A. L., Lin A. W., Chen L. Q., Safer D., Cain S. M., Hasson T., Carragher B. O., Milligan R. A., Sweeney H. L. (1999) Nature 401, 505–508 [DOI] [PubMed] [Google Scholar]
- 2. Sweeney H. L., Houdusse A. (2007) Curr. Opin. Cell Biol. 19, 57–66 [DOI] [PubMed] [Google Scholar]
- 3. Ménétrey J., Bahloul A., Wells A. L., Yengo C. M., Morris C. A., Sweeney H. L., Houdusse A. (2005) Nature 435, 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rock R. S., Rice S. E., Wells A. L., Purcell T. J., Spudich J. A., Sweeney H. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13655–13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nishikawa S., Homma K., Komori Y., Iwaki M., Wazawa T., Hikikoshi Iwane A., Saito J., Ikebe R., Katayama E., Yanagida T., Ikebe M. (2002) Biochem. Biophys. Res. Commun. 290, 311–317 [DOI] [PubMed] [Google Scholar]
- 6. Park H., Ramamurthy B., Travaglia M., Safer D., Chen L. Q., Franzini-Armstrong C., Selvin P. R., Sweeney H. L. (2006) Mol. Cell 21, 331–336 [DOI] [PubMed] [Google Scholar]
- 7. Phichith D., Travaglia M., Yang Z., Liu X., Zong A. B., Safer D., Sweeney H. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17320–17324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rosenfeld S. S., Sweeney H. L. (2004) J. Biol. Chem. 279, 40100–40111 [DOI] [PubMed] [Google Scholar]
- 9. Veigel C., Wang F., Bartoo M. L., Sellers J. R., Molloy J. E. (2002) Nat. Cell Biol. 4, 59–65 [DOI] [PubMed] [Google Scholar]
- 10. Purcell T. J., Sweeney H. L., Spudich J. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13873–13878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Veigel C., Schmitz S., Wang F., Sellers J. R. (2005) Nat. Cell Biol. 7, 861–869 [DOI] [PubMed] [Google Scholar]
- 12. Sweeney H. L., Park H., Zong A. B., Yang Z., Selvin P. R., Rosenfeld S. S. (2007) EMBO J. 26, 2682–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De La Cruz E. M., Ostap E. M., Sweeney H. L. (2001) J. Biol. Chem. 276, 32373–32381 [DOI] [PubMed] [Google Scholar]
- 14. Lumb K. J., Carr C. M., Kim P. S. (1994) Biochemistry 33, 7361–7367 [DOI] [PubMed] [Google Scholar]
- 15. Sweeney H. L., Rosenfeld S. S., Brown F., Faust L., Smith J., Xing J., Stein L. A., Sellers J. R. (1998) J. Biol. Chem. 273, 6262–6270 [DOI] [PubMed] [Google Scholar]
- 16. De La Cruz E. M., Wells A. L., Rosenfeld S. S., Ostap E. M., Sweeney H. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13726–13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De La Cruz E. M., Sweeney H. L., Ostap E. M. (2000) Biophys. J. 79, 1524–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 19. Diederichs K., McSweeney S., Ravelli R. B. G. (2003) Acta Crystallogr. D 59, 903–909 [DOI] [PubMed] [Google Scholar]
- 20. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 21. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 22. Coureux P. D., Sweeney H. L., Houdusse A. (2004) EMBO J. 23, 4527–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geeves M. A., Jeffries T. E. (1988) Biochem. J. 256, 41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ménétrey J., Llinas P., Cicolari J., Squires G., Liu X., Li A., Sweeney H. L., Houdusse A. (2008) EMBO J. 27, 244–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosenfeld S. S., Houdusse A., Sweeney H. L. (2005) J. Biol. Chem. 280, 6072–6079 [DOI] [PubMed] [Google Scholar]
- 26. Menz R. I., Walker J. E., Leslie A. G. (2001) Cell 106, 331–341 [DOI] [PubMed] [Google Scholar]
- 27. Miller B. M., Bloemink M. J., Nyitrai M., Bernstein S. I., Geeves M. A. (2007) J. Mol. Biol. 368, 1051–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Driest S. L., Jaeger M. A., Ommen S. R., Will M. L., Gersh B. J., Tajik A. J., Ackerman M. J. (2004) J. Am. Coll. Cardiol. 44, 602–610 [DOI] [PubMed] [Google Scholar]
- 29. Havndrup O., Bundgaard H., Andersen P. S., Allan Larsen L., Vuust J., Kjeldsen K., Christiansen M. (2003) Cardiovasc. Res. 57, 347–357 [DOI] [PubMed] [Google Scholar]
- 30. Risal D., Gourinath S., Himmel D. M., Szent-Györgyi A. G., Cohen C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8930–8935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reifenberger J. G., Toprak E., Kim H., Safer D., Sweeney H. L., Selvin PR. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]