Abstract
Neutrophil (polymorphonuclear leukocyte; PMN) inflammatory functions, including cell adhesion, diapedesis, and phagocytosis, are dependent on the mobilization and release of various intracellular granules/vesicles. In this study, we found that treating PMN with damnacanthal, a Ras family GTPase inhibitor, resulted in a specific release of secondary granules but not primary or tertiary granules and caused dysregulation of PMN chemotactic transmigration and cell surface protein interactions. Analysis of the activities of Ras members identified Ral GTPase as a key regulator during PMN activation and degranulation. In particular, Ral was active in freshly isolated PMN, whereas chemoattractant stimulation induced a quick deactivation of Ral that correlated with PMN degranulation. Overexpression of a constitutively active Ral (Ral23V) in PMN inhibited chemoattractant-induced secondary granule release. By subcellular fractionation, we found that Ral, which was associated with the plasma membrane under the resting condition, was redistributed to secondary granules after chemoattractant stimulation. Blockage of cell endocytosis appeared to inhibit Ral translocation intracellularly. In conclusion, these results demonstrate that Ral is a critical regulator in PMN that specifically controls secondary granule release during PMN response to chemoattractant stimulation.
Keywords: Cell Adhesion, Chemotaxis, Endocytosis, Exocytosis, Neutrophil
Introduction
Neutrophils, or polymorphonuclear leukocytes (PMN),2 occupy the largest population in peripheral leukocytes and play a crucial role in innate immunity. During acute infection and inflammation, PMN sense chemoattractant signals and swiftly migrate across the microvasculature and underlying tissues to accumulate at inflammatory foci, where these cells execute forceful antimicrobial and tissue-damaging functions. Therefore, the timely and precise response of PMN constitutes an essential first line defense against invading microbials. On the other hand, PMN-mediated inflammation is also a central component of many pathophysiological processes and diseases in which dysregulated PMN transmigration and activation often result in severe tissue injuries and organ dysfunction (1, 2).
Different from most of other cell types, PMN contain a highly granular cytoplasm. At least four types of granules and vesicles have been classified in PMN, including peroxidase-positive primary granules (also termed azurophil granules), peroxidase-negative secondary and tertiary granules (also termed as specific and gelatinase granules, respectively), and a group of highly mobilizable secretory vesicles (3, 4). These various granules and vesicles in PMN constitute an important reservoir of antimicrobial peptides, proteases, respiratory burst oxidases, and many membrane-bound receptors and adhesion molecules. PMN functions in response to inflammatory stimuli are largely dependent on the mobilization and release (degranulation) of these cytoplasmic granules and vesicles. In general, the secretory vesicles have the highest propensity for extracellular release. These vesicles provide the PMN cell surface with essential but low levels of receptors and molecules that are important in PMN sensing and triggering activation (3–5). Upon chemoattractant stimulation, secondary and tertiary granules are released, and this vital degranulation process enables PMN to adhere to and migrate across vascular endothelium and tissue layers (3–8). PMN secondary granules contain a large number of key cell adhesion molecules, such as the β2 integrin CD11b/CD18 (9, 10) and the Ig superfamily members CD47 and SIRPα (11–14), all of which are required for PMN adhesion and chemotaxis. Degranulation of secondary granules also releases the anti-pathogen protein lactoferrin. Tertiary granules have a high content of gelatinases; release of these collagenolytic metalloproteases plays a critical role in degradation of the extracellular matrix during PMN extravasation. In addition to these granules/vesicles, studies have also found that certain primary granules are also released during PMN activation and that some of the primary granule components, such as elastase, are indispensable for PMN migration in the tissues.
The molecular mechanisms that control PMN degranulation are, however, still unclear. The signaling events elicited by chemoattractant stimulation leading to PMN degranulation of each type of granule are complex and are only inadequately understood. In other cell types, the Ras family small GTPase Ral has been shown to be required in intracellular vesicle transportation, including both endocytosis and exocytosis (15–19). Here, we report our experimental findings, which reveal for the first time that Ral controls PMN release of secondary granules. In particular, our studies suggest that active Ral serves as an essential gatekeeper that prevents secondary granule release. Disruption of Ral regulation, either by inhibition of Ral activity in resting PMN or overexpression of a constitutively active Ral during chemoattractant stimulation, results in dysregulation of secondary granule mobilization and impairment of PMN chemotactic function.
EXPERIMENTAL PROCEDURES
Cells and Antibodies
Human whole blood drawn from healthy volunteers was centrifuged (200 × g, 10 min) to separate the platelet-rich plasma and blood cells, followed by isolation of PMN and platelets. PMN were isolated by dextran sedimentation and Ficoll-Paque density gradients as described previously (12, 20) To isolate platelets, the collected plasma was supplemented with 0.1 volume of ACD (2.5% trisodium citrate, 1.5% citric acid, 2% d-glucose), followed by centrifugation at 3000 rpm for 15 min. The pelleted platelets were washed with a citrate buffer (120 mm NaCl, 4.26 mm NaH2PO4, 5.5 mm glucose, 4.77 mm sodium citrate, and 2.35 mm citric acid, pH 6.5) and finally resuspended in HBSS (4 °C) (21).
A hybridoma cell line that produces anti-CD11b antibody LM2/1 was obtained from the American Type Culture Collection (ATCC). Monoclonal antibody against the extracellular domain of CD47 (PF3.1) was generated previously (12). Rabbit polyclonal antibodies against the extracellular domains of SIRPα (anti-SIRPα.ex) and JAM-A (anti-JAM-A.ex) were raised by immunizing rabbits with purified fusion proteins composed of rabbit Fc and the entire extracellular domains of SIRPα or JAM-A (22, 23). Antibodies against Ral, Ras, and Rap1 were purchased from Millipore. Anti-β-actin mAb was purchased from Sigma. Rabbit anti-α-adaptin was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Treatment of PMN with Inhibitors and Agents
For treating PMN, damnacanthal (Calbiochem) was freshly dissolved in DMSO, followed by further dilution in HBSS(−) at concentrations ranging from 0.1 to 18 μm before incubation with PMN (5 × 106 cells) at 25 °C for 15 min. Other inhibitors, such as Bruton tyrosine kinase inhibitor LMF-A13, Src family inhibitors PP1 and genistein, Syk inhibitor piceatannol, mitogen-activated protein kinase (MAPK) inhibitor SB203580, and MEK inhibitor PD98059 (all from Biomol), PI3K inhibitor LY294002 (from EMD4Biosciences), and others, were used as described previously (20, 24). Reagents that block general endocytosis included chlorpromazine hydrochloride (25) (Sigma), prepared in HBSS to 50 μm, and succinic acid (26, 27), prepared in HBSS to 20 mm, followed by adjusting pH to 5.5. To block the clathrin-mediated endocytosis, dynamin inhibitor dynasore (28) and an AP-2-binding peptide fragment of the EGFR protein-tyrosine kinase substrate 15 (Eps15-DIII) (29, 30) were used as described previously. Eps15-DIII was delivered into PMN by tagging with HIV Tat. PMN were pretreated with these reagents for 5–15 min before stimulation with 1 μm fMLF at 37 °C.
PMN Degranulation Assays
To test the effect of inhibitors on resting PMN, PMN (5 × 106) were incubated in 400 μl of HBSS(−) with different inhibitors or the same dilution of DMSO at 25 °C for 15 min, followed by centrifugation. Cell-free supernatants were collected and assayed for specific granular markers, including myeloperoxidase (MPO), lactoferrin, and gelatinase, correlating to the release of primary, secondary, and tertiary granules, respectively (12, 20). MPO activity was assayed using 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid. Lactoferrin was assayed by ELISA using anti-lactoferrin antibody (Jackson ImmunoResearch Laboratories). Gelatinase was assayed by zymogram using gelatin-embedded gels (Bio-Rad). To assay PMN degranulation after chemoattractant stimulation, PMN, untreated or treated with Tat-tagged Ral peptides or endocytic inhibitors, were stimulated with 1 μm fMLF in HBSS for 20 min (37 °C), followed by assaying for the release of granular markers. Positive degranulation controls were performed by stimulating the same amount of PMN with fMLF plus 10 μm cytochalasin B (CB) (31).
PMN Transmigration Assay
PMN migration across collagen-coated transfilters was performed as described previously (12, 20). Briefly, freshly isolated PMN (106), non-treated or treated with inhibitors (15 min, 25 °C) or Tat-tagged Ral peptides (30 min, 4 °C), were loaded into the upper chambers of the transmigration setups in 150 μl of HBSS. Transmigration was initiated by adding 0.1 μm fMLF in the lower chambers containing 500 μl of HBSS and incubation at 37 °C. PMN that migrated into the lower chambers were quantified by MPO assay.
Ras Family GTPase Activation Assay
The GTPase activities of Ras, Rap, and Ral in PMN and platelets were assayed using Ras, Rap, and Ral activation assay kits (Millipore) with modifications. Briefly, PMN (2 × 107 cells/condition) or platelets (3 × 108 cells/condition), with or without different treatment, were lysed with a buffer containing 25 mm Hepes, pH 7.5, 150 mm NaCl, 2 mm MgCl2, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1% Triton X-100, 2% glycerol, and protease inhibitor (mixture (Sigma) and 10 mm PMSF), followed by incubation with Raf-1 RBD, RalGDS-RBD, or RalBP1 (residues 397–519 of rat RalBP1)-conjugated agarose for 45 min at 4 °C. After washing the agarose, the pulled down active Ras, Rap, and Ral were detected by Western blot. Alternatively, active Ral in PMN was also pulled down using a RalBP1 of human origin (amino acids 397–518 of human RalBP1) and GST fusion protein, a gift from Dr. Johannes Bos (Utrecht University, Netherlands) (32). Our results indicated that RalBP1 of either human or rat origin worked equally well to pull down human Ral.
Generation of HIV Tat-tagged Recombinant Proteins
To generate Tat-tagged Ral proteins, the entire cDNA of wild-type Ral was RT-PCR-amplified from leukocytes (Marathon Ready cDNAs, Clontech) using a sense primer, 5′- atatggatcccctacggccgcaagaaacgccgccagcgccgccgcatggctgcaaataagcccaagggt, which also contained a sequence (underlined) coding for the 11 residues of HIV Tat (YGRKKRRQRRR) (33–35), and an antisense primer, 5′-atatctcgagttataaaatgcagcatctttc, followed by cloning into pGEX-5X1 vector. To produce the constitutively active (Ral23V) and dominant negative (Ral28N) mutants (15, 36, 37), Gly23 and Ser28 in wild-type Tat-Ral were mutated to Val and Asn, respectively. Two Tat-tagged Eps15 C-terminal peptides were produced, the AP-2-binding peptide (amino acids 563–885, designated as Eps15-DIII) and non-binding peptide (amino acids 740–885, designated as Eps15-Δ) (29, 30). To construct these proteins, RT-PCR was performed using an antisense primer, 5′-tagccagttctaagtcttc, and Tat sequence (underlined)-containing sense primers, 5′-atatggatcccctacggccgcaagaaacgccgccagcgccgccgccctgaactactgccttctggtgtga and 5′-atatggatcccctacggccgcaagaaacgccgccagcgccgccgctcagccacatcgagctctgtcagcaac, respectively. After production of the recombinant proteins, GST within the fusion was removed by Factor Xa (New England Biolabs), and the obtained Tat-tagged Ral and Eps15 proteins were obtained, followed by filtering through 0.22-μm filters. To label peptides with FITC, an EZ-Label FITC protein labeling kit (Pierce) was used. For transduction into PMN, Tat-tagged protein (10–100 nm) was directly incubated with freshly isolated PMN in HBSS(−) for 10–30 min at 4 °C.
Subcellular Fractionation of PMN
Freshly isolated PMN (108 cells/condition), with or without fMLF stimulation or inhibitor treatment, were lysed in a buffer containing 25 mm Hepes, pH 7.5, 150 mm NaCl, 10% sucrose, 2 mm MgCl2, and protease inhibitors using a Dounce homogenizer, followed by centrifugation at 200 × g for 5 min. Meanwhile, discontinuous sucrose density gradients were prepared by layering five layers of successive decreasing sucrose density solutions (5 ml/layer) upon one another (60, 50, 40, 30, and 20%). PMN lysates (10 ml) were then layered on top of the gradients. After centrifugation with a Beckman SW28 swinging bucket ultracentrifuge rotor for 3 h at 28,000 rpm (141,000 × g) (4 °C), the gradients were fractionated (1 ml/fraction), followed by analysis for plasma membrane (alkaline phosphatase), primary granule (myeloperoxidase), secondary granule (lactoferrin), and tertiary granule (gelatinase) (12, 13).
RESULTS
Ras Family Inhibitor Damnacanthal Triggers Release of PMN Secondary Granules
PMN surface adhesive proteins, such as CD47 (12, 14), CD11b/CD18 (9, 10), and SIRPα (11, 38), play important roles in PMN adhesion, transmigration, and phagocytosis. Instead of being constitutively expressed on the cell surface, these essential proteins are mainly stored in intracellular granules under resting conditions but are rapidly up-regulated onto the cell surface in response to inflammatory chemoattractants following the process of PMN degranulation (12, 13). To reveal the mechanisms involved in control of PMN storage of these important cell surface proteins and PMN degranulation, we tested a panel of pharmacological probes/inhibitors targeting different signaling molecules and pathways for their effects on the cell surface protein expression. We found that treatment of PMN with damnacanthal, a membrane-permeable inhibitor of the Ras family GTPases and the Src family tyrosine kinase Lck (39–41), resulted in a dramatic increase of CD11b/CD18, CD47, and SIRPα on the PMN cell surface despite the lack of chemoattractant stimulation. As shown in Table 1, after damnacanthal treatment, FACS analysis showed increases of the mean fluorescence intensities of cell surface CD11b/CD18, CD47, and SIRPα labeling from the basal level of 90.7, 35.2, and 14.2 up to 203.8, 112.8, and 67.8, respectively. In contrast, treating PMN with the vehicle control or other inhibitors, such as genistein, piceatannol, LFM-A13, PP1, PD98059, LY294002, and others, did not change the levels of these proteins on PMN cell surface. Analysis of the cell shape and/or sizes by FSC and SSC parameters also indicated a visible shift of the PMN population toward lower FSC/SSC after damnacanthal treatment (data not shown), suggesting a potential of PMN degranulation.
TABLE 1.
Effects of pharmacological inhibitors on PMN cell surface protein expression
Damnacanthal treatment induces an up-regulation of CD11b/CD18, CD47, and SIRPα on the PMN surface. In these experiments, freshly isolated PMN (5 × 106 cells) were incubated with damnacanthal or other inhibitors in their working concentrations for 15 min at 25 °C. After blocking with 5% normal goat serum, PMN cell surface CD11b, CD47, and SIRPα were labeled with LM2/1, PF3.1 (10 μg/ml each), and anti-SIRPα.ex (1:2000 dilution), respectively, for 1 h (4 °C). After washing and fixation with 3% paraformaldehyde (5 min, 20 °C), cells were incubated with Alexa Fluor-conjugated secondary antibodies followed by washing and analysis by flow cytometry. PMN labeled with normal mouse IgG served as base line. The table shows the mean fluorescence intensity ± S.E. (S.D. divided by the square root of the sample size).
| Inhibitors | PMN surface fluorescence intensity (mean ± S.E.) |
|||
|---|---|---|---|---|
| Control IgG | CD11b/CD18 | CD47 | SIRPα | |
| Vehicle control (DMSO) | 3.2 ± 0.08 | 90.7 ± 0.11 | 35.2 ± 0.038 | 14.2 ± 0.09 |
| Damnacanthal (10 μm) | 2.5 ± 0.18 | 203.8 ± 0.22 | 112.8 ± 0.19 | 67.8 ± 0.16 |
| Genistein (100 μg/ml) | 4.3 ± 0.13 | 76.6 ± 0.28 | 30.0 ± 0.13 | 19.6 ± 0.14 |
| Piceatannol (40 μm) | 5.5 ± 0.26 | 94.0 ± 0.17 | 28.2 ± 0.14 | 13.5 ± 0.16 |
| PP1 (18 μm) | 2.7 ± 0.33 | 85.2 ± 0.43 | 29.3 ± 0.46 | 11.6 ± 0.31 |
| LFM-A13 (140 μm) | 4.4 ± 0.51 | 89.3 ± 0.44 | 28.5 ± 0.69 | 9.7 ± 0.42 |
| Herbimycin A (2 μm) | 5.2 ± 0.26 | 95.7 ± 0.25 | 30.3 ± 0.15 | 16.3 ± 0.45 |
| Lavendustin A (10 μm) | 5.5 ± 0.16 | 91.4 ± 0.32 | 32.2 ± 0.21 | 20.1 ± 0.25 |
| Erbstatin analog (25 μm) | 4.8 ± 0.17 | 81.6 ± 0.20 | 29.8 ± 0.19 | 18.3 ± 0.23 |
| SB203580 (1 μm) | 6.1 ± 0.30 | 83.8 ± 0.40 | 28.8 ± 0.15 | 15.2 ± 0.23 |
| PD98059 (80 nm) | 3.7 ± 0.16 | 89.9 ± 0.24 | 26.7 ± 0.18 | 12.5 ± 0.25 |
| LY294002 (100 nm) | 3.0 ± 0.14 | 79.6 ± 0.13 | 31.9 ± 0.30 | 19.1 ± 0.15 |
| AG126 (150 μm) | 4.3 ± 0.17 | 97.5 ± 0.19 | 30.7 ± 0.14 | 14.6 ± 0.24 |
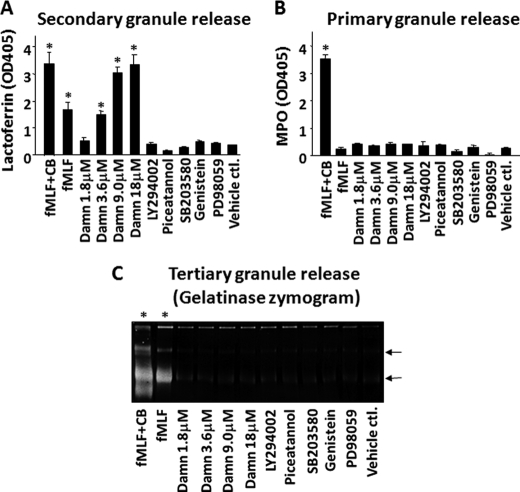
Because PMN contain multiple types of intracellular granules, and CD47, CD11b/CD18, and SIRPα are mainly stored in secondary granules (12, 13), it was highly possible that the increase of these proteins on the cell surface by damnacanthal treatment was through PMN degranulation. Therefore, we analyzed the release of primary, secondary, and tertiary granules after damnacanthal treatment. As shown in Fig. 1, compared with other inhibitors that did not cause PMN degranulation, damnacanthal dose-dependently induced PMN release of secondary granules. In contrast, no apparent release of primary or tertiary granules was observed by treating PMN with damnacanthal. In the experiments, PMN were also treated with chemoattractant fMLF and a combination of fMLF and actin filament disruption reagent CB to serve as positive degranulation controls (31). As shown in Fig. 1, as predicted, treating PMN with only fMLF resulted in significant degranulation of secondary and tertiary granules but not primary granules, whereas treatment with an fMLF-CB combination induced a robust and complete degranulation of all of the granules. At a concentration of 18 μm, damnacanthal treatment could generate amounts of lactoferrin release comparable with the combined fMLF and CB treatment, indicating a nearly complete degranulation of secondary granules (Fig. 1). In conclusion, these studies found that treatment of PMN with damnacanthal selectively induced the degranulation of secondary granules.
FIGURE 1.
Damnacanthal treatment induces specific release of PMN secondary granules. Freshly isolated PMN were treated with damnacanthal (Damn) of varied concentrations and other inhibitors for 15 min at 25 °C. A control treatment was performed using vehicle (DMSO) of the same dilution (0.1%). After centrifugation to pellet cells, PMN degranulation was assessed by assaying different granular markers in the cell-free supernatants. Positive degranulation controls were performed by stimulation of PMN with 1 μm fMLF with and without an addition of 10 μm CB. Concentrations of LY94002, piceatannol, SB203580, genistein, and PD98059 used in the treatment were the same as those in Table 1. A, assaying secondary granule release by measuring lactoferrin in the supernatants. B, assaying primary granule release by measuring MPO activity in the supernatants. C, assaying tertiary granule release by zymogram detecting gelatinase in the supernatants. The arrows indicate the two major forms of gelatinase (92 and 56 kDa, respectively) in PMN. As shown, damnacanthal specifically and dose-dependently induced secondary granule degranulation. Data (mean ± S.D.) represent more than five independent experiments. *, significant degranulation.
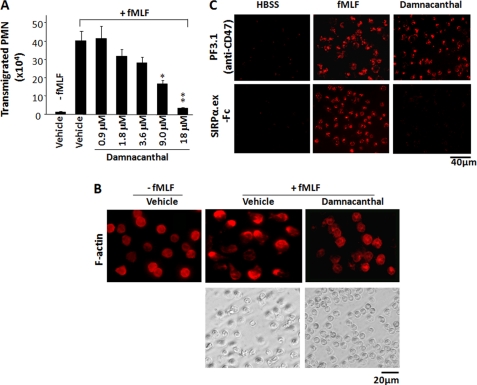
Damnacanthal Treatment Results in Defect of PMN Function
In general, PMN degranulation of secondary granules is triggered by chemoattractants binding to the cell surface receptors, followed by signaling relays involving G protein and a series of downstream signaling molecules (42). Because damnacanthal caused the release of secondary granules in the absence of chemoattractant stimulation, we questioned whether this kind of “dysregulated” degranulation would affect PMN function and thus assayed chemoattractant-induced PMN transmigration and adhesion/spreading. As shown in Fig. 2A, in contrast to the finding that PMN vigorously transmigrated toward fMLF in the control conditions, damnacanthal dose-dependently inhibited PMN transmigration and completely abolished PMN migration at a concentration of 18 μm. Also as shown in Fig. 2B, compared with control PMN, which responded to fMLF stimulation and quickly spread on collagen-coated surfaces displaying a polarized F-actin labeling pattern, damnacanthal treatment impaired PMN spreading. Indeed, PMN adhesion to collagen induced by fMLF was significantly weakened in the presence of damnacanthal (data not shown). Next we examined whether damnacanthal treatment affected cell surface protein function. Because CD47 and SIRPα are extracellular counterligands (11, 22), we tested cell surface CD47 direct binding to a SIRPα extracellular domain fusion protein, SIRPα1.ex-Fc (13, 22). As shown in Fig. 2C, although damnacanthal induced a significant increase of CD47 on the PMN cell surface, these up-regulated CD47 were not binding to its ligand SIRPα. In contrast, CD47 was also increased on the PMN surface after fMLF stimulation, and these CD47 directly bound to SIRPα1.ex-Fc (Fig. 2C).
FIGURE 2.
Damnacanthal inhibits chemoattractant-triggered PMN function. A, damnacanthal inhibits PMN chemotaxis. PMN (106) were treated with damnacanthal of varying concentrations for 15 min at 25 °C, followed by assaying transmigration across collagen-coated filters toward fMLF (0.1 μm) in 1 h. Data (mean ± S.D. (error bars)) represent three independent experiments with triplicates in each condition (*, p < 0.05; **, p < 0.01). B, damnacanthal inhibits PMN spreading and F-actin polarization. PMN (106), untreated or treated with damnacanthal (10 μm), were stimulated with fMLF (1 μm, 37 °C) in collagen-coated wells, followed by observation of PMN adhesion and spreading (phase-contrast images). After stimulation for 20 min, the cells were then fixed and permeabilized, followed by labeling F-actin with rhodamine-conjugated phalloidin. C, cell surface CD47 binding to SIRPα. An extracellular domain fusion protein, SIRPα1.ex-Fc, was used to test PMN cell surface CD47 binding after treatment with either fMLF (1 μm) or damnacanthal (10 μm). Total amount of CD47 on cell surface was confirmed by labeling non-permeabilized cells with anti-CD47 mAb PF3.1.
Identification of the Specific GTPase Associated with Secondary Granule Release
Efforts have been made to identify the target molecule of damnacanthal that plays an important role in preventing secondary granule release in PMN prior to chemoattractant stimulation. We have found that, although damnacanthal is a potent inhibitor of the tyrosine kinase Lck, its effect on PMN degranulation was not through targeting Lck. In agreement with many studies by others, we observed that Lck is mainly expressed in T cells but not in PMN (supplemental Fig. 1). In addition, we obtained Lck-knock-out mice (43) from Dr. Tak Mak (Ontario Cancer Institute). Comparing PMN isolated from Lck knock-out mice with the cells from their wild-type littermates yielded no difference in the levels of cell surface protein expression, PMN response to chemoattractant stimulation, degranulation, and chemotactic transmigration (data not shown), suggesting that Lck is dispensable in PMN function. A previous study by Brumell et al. (44) also indicated no Lck activity in PMN. In addition to Lck, damnacanthal is also an established inhibitor of Ras family small GTPases (40, 41). Because some of these GTPases, such as Ras family member Ral, have been demonstrated to play important roles in vesicle transportation, it was likely that damnacanthal affected PMN through inhibition of Ras family members.
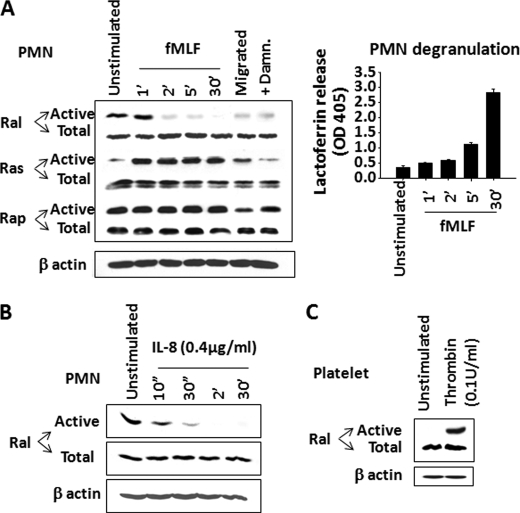
The Ras family small GTPases are classified into three major subfamilies, Ras (H, R, etc.), Rap (1/2), and Ral (A/B) (45–47). To identify which is potentially involved in the degranulation of secondary granules, we screened the Ras members for activity changes before and after chemoattractant stimulation. Given that damnacanthal treatment caused secondary granule release in the absence of stimulation, we speculated that the candidate Ras member must be in its active form in resting PMN. Also, because chemoattractant stimulation triggers secondary granule release, we reasoned that this specific Ras should be deactivated in order to coordinate secondary granule release. Based on this rationale, we assayed the activities of Ras, Rap, and Ral in PMN before and after fMLF stimulation and with or without damnacanthal treatment. As shown in Fig. 3A, Ras, Rap, and Ral were all expressed in PMN. Among the three family members, Ras was found mainly in its inactive form in unstimulated PMN and was quickly activated after fMLF stimulation. Rap, on the other hand, showed minor activity changes before or after fMLF stimulation and, to our surprise, was insensitive to damnacanthal treatment (Fig. 3A). Therefore, neither Ras nor Rap was likely to be the candidate that regulates secondary granule release.
FIGURE 3.
Ral deactivation correlates secondary granule release in PMN. A, detecting activity changes of Ras family small GTPases, including Ral, Ras, and Rap, in PMN. After time course stimulation with fMLF (1 μm), PMN were lysed, and active Ras, Ral, and Rap were pulled down using Raf-1 RBD, RalBP1, and RalGDS-RBD, respectively. Right, in parallel, fMLF-induced PMN degranulation of secondary granules was assayed simultaneously by detecting lactoferrin in the cell-free supernatants. PMN that transmigrated across collagen-coated filters into the fMLF-containing lower chambers or PMN that were treated with damnacanthal were also assayed for active Ral, Ras, and Rap. Cell lysates before pull-down were used to detect total Ral, Ras, and Rap as well as actin using antibodies against Ral, Ras, Rap, and β-actin. B, deactivation of Ral by IL-8 stimulation. Recombinant IL-8 (0.4 μg/ml) (Sigma) was used to stimulate PMN for different time periods, followed by RalBP1 pull-down to detect active Ral. C, Ral activities in unstimulated and thrombin (0.1 unit/ml) (Sigma)-stimulated platelets. Data (mean ± S.D. (error bars)) represent three independent experiments with triplicates in each condition.
In contrast to Ras and Rap, Ral demonstrated a dynamic activity change that correlated with chemoattractant stimulation and PMN degranulation. As shown in Fig. 3A, Ral appeared to be in its active form in unstimulated PMN and quickly became inactive following fMLF stimulation. Pull-down experiments using RalBP1, a downstream effector that specifically binds to active Ral, recovered a significant amount of active Ral from resting PMN. On the other hand, stimulation with fMLF resulted in a quick diminishing of RalBP1 pull-down within 2–5 min, a time course paralleled with secondary granule release that was assayed simultaneously (Fig. 3A, right). The same results were obtained when PMN were stimulated by another chemoattractant, IL-8 (Fig. 3B). Deactivation of Ral was also observed in PMN that had chemotactically transmigrated across collage-coated filters toward fMLF (Fig. 3A, Migrated). In addition, the activity of Ral in resting PMN was inhibited after damnacanthal treatment (Fig. 3A), which correlated with damnacanthal-induced secondary granule release. Thus, these results suggested Ral to be the regulator that controls secondary granule release in PMN.
To confirm our studies in PMN and verify the pull-down assays, in parallel experiments, we simultaneously assayed Ral activity in platelets. As shown in Fig. 3C, our RalBP1 pull-down results agreed with the previous report by others showing that Ral is inactive in resting platelets and activated after thrombin stimulation (32). The different Ral activity profiles in PMN and platelets thus suggest that these cells employ distinctive signaling mechanisms to regulate Ral activity and that Ral may have different functional aspects in PMN and platelets.
Active Ral Blocks Secondary Granule Release in PMN
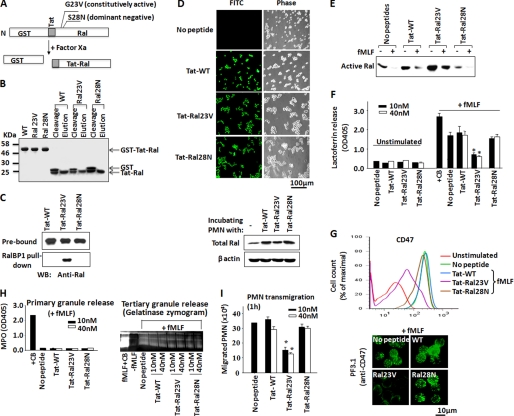
To test that active Ral in resting PMN serves to prevent secondary granule release and that Ral deactivation in response to chemoattractant stimulation is required for degranulation, we designed experiments to overexpress a constitutively active Ral, Ral23V, in PMN and tested its effect on fMLF-induced degranulation. Because freshly isolated PMN are short lived and not suitable for protein expression by conventional gene transfection methods, we employed protein transduction technology and directly delivered purified recombinant Ral into PMN by tagging the proteins with an HIV Tat peptide (YGRKKRRQRRR) (34, 48, 49). As demonstrated by previous studies, HIV Tat and other polyarginine peptides have been widely used to transduce proteins of up to 120 kDa into live cells (34, 35, 49). Although PMN are highly sensitive and difficult to handle, using HIV Tat to study important intracellular molecules has been very successful (48, 50, 51). In this study, three Tat-tagged Ral proteins were generated, including a wild-type Ral (WT), a constitutively active mutant Ral (Ral23V), and a dominant negative mutant Ral (Ral28N) (15, 36, 37). As depicted in Fig. 4A, these proteins were first produced as recombinant GST fusions, followed by removal of GST, yielding Tat-tagged Ral. Fig. 4, B and C, shows the stepwise production of Tat-tagged WT, Ral23V, and Ral28N and RalBP1 pull-down, confirming that Ral23V was a constitutively active form.
FIGURE 4.
Constitutively active Ral (Ral23V) inhibits chemoattractant-induced secondary granule release in PMN. A, a schematic depiction of Tat-tagged Ral production. The recombinant fusion proteins containing GST and Tat-Ral were produced, followed by cleavage of GST with Factor Xa to yield Tat-Ral peptides. The figure also shows that two point mutations, G23V and S28N, were created in Ral to produce constitutively active Ral (23V) and dominant negative Ral (28N). B, a stepwise analysis of Tat-tagged Ral protein production by SDS-PAGE and Coomassie Blue staining. C, RalBP1 pull-down confirming that Tat-Ral23V was a constitutively active GTPase. Purified Tat-tagged wild-type Ral, Ral23V, and Ral28N (0.2 μg each) were incubated with GST-RalBP1 (1 μg) and glutathione-Sepharose in HBSS containing 5% nonfat dry milk, 2% BSA, and 0.01% Tween 20 for 1 h. After washing the precipitates, Western blot (WB) was performed to detect active Ral pulled down by RalBP1. D, assaying Tat-tagged Ral transduction into PMN. Tat-tagged wild-type Ral (WT), Ral23V, and Ral28N (0.1 μm each) were labeled with FITC, followed by incubation with PMN for 10 min at 4 °C. Effective transduction into PMN was then analyzed by fluorescence microscopy and Western blot. E, constitutively active Ral in PMN. After transduction of Tat-tagged Ral into PMN, PMN were stimulated with fMLF (1 μm) for 20 min at 37 °C, followed by cell lysis. RalBP1 pull-down was then performed to detect active Ral. As shown, the result confirmed that transduction of Ral23V in PMN resulted in a sustained Ral activity even with fMLF stimulation. F and G, effects of Tat-tagged Ral proteins on fMLF-induced secondary granule release in PMN. PMN were incubated with purified Tat-tagged wild-type Ral (WT), the constitutive active Ral (Ral23V), and dominant negative Ral (Ral28N) at concentrations of 10 and 40 nm or control buffer (No peptide) (30 min, 4 °C), followed by stimulation with fMLF (1 μm) for 20 min at 37 °C. Degranulation of secondary granules was analyzed by measuring lactoferrin release in the cell-free supernatants (F) and cell surface labeling of CD47 by anti-CD47 antibody PF3.1 (G), followed by flow cytometry (FACS). H, Tat-Ral23V has no effect on the mobilization of primary and tertiary granules. I, transduction of Tat-Ral23V into PMN inhibited PMN chemotactic transmigration. Data (mean ± S.D.) represent three independent experiments with triplicates in each condition (*, p < 0.01).
To determine that Tat-tagged Ral proteins were capable of directly delivery into PMN, we chemically labeled the purified proteins with FITC, followed by incubation with PMN. As shown in Fig. 4D, fluorescence microscopy revealed that the three Tat-tagged Ral proteins (Tat-WT, Tat-Ral23V, and Tat-Ral28N) are all rapidly delivered into PMN after only a short incubation (10 min) at a low temperature (4 °C). Subsequent Western blot detected an over 1-fold increase of Ral in PMN after incubating with each Tat-tagged Ral protein (Fig. 4D). We also performed RalBP1 pull-down assays to test Ral activity after delivery of each Tat-tagged Ral protein into PMN. As shown in Fig. 4E, transduction of exogenous Ral23V into PMN led to a sustained Ral activity in PMN even in the presence of fMLF stimulation, a condition in which the endogenous Ral activity was completely diminished as that observed in the control, non-peptide transduction PMN. Transduction of WT Ral or Ral28N, on the other hand, only slightly increased (WT) or did not increase (Ral28N) Ral activity in PMN (Fig. 4E).
We next investigated whether Ral activity blocks fMLF-induced PMN degranulation. In these experiments, PMN were first loaded with Tat-tagged WT Ral, Ral23V, or Ral28N and then stimulated with fMLF. As shown in Fig. 4F, no spontaneous PMN degranulation was detected by just loading Tat-tagged Ral proteins (unstimulated). Prominent release of secondary granules was induced by fMLF in PMN loaded with WT Ral or Ral28N or without peptide delivery. On the contrary, transduction of the constitutively active Ral, Ral23V, into PMN strikingly blocked secondary granule release in the presence of fMLF. As shown in Fig. 4, both lactoferrin release (Fig. 4F) and CD47 translocation to the cell surface plasma membrane (Fig. 4G) were inhibited by Ral23V in PMN despite fMLF stimulation. However, Ral23V had no effect on fMLF-induced tertiary granule release or the retention of primary granules (Fig. 4H), further suggesting that Ral selectively regulates secondary granule mobilization.
Given that PMN degranulation of secondary granules is crucial for chemotaxis, we thus performed PMN transmigration assays and examined the effect of Ral23V. As shown in Fig. 4I, as expected, transduction of Ral23V into PMN significantly reduced PMN transmigration toward fMLF, whereas WT Ral or Ral28N had no inhibition. In conclusion, these results demonstrated that Ral activity blocks secondary granule release from PMN. Therefore, chemoattractant-induced Ral deactivation is essential for PMN degranulation, activation, and chemotaxis.
Ral Translocation between Plasma Membranes and Secondary Granules
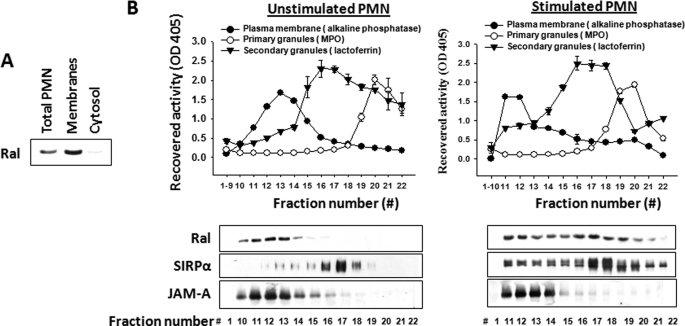
Like Ras family GTPases, Ral contains a lipid modification site at the carboxyl terminus, suggesting that Ral may be associated with the cell membranes. To test this, we separated PMN membranes from the cytosol by ultracentrifugation in the absence of detergent. As shown in Fig. 5A, Western blot indicated that Ral is exclusively associated with the cell membranes in PMN. To further localize Ral and define whether Ral is dynamically coupled with secondary granules, we performed subcellular fractionation using sucrose density gradients. As shown in Fig. 5B, the fractionations successfully separated PMN plasma membranes (alkaline phosphatase) from secondary granules (lactoferrin) and primary granules (MPO). To our surprise, detection of Ral in the fractions by Western blots revealed that Ral is entirely associated with the plasma membrane in unstimulated PMN that is co-localized with another plasma membrane protein, JAM-A. Stimulation with fMLF, on the other hand, resulted in translocation of a considerable amount of Ral to secondary granules (Fig. 5B). In agreement with previous observations (11, 13), proteins initially stored in secondary granules, such as SIRPα, were redistributed to the plasma membrane after fMLF stimulation following degranulation.
FIGURE 5.
Localization of Ral in PMN. A, Ral is associated with membranes. PMN (2.5 × 107) were Dounce-lysed, followed by ultracentrifugation to separate cytosol from cell membranes. SDS-PAGE and Western blot were performed to detect Ral in the total lysate before centrifugation (total PMN) and in the separated cytosol and membranes. B, subcellular localization of Ral in PMN. PMN (108/condition), unstimulated and fMLF-stimulated, were lysed and subjected to sucrose density gradient centrifugation to separate the plasma membrane and intracellular granules, followed by fractionation. Western blots (lower panel) were performed to detect Ral, SIRPα, and JAM-A in the fractions using antibodies against Ral, anti-SIRPα.ex, and anti-JAM-A.ex. JAM-A is a transmembrane protein consistently expressed in the plasma membranes.
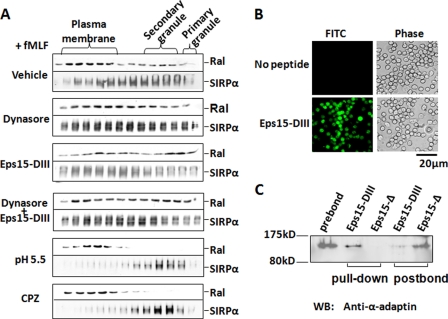
Because one major mechanism of plasma membrane internalization in PMN after chemoattractant stimulation is through clathrin-mediated cell surface receptor endocytosis (42, 52), we further tested whether fMLF-induced Ral translocation from the plasma membrane to intracellular granules followed this pathway. Two inhibitors, dynasore (28) and an AP-2-binding Eps15 C-terminal peptide (Eps15-DIII) (29, 30), which specifically act on the clathrin-dependent endocytosis, were examined. As shown in Fig. 6A, pretreatment of PMN with dynasore (80 μm), recombinant Eps15-DIII (40 nm), or their combination appeared to have no inhibition on Ral translocation to secondary granules after fMLF stimulation. Dynasore inhibits dynamin, an essential GTPase regulating clathrin-coated pit fission, whereas Eps15-DIII inhibits clathrin-mediated endocytosis through sequestration of AP-2 and thus interferes with formation of the clathrin coat. To effectively use Eps15-DIII in the study, this peptide was tagged with HIV Tat, which significantly facilitated protein entering PMN (Fig. 6B). We also confirmed the function of Eps15-DIII by protein pull-down assays, which showed that Eps15-DIII successfully co-precipitated AP-2 from PMN lysates. However, despite the fact that dynasore and Eps15 peptide failed to exhibit an inhibitory effect, other endocytosis inhibitors remarkably blocked Ral translocation. As shown in Fig. 6A, treatment of PMN with chlorpromazine (25) or cytosolic acidification (succinic acid, pH 5.5) (26, 27) eliminated Ral translocation from plasma membranes to secondary granules in response to fMLF. These inhibitors also effectively blocked secondary granule release, as indicated by a failure of translocation of secondary granule proteins (e.g. SIRPα) to the plasma membrane.
FIGURE 6.
Ral translocation from the plasma membrane to secondary granules may follow fMLF-induced endocytic process. A, effect of clathrin-dependent endocytic inhibitors on Ral redistribution in PMN. In these experiments, PMN were preincubated with dynasore (80 μm), Tat-tagged Eps15 peptide (Eps15-DIII) (40 nm), or a combination of the two reagents for 15 min (25 °C) before fMLF stimulation (20 min at 37 °C). After cell lysis, subcellular fractionation was performed, followed by detection of Ral by Western blots. The effect of other endocytosis inhibitors, chlorpromazine hydrochloride (CPZ) (50 μm) and succinic acid (20 mm, pH 5.5), on Ral distribution was also tested. B, protein transduction of Tat-tagged Eps15-DIII into PMN. Purified Tat-tagged Eps15-DIII was fluorescence-labeled, followed by incubation with PMN for 10 min (4 °C). The image shows that Eps15-DIII was successfully delivered into the cells. C, pull-down assay confirmed that Eps15-DIII directly binds to AP-2 complexes. PMN (1.5 × 107) were lysed in a buffer containing 1% Triton X-100 and protease inhibitors, followed by incubation with 10 μg of purified Tat-tagged Eps15-DIII in the form of GST fusion protein or its non-binding control peptide Tat-tagged Eps15-Δ for 3 h (4 °C). After pull-down with glutathione-Sepharose, the precipitated protein complexes were washed and analyzed by Western blot (WB) using antibody against α-adaptin, an important component of AP-2 complexes.
DISCUSSION
As a type of highly specialized and fast responsive cells, PMN store a large number of functionally important molecules in the intracellular granules and vesicles. Upon stimulation, PMN promptly mobilize these granules/vesicles and rapidly release arrays of cell adhesion molecules, receptors, and proteases to the cell surfaces to facilitate PMN adhesion, chemotaxis, and microbial killing. At least four types of granules and vesicles varied in size/shape and content have been identified in PMN (1, 2), but the mechanisms that control their mobilization and release during PMN response to inflammatory stimulation remain largely undefined. Among these granules and vesicles, secondary granules are defined as being mostly rich in leukocyte integrins and adhesive receptor-like proteins. Degranulation of secondary granules thus serves as a major means to furnish the cell surface plasma membrane with the essential cell adhesion and migration molecules. Here, we report our studies of PMN degranulation, which reveal, for the first time, that the Ras family member Ral is a critical regulator specifically in the degranulation of secondary granules.
As shown by our results, Ral exits as an active GTPase in freshly isolated PMN, and its activity plays a key role in keeping secondary granules in the cell. Inhibition of Ral in resting PMN by damnacanthal, a Ras family GTPase inhibitor with unclear mechanism (40, 41), causes “premature” liberation of secondary granules in the absence of chemoattractant stimulation. However, this dysregulated degranulation has no benefit to but instead impeded PMN function. As we observed, although damnacanthal increased cell surface integrins and adhesive molecules, these up-regulated molecules failed to mediate effective PMN interactions, adhesion, and migration. In particular, our results (Fig. 2) showed that, in contrast to fMLF-induced CD47 on the cell surface that mediates binding interactions with its extracellular ligand, damnacanthal-induced CD47 on PMN were incapable of binding interaction. Similarly, damnacanthal-mediated increased cell surface CD11b/CD18 did not appear to mediate strong extracellular interactions, which count, in part, for the decrease of PMN adhesion and spreading in response to fMLF. Thus, these results suggest that a critical mechanism, which is elicited by chemoattractant stimulation, controls secondary granule degranulation, and also converts essential cell surface proteins to be functional. These studies thus further signify the important role of Ral in PMN. By preventing irregular degranulation in the absence of stimuli, the Ral-mediated mechanism preserves PMN functional capability for potent and effective response during inflammation.
In this study, we also obtained the important finding that chemoattractant induces a rapid deactivation of Ral in PMN, which explains a dynamic regulation required for secondary granule release. In contrast, experimental overexpression of a constitutively active Ral, Ral23V (15, 36, 37), into PMN to sustain Ral activity even in the presence of fMLF resulted in prohibition of secondary granule release and hence hampered PMN chemotactic transmigration. Although its role in secondary granule mobilization is evident, Ral has no control of the mobilization of primary and tertiary granules, and both damnacanthal and Ral23V only had an effect on the degranulation of secondary granules.
The mechanism by which Ral regulates secondary granule storage and release is, however, unclear. By subcellular fractionation, we found that Ral is associated with the plasma membrane under resting conditions, whereas chemoattractant stimulation triggers a translocation of Ral to secondary granules. Presumably, chemoattractant-induced cell surface endocytosis plays an important role in Ral intracellular translocation and thus PMN degranulation. Because PMN chemoattractant receptors are G protein-coupled and clathrin-coated pits are generally employed to internalize these receptors after ligand binding, one possible hypothesis would be that Ral is in complex with the chemoattractant receptor and assembled into the clathrin pits after chemoattractant stimulation. However, testing this hypothesis using two reagents, dynasore and an AP-2-binding Eps15 peptide, both specifically blocking clathrin-dependent endocytosis, failed to inhibit Ral translocation to the pool of secondary granules. Interestingly, several other inhibitors that generally inhibit endocytosis, vesicle transportation, and/or cytoskeleton structures blocked Ral intracellular translocation in the experiments.
In other cell types, Ral has been extensively implicated in different vesicle transportations, including both endocytosis and exocytosis (15–19). In platelets, it was found that Ral is inactive under resting conditions but activated after stimulation (32). Contrary to our observation in this study, scientists in another study also showed that Ral is inactive in resting PMN (53), a variation that was probably caused by different methods of cell preparation. A guanine nucleotide dissociation stimulator, RalGDS, was also shown to activate Ral, and such regulation involves Ras. In particular, RalGDS was shown to directly bind to Ras, suggesting that Ral acts as a downstream effector in the Ras network (36, 54). However, we found that in PMN, Ral has a completely different activity profile from that of Ras, suggesting a dissociation of Ral from the Ras pathway. Although it is important to render Ral activation, our study suggests that the mechanism that deactivates Ral is crucial for PMN degranulation and conversion of PMN to be functional inflammatory leukocytes. In addition to Ral, other small GTPases, such as Ras family members Ras and Rap, and Rho family members Rho, Rac, and Cdc42 are also expressed in PMN (55, 56), and some of these are found to be associated with granules (57–59). Thus, detailed investigation of these small GTPases in PMN and correlation of their activity changes with PMN activation and degranulation will shed new light on the understanding of PMN inflammatory functions. Given the critical role of PMN in innate immunity and inflammation, our findings illustrate a novel mechanism that modulates PMN inflammatory response, which may provide a potential drug design for treatment against various inflammatory conditions.
Supplementary Material
Acknowledgment
We thank Alex Guïle for critical proofreading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health R21 Grant AI073622. This study was also supported in part by Beginning Grant-in-aid 0565251B from the American Heart Association and Research Scholar Grant RSG-09-193-01-LIB from the American Cancer Society.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- PMN
- polymorphonuclear leukocyte(s)
- fMLF
- formyl-methionyl-leucyl-phenylalanine
- HBSS
- Hanks' balanced salt buffer
- HBSS(−)
- Hanks' balanced salt buffer devoid of Ca2+ and Mg2+
- CB
- cytochalasin B
- MPO
- myeloperoxidase
- Raf-1 RBD
- Ras binding domain of Raf-1
- RalGDS-RBD
- Ral GDS-Rap binding domain
- RalBP1
- Ral-binding protein 1
- RalGDS
- Ral guanine nucleotide dissociation stimulator.
REFERENCES
- 1. Kobayashi S. D., Voyich J. M., DeLeo F. R. (2003) Microbes Infect. 5, 1337–1344 [DOI] [PubMed] [Google Scholar]
- 2. Butterfield T. A., Best T. M., Merrick M. A. (2006) J. Athl. Train 41, 457–465 [PMC free article] [PubMed] [Google Scholar]
- 3. Borregaard N., Cowland J. B. (1997) Blood 89, 3503–3521 [PubMed] [Google Scholar]
- 4. Borregaard N., Sørensen O. E., Theilgaard-Mönch K. (2007) Trends Immunol. 28, 340–345 [DOI] [PubMed] [Google Scholar]
- 5. Faurschou M., Borregaard N. (2003) Microbes Infect. 5, 1317–1327 [DOI] [PubMed] [Google Scholar]
- 6. Nourshargh S., Marelli-Berg F. M. (2005) Trends Immunol. 26, 157–165 [DOI] [PubMed] [Google Scholar]
- 7. Kelly M., Hwang J. M., Kubes P. (2007) J. Allergy Clin. Immunol. 120, 3–10 [DOI] [PubMed] [Google Scholar]
- 8. Ley K., Laudanna C., Cybulsky M. I., Nourshargh S. (2007) Nat. Rev. Immunol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 9. Mayadas T. N., Cullere X. (2005) Trends Immunol. 26, 388–395 [DOI] [PubMed] [Google Scholar]
- 10. Parkos C. A. (1997) Am. J. Physiol. 273, G763–G768 [DOI] [PubMed] [Google Scholar]
- 11. Liu Y., Bühring H. J., Zen K., Burst S. L., Schnell F. J., Williams I. R., Parkos C. A. (2002) J. Biol. Chem. 277, 10028–10036 [DOI] [PubMed] [Google Scholar]
- 12. Liu Y., Merlin D., Burst S. L., Pochet M., Madara J. L., Parkos C. A. (2001) J. Biol. Chem. 276, 40156–40166 [DOI] [PubMed] [Google Scholar]
- 13. Liu Y., Soto I., Tong Q., Chin A., Bühring H. J., Wu T., Zen K., Parkos C. A. (2005) J. Biol. Chem. 280, 36132–36140 [DOI] [PubMed] [Google Scholar]
- 14. Parkos C. A., Colgan S. P., Liang T. W., Nusrat A., Bacarra A. E., Carnes D. K., Madara J. L. (1996) J. Cell Biol. 132, 437–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vitale N., Mawet J., Camonis J., Regazzi R., Bader M. F., Chasserot-Golaz S. (2005) J. Biol. Chem. 280, 29921–29928 [DOI] [PubMed] [Google Scholar]
- 16. Polzin A., Shipitsin M., Goi T., Feig L. A., Turner T. J. (2002) Mol. Cell Biol. 22, 1714–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brymora A., Valova V. A., Larsen M. R., Roufogalis B. D., Robinson P. J. (2001) J. Biol. Chem. 276, 29792–29797 [DOI] [PubMed] [Google Scholar]
- 18. Balasubramanian N., Meier J. A., Scott D. W., Norambuena A., White M. A., Schwartz M. A. (2010) Curr. Biol. 20, 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jullien-Flores V., Mahé Y., Mirey G., Leprince C., Meunier-Bisceuil B., Sorkin A., Camonis J. H. (2000) J. Cell Sci. 113, 2837–2844 [DOI] [PubMed] [Google Scholar]
- 20. Zen K., Reaves T. A., Soto I., Liu Y. (2006) J. Immunol. Methods 309, 86–98 [DOI] [PubMed] [Google Scholar]
- 21. Franke B., Akkerman J. W., Bos J. L. (1997) EMBO J. 16, 252–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y., Tong Q., Zhou Y., Lee H. W., Yang J. J., Bühring H. J., Chen Y. T., Ha B., Chen C. X., Yang Y., Zen K. (2007) J. Mol. Biol. 365, 680–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y., Nusrat A., Schnell F. J., Reaves T. A., Walsh S., Pochet M., Parkos C. A. (2000) J. Cell Sci. 113, 2363–2374 [DOI] [PubMed] [Google Scholar]
- 24. Zen K., Liu Y. (2008) Immunobiology 213, 13–23 [DOI] [PubMed] [Google Scholar]
- 25. Wang L. H., Rothberg K. G., Anderson R. G. (1993) J. Cell Biol. 123, 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sandvig K., Olsnes S., Petersen O. W., van Deurs B. (1987) J. Cell Biol. 105, 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sandvig K., Olsnes S., Petersen O. W., van Deurs B. (1988) J. Cell Biochem. 36, 73–81 [DOI] [PubMed] [Google Scholar]
- 28. Macia E., Ehrlich M., Massol R., Boucrot E., Brunner C., Kirchhausen T. (2006) Dev. Cell 10, 839–850 [DOI] [PubMed] [Google Scholar]
- 29. van Bergen En Henegouwen P. M. (2009) Cell Commun. Signal. 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benmerah A., Lamaze C., Bègue B., Schmid S. L., Dautry-Varsat A., Cerf-Bensussan N. (1998) J. Cell Biol. 140, 1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mócsai A., Jakus Z., Vántus T., Berton G., Lowell C. A., Ligeti E. (2000) J. Immunol. 164, 4321–4331 [DOI] [PubMed] [Google Scholar]
- 32. Wolthuis R. M., Franke B., van Triest M., Bauer B., Cool R. H., Camonis J. H., Akkerman J. W., Bos J. L. (1998) Mol. Cell Biol. 18, 2486–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joliot A., Prochiantz A. (2004) Nat. Cell Biol. 6, 189–196 [DOI] [PubMed] [Google Scholar]
- 34. Schwarze S. R., Dowdy S. F. (2000) Trends Pharmacol. Sci. 21, 45–48 [DOI] [PubMed] [Google Scholar]
- 35. Schwarze S. R., Hruska K. A., Dowdy S. F. (2000) Trends Cell Biol. 10, 290–295 [DOI] [PubMed] [Google Scholar]
- 36. Hinoi T., Kishida S., Koyama S., Ikeda M., Matsuura Y., Kikuchi A. (1996) J. Biol. Chem. 271, 19710–19716 [DOI] [PubMed] [Google Scholar]
- 37. Kawai M., Kawashima S., Sakoda T., Toh R., Kikuchi A., Yamauchi-Takihara K., Kunisada K., Yokoyama M. (2003) Hypertension 41, 956–962 [DOI] [PubMed] [Google Scholar]
- 38. de Vries H. E., Hendriks J. J., Honing H., De Lavalette C. R., van der Pol S. M., Hooijberg E., Dijkstra C. D., van den Berg T. K. (2002) J. Immunol. 168, 5832–5839 [DOI] [PubMed] [Google Scholar]
- 39. Faltynek C. R., Schroeder J., Mauvais P., Miller D., Wang S., Murphy D., Lehr R., Kelley M., Maycock A., Michne W. (1995) Biochemistry 34, 12404–12410 [DOI] [PubMed] [Google Scholar]
- 40. Hiramatsu T., Imoto M., Koyano T., Umezawa K. (1993) Cancer Lett. 73, 161–166 [DOI] [PubMed] [Google Scholar]
- 41. Hiwasa T., Kondo K., Hishiki T., Koshizawa S., Umezawa K., Nakagawara A. (1997) Neurosci. Lett. 238, 115–118 [DOI] [PubMed] [Google Scholar]
- 42. Liu Y., Shaw S. K., Ma S., Yang L., Luscinskas F. W., Parkos C. A. (2004) J. Immunol. 172, 7–13 [DOI] [PubMed] [Google Scholar]
- 43. Molina T. J., Kishihara K., Siderovski D. P., van Ewijk W., Narendran A., Timms E., Wakeham A., Paige C. J., Hartmann K. U., Veillette A. (1992) Nature 357, 161–164 [DOI] [PubMed] [Google Scholar]
- 44. Brumell J. H., Burkhardt A. L., Bolen J. B., Grinstein S. (1996) J. Biol. Chem. 271, 1455–1461 [DOI] [PubMed] [Google Scholar]
- 45. Colicelli J. (2004) Sci. STKE 2004, RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Macaluso M., Russo G., Cinti C., Bazan V., Gebbia N., Russo A. (2002) J. Cell Physiol. 192, 125–130 [DOI] [PubMed] [Google Scholar]
- 47. Oxford G., Theodorescu D. (2003) Cancer Lett. 189, 117–128 [DOI] [PubMed] [Google Scholar]
- 48. Bruyninckx W. J., Comerford K. M., Lawrence D. W., Colgan S. P. (2001) Blood 97, 3251–3258 [DOI] [PubMed] [Google Scholar]
- 49. Schwarze S. R., Ho A., Vocero-Akbani A., Dowdy S. F. (1999) Science 285, 1569–1572 [DOI] [PubMed] [Google Scholar]
- 50. Choi M., Rolle S., Wellner M., Cardoso M. C., Scheidereit C., Luft F. C., Kettritz R. (2003) Blood 102, 2259–2267 [DOI] [PubMed] [Google Scholar]
- 51. Gao X. P., Zhu X., Fu J., Liu Q., Frey R. S., Malik A. B. (2007) J. Biol. Chem. 282, 6116–6125 [DOI] [PubMed] [Google Scholar]
- 52. Wolfe B. L., Trejo J. (2007) Traffic 8, 462–470 [DOI] [PubMed] [Google Scholar]
- 53. M'Rabet L., Coffer P. J., Wolthuis R. M., Zwartkruis F., Koenderman L., Bos J. L. (1999) J. Biol. Chem. 274, 21847–21852 [DOI] [PubMed] [Google Scholar]
- 54. Jullien-Flores V., Dorseuil O., Romero F., Letourneur F., Saragosti S., Berger R., Tavitian A., Gacon G., Camonis J. H. (1995) J. Biol. Chem. 270, 22473–22477 [DOI] [PubMed] [Google Scholar]
- 55. Makino A., Glogauer M., Bokoch G. M., Chien S., Schmid-Schönbein G. W. (2005) Am. J. Physiol. Cell Physiol. 288, C863–C871 [DOI] [PubMed] [Google Scholar]
- 56. Cicchetti G., Allen P. G., Glogauer M. (2002) Crit. Rev. Oral Biol. Med. 13, 220–228 [DOI] [PubMed] [Google Scholar]
- 57. Abdel-Latif D., Steward M., Macdonald D. L., Francis G. A., Dinauer M. C., Lacy P. (2004) Blood 104, 832–839 [DOI] [PubMed] [Google Scholar]
- 58. Mollinedo F., Perez-Sala D., Gajate C., Jimenez B., Rodriguez P., Lacal J. C. (1993) FEBS Lett. 326, 209–214 [DOI] [PubMed] [Google Scholar]
- 59. Maridonneau-Parini I., de Gunzburg J. (1992) J. Biol. Chem. 267, 6396–6402 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.