Abstract
Malaria parasites are subjected to high levels of oxidative stress during their development inside erythrocytes and the ability of the parasite to defend itself against this assault is critical to its survival. Therefore, Plasmodium possesses an effective antioxidant defense system that could potentially be used as a target for the development of inhibitor-based therapy. We have identified an unusual peroxiredoxin protein that localizes to the nucleus of Plasmodium falciparum and have renamed it PfnPrx (PF10_0268, earlier called MCP1). Our work reveals that PfnPrx has a broad specificity of substrate being able to utilize thioredoxin and glutaredoxin as reductants and having the ability to reduce simple and complex peroxides. Intriguingly, chromatin immunoprecipitation followed by deep sequencing reveals that the enzyme associates with chromatin in a genome-wide manner with a slight enrichment in coding regions. Our results represent the first description of a dedicated chromatin-associated peroxiredoxin and potentially represent an ingenious way by which the parasite can survive the highly oxidative environment within its human host.
Keywords: Chromatin, Enzymes, Nucleus, Oxidation Reduction, Parasitology
Introduction
Malaria is one of the most common infectious diseases in the world with ∼500 million cases each year and one to three million deaths (1). The disease has an enormous burden on human health and causes decreased productivity and economic growth and increased poverty (2). The lack of an effective vaccine, the emergence of resistance to first-line drugs such as chloroquine and antifolates and recent reports of clinical cases of reduced susceptibility to artemisinin in Cambodia (reviewed in Ref. 3), combined with the small number of suitable new drugs against the malaria parasite demonstrate the need to identify potential new targets.
The malaria-infected erythrocyte is under constant attack from reactive oxygen and nitrogen species. These are produced exogenously by the host immune system in response to infection, or endogenously by the generation of redox-active by-products resulting from the high metabolic rates of the multiplying parasite and the degradation of large quantities of hemoglobin (4). The ability of Plasmodium falciparum to protect itself against oxidative damage is thus critical and the parasite possesses multiple biochemical pathways able to mediate antioxidant defense and redox regulation (5, 6). P. falciparum lacks catalase and a genuine glutathione peroxidase and therefore relies heavily on peroxiredoxins for the reduction of reactive oxygen species and reactive nitrogen species (7).
Peroxiredoxins (Prx) are a family of thiol-dependent peroxidases known to reduce and scavenge hydroperoxides and thus form an important part of the cellular antioxidant machinery. Most peroxiredoxins contain at least one cysteine residue in their active site and because of its ability to react with the peroxide substrate it is referred to as the peroxidatic cysteine. This residue is the defining feature of this class of enzymes also referred to as AhpC-TSA5 family (8) The reaction results in the formation of a sulfenic acid intermediate that is usually reduced by the resolving cysteine present in the 2-Cys peroxiredoxins. The inter- or intramolecular disulfide bond formed can be reduced by thioredoxin, glutaredoxin or glutathione depending on the substrate specificity of the respective peroxiredoxin. For instance, mammalian 2-Cys peroxiredoxin 1 can only react with thioredoxin and does not accept glutaredoxin or thioredoxin as substrates (9, 10).
Apart from their role as antioxidants, the proteins are also known to function as redox sensors in response to oxidative stress. They have also been implicated in cellular processes such as apoptosis, cell proliferation and differentiation (11). This functional diversity can be attributed to their structural flexibility, which leads to the association of the dimers to high Mr oligomeric proteins or vice versa, the susceptibility of one of their active site cysteines to hyperoxidation by their peroxide substrates and their high abundance in the cell (12). In addition, it was shown that the phosphorylation of peroxiredoxins affects their subcellular distribution and enzymatic activity (13).
Usually organisms contain more than one peroxiredoxin, which are found in different subcellular locations such as cytosol, mitochondria, peroxisomes, and nucleus (8). This is also the case for Plasmodium. To date, five malarial peroxiredoxins have been characterized: cytosolic PfTrx-Px1 and mitochondrial PfTrx-Px2 (14–16), cytosolic PfTrx-Px3 (17, 18), PfAOP, potentially localizing to the apicoplast (19), and finally a glutathione-dependent-like peroxidase with a preference for thioredoxin as reducing substrate (7). P. falciparum also possesses several proteins belonging to the thioredoxin superfamily, which provide reducing equivalents to the peroxiredoxins (reviewed in Ref. 6).
In addition to the five characterized peroxiredoxins, the malaria parasite possesses an additional protein with a conserved AhpC-TSA domain called MCP1 (merozoite capping protein-1, PF10_0268) (20, 21). MCP1 was described as a cytosolic protein localizing to the invasion tight junction, a zone of tight apposition between the merozoite and the red blood cell membrane formed during the invasion process (20). However, it is not known whether the AhpC-TSA domain of MCP1 is functional (20, 21). Additionally, MCP1 has two other defined domains with a negatively charged middle region enriched in glutamate and a C-terminal positively charged section enriched in lysine. It was speculated from the solubility properties of MCP1 and the C-terminal lysine-rich domain that this region may be required for binding to the cytoskeleton within the invading merozoite (21).
In this work, we show that MCP1 is a peroxiredoxin with unusual biochemical characteristics. As this enzyme exclusively localizes to the nucleus (in contrast to previous reports) we renamed it PfnPrx (P. falciparum nuclear peroxiredoxin). In addition, we demonstrate that PfnPrx is associated with the chromatin of the parasite in a genome-wide manner, suggesting a potentially essential role in the protection of nuclear DNA against oxidative stress.
EXPERIMENTAL PROCEDURES
Parasite Cultures
P. falciparum asexual stage parasites were maintained in human erythrocytes (blood group O+) at a hematocrit of 4% with 0.5% (w/v) AlbumaxTM (Invitrogen) (22). P. falciparum 3D7 parasites were originally obtained from David Walliker at Edinburgh University. Cultures were synchronized as described previously (23).
Antisera
Rabbit and mouse antibodies were generated against PfnPrx1–100, containing the conserved oxidoreductase domain, from a GST fusion protein expressed from a plasmid construct using the following primers: PfnPrxF (5′-CGCGGATCCATGGCTCAATTAGCAGAAAATAC-3′) and PfnPrxR (5′-CCGCTCGAGTAAGGAAGTTCTAGCTTTTC-3′. PCR products were digested with BamH1/Xho1 (underlined letters in primers), purified and cloned into the plasmid pGEX4T-1 (Amersham Biosciences). For immunoblots, saponin-lysed parasite pellets from highly synchronous 3D7 parasites were separated in sample buffer on 4–12% (w/v) SDS-NuPAGE gels (Invitrogen) under reducing conditions and transferred to nitrocellulose membranes (Schleicher & Schuell). Affinity purified PfnPrx rabbit anti-serum and anti-PfnPrx mouse monoclonal were diluted in 0.1% (v/v) Tween 20-phosphate-buffered saline with 1% (w/v) skim milk. Appropriate secondary antibodies were used, and immunoblots were developed by ECL (Amersham Biosciences). For the time course of expression analysis, proteins extracted from an equal number of cells were used for each time point.
Fluorescence Imaging
Fluorescence images of parasites were captured using a Carl Zeiss Axioskop microscope with a PCO Sensicam and Axiovision software (version 2). For immunofluorescence assays of free and/or invading merozoites, highly synchronous schizont stage 3D7 parasites in the process of rupture/reinvasion were smeared and fixed in 100% methanol at −20 °C. After blocking in 3% (w/v) bovine serum albumin (Sigma), the cells were incubated for 1 h with the appropriate antisera: rabbit anti-PfRON4 (1/200) (24, 25), mouse monoclonal anti-PfnPrx (1/200). Bound antibodies were then visualized with Alexa Fluor 488/594 anti-rabbit IgG or anti-mouse IgG (Molecular Probes) diluted 1:1000. Parasites were mounted in Vectashield (Vecta Laboratories) containing DAPI (Roche Molecular Biochemicals).
Immunoelectron Microscopy
Parasites for electron microscopy immunolabeling were fixed and prepared as described previously (26). The primary antibody used was the rabbit anti-PfnPrx. Samples were washed and incubated with secondary antibodies conjugated to 15-nm colloidal gold (BB International). Samples were then post-stained with 2% aqueous uranyl acetate then 5% triple lead and observed at 120 kV on a Philips CM120 BioTWIN Transmission Electron Microscope.
Vector Construction, Transfection, and Southern Blotting
To create the plasmid used for the integration of GFP at the 3′ end of the nPrx gene by single crossover, a PCR fragment containing exon 3 of nPrx without a stop codon was cloned into the BglII-AvrII sites of the pARL1-GFP vector (27, 28). Parasites were transfected, and integrants were selected as described previously (27). Integration was monitored by Southern blots according to standard procedures.
Protein Purification and Enzymatic Assays
A PCR fragment containing nucleotides 1 to 492 of PfnPrx was amplified from P. falciparum cDNA and cloned in the BamH1-Xho1 site of the pET-45b expression vector (Novagen). The latter directs expression of the protein with an N-terminal His6 tag to facilitate purification of the recombinant protein. Generation of the nPrxC56S mutant was performed by site-directed mutagenesis with the QuikChange II XL kit from Agilent Technologies. Induction and purification of His6-nPrx1–164 were performed according to standard procedures. P. falciparum TrxR and PfTrx1 were generated as described (29, 30). P. falciparum glutaredoxin 1 (PfGrx1, PFC0271c) was amplified from 3D7 genomic DNA and cloned in the BamH1-Xho1 site of the pET-45b expression vector (Novagen). Induction of the protein was performed at 27 °C overnight.
The overall antioxidant activity of PfnPrx was estimated by analyzing its ability to protect Escherichia coli glutamine synthetase (Sigma) from inactivation by a thiol-metal catalyzed oxidation system (DTT/Fe3+/O2) (31). The P. falciparum nPrx kinetic parameters were determined (30) using a spectrophotometric assay to determine the initial rates of the peroxidase reaction as described previously for Toxoplasma gondii Trx-Px2 (30, 32). For the assays using the thioredoxin system, the reaction mix contained 100 mm HEPES, pH 7.6, 1 mm EDTA, 0.25 units of P. falciparum thioredoxin reductase, 2 μm PfnPrx, 200 μm NADPH, varying concentrations of P. falciparum cytosolic thioredoxin PfTrx1 (20–100 μm) at a constant concentration of 20 μm hydrogen peroxide.
For assays using the GSH system with P. falciparum Grx1, the reaction mix contained 100 mm HEPES, pH 7.6, 1 mm EDTA, 0.5 units glutathione reductase, 1 mm GSH, 2 μm PfnPrx, 200 μm NADPH, varying concentrations of P. falciparum cytosolic glutaredoxin PfGrx1 (1–30 μm) at a constant concentration of 20 μm hydrogen peroxide or varying concentrations of hydrogen peroxide (1–50 μm) and cumene hydroperoxide (1–100 μm) at a constant concentration of 10 μm PfGrx1. No NADPH oxidation was observed without addition of PfGrx1 indicating that PfnPrx cannot be reduced by GSH itself. The decrease in absorbance at 340 nm due to NADPH oxidation was determined over 30 s with time points taken every 10 ms. Assays were performed at 25 °C. The linear rates were determined and fitted to the Michaelis-Menten equation using Graphit (version 5.0; Erithracus).
Chromatin Immunoprecipitation and Deep Sequencing
Chromatin immunoprecipitation of nPrx-GFP from formaldehyde cross-linked and sonicated chromatin was performed as described (33) using an anti-GFP antibody (AbCam 290). Immunoprecipitated as well as input chromatin was decross-linked at 45 °C in the presence of 0.5 m NaCl and tested by quantitative PCR (for primer list, see supplemental Table 1) or used for Illumina sequencing library preparation according to the linear amplification protocol (34). Sequencing libraries were loaded on the Illumina Genome Analyzer IIx and sequenced for 36 cycles from one side of the fragments. The quality filtered 35-bp sequence reads were mapped against the P. falciparum genome assembly (PlasmoDB version 6.1) using the standard Illumina pipeline. Coverage plots were generated using uniquely mapped sequence reads by counting the number of overlapping tags in 100-bp windows and visualized in SignalMap (NimbleGen). The ratio track was obtained by dividing the ChIP-seq tag counts with the input tag counts and displayed on a log2 scale. For generation of average gene profile the coding body of all “normal size” genes (1–10 Kb) were divided into 20 equal size windows and five 150-bp windows immediately up- or downstream represented flanking sequences. The ratios of the tag counts in the ChIP versus input data set has been computed in each individual windows and averaged in the corresponding window of all genes. For scatter plot analysis, the ratios of ChIP and input tag counts have been calculated in the coding body of each gene have been plotted against the RNAseq tag density (tag/1000 bp transcript) of an independent 3D7 schizont population (34).
RESULTS
Identification of PfnPrx (Earlier MCP1) as a Nuclear Peroxiredoxin
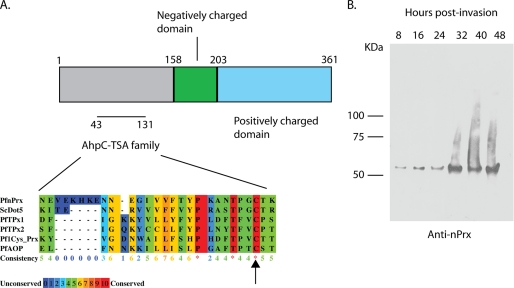
To investigate the role of PfnPrx (MCP1) in the merozoite invasion process, we generated an antibody against the conserved oxidoreductase domain (Fig. 1A) (20). Western blots on schizont stage protein extracts showed a single specific band of ∼55 kDa, which was slightly higher than the predicted 45 kDa (Fig. 1B, 48 h lane) and is in agreement with previous results (20). All of the other P. falciparum peroxiredoxins having sizes between 22 and 28 kDa, we can be confident that our antibody is not cross-reacting with any of them. We next performed a time course analysis for expression of PfnPrx (MCP1) using parasite protein extracts taken at 8-h intervals throughout the erythrocytic asexual cycle. The PfnPrx (MCP1) protein was present as a single 55 kDa band in all samples analyzed with the highest expression between 32 to 48 h (Fig. 1B) which corresponds to the schizont stage where the parasite undergoes nuclear division and also when proteins involved in merozoite invasion are expressed. This pattern is also in agreement with the RNA expression analyses published previously (35).
FIGURE 1.
PfnPrx is expressed throughout the erythrocytic stage. A, domain organization of PfnPrx PF10_0268. The alignment shows the conserved AhpC-TSA family domain with the peroxidatic cysteine. Comparison was realized with Praline software. The GenbankTM accession numbers of the aligned sequences are as follows: PfnPrx, AAC46600; Saccharomyces cerevisiae DOT5, P40553; PfTPx1, XP_001348542; PfTPx2, XP_001350554; Pf1-Cys_Prx, AAG14353; and PfAOP, AY306209. The asterisks represent conserved residues, and the arrow highlights the peroxidatic cysteine. B, time course of PfnPrx expression during the blood stage using an affinity-purified rabbit anti-serum raised against the first 100 residues of PfnPrx.
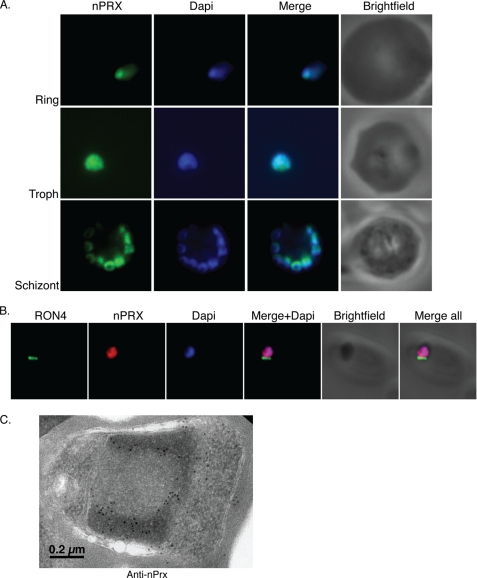
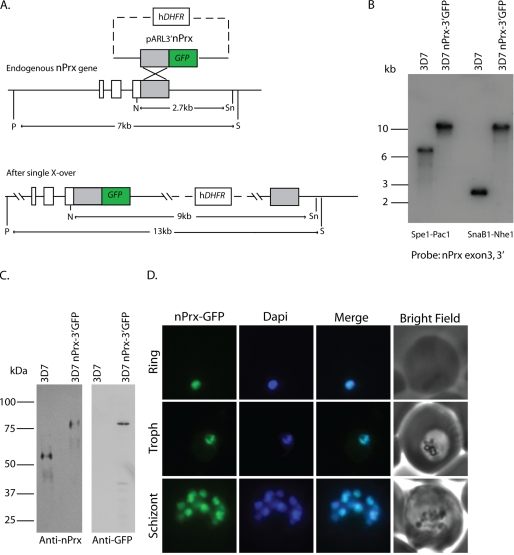
To determine the subcellular localization of the PfnPrx (MCP1) protein, we performed immunofluorescence assays on parasites from different stages of the erythrocytic cycle. The anti-nPrx (MCP1) monoclonal antibody overlapped almost completely with the DAPI-stained parasite DNA in rings, trophozoites, and schizonts, suggesting that PfnPrx was located in the nucleus (Fig. 2A), a subcellular localization different to that described previously (20). Analysis of the protein sequence with the PredictProtein software (36) also revealed a potential nuclear localization signal in the C terminus of PfnPrx (KKPAKKVVKKK). To analyze the subcellular localization of PfnPrx (MCP1) in more detail, we performed immunofluorescence experiments on merozoites during erythrocyte invasion in which we observed that PfnPrx also localized to the nucleus of the parasite. No anti-PfnPrx (MCP1) signal was found at the tight junction, visualized by staining with an antibody against the RON4 protein, a well characterized marker of this transient structure (25). To determine whether this localization was due to our antibody recognizing a protein other than PfnPrx (MCP1), we engineered a parasite line in which the green fluorescent protein (GFP) gene was integrated by single crossover at the 3′ end of the PfnPrx (MCP1) gene leading to the production of a fluorescent PfnPrx (MCP1) (Fig. 3, A and B). Western blot analysis showed that PfnPrx (MCP1) was detected as an 80-kDa protein with both the PfnPrx (MCP1) mouse monoclonal antibody or an anti-GFP monoclonal antibody confirming expression of the chimeric protein and providing additional evidence for the specificity of our mouse monoclonal anti-nPrx antibody (Fig. 3C). To determine the localization of the PfnPrx-GFP fusion, the 3D7 PfnPrx-GFP line was analyzed by epifluorescence microscopy. As shown in Fig. 3D, the PfnPrx-GFP fluorescence overlaps with the DAPI-stained parasite DNA in rings, trophozoites, and schizonts confirming that nPrx (MCP1) is found in the nucleus of the parasite throughout the blood stage cycle. To determine more precisely the localization of PfnPrx in the nucleus, we performed immunoelectron microscopy on P. falciparum schizont stage sections. PfnPrx localizes predominantly to the more electron dense regions at the nuclear periphery, and this is believed to represent heterochromatin (Fig. 2C) (37). Consequently, because of the nuclear localization and functional data presented below, we renamed this protein PfnPrx (P. falciparum nuclear peroxiredoxin).
FIGURE 2.
PfnPrx is a nuclear protein. A, immunofluorescence using the mouse monoclonal anti-PfnPrx antibody demonstrates localization of PfnPrx in the nucleus throughout the erythrocytic stage. Troph, trophozoite. B, PfnPrx does not localize to the tight junction of merozoites in the process of invasion. Immunofluorescence with mouse monoclonal anti-nPrx and rabbit anti-RON4 antibody shows that PfnPrx is restricted to the nucleus in parasites invading a red blood cell. C, immunoelectron microscopy using the rabbit anti-PfnPrx antibody on schizont stage parasites reveals that PfnPrx is in the electron dense nuclear periphery. The image shows a single merozoite located within a schizont.
FIGURE 3.
Endogenously tagging nPrx confirms its status as a nuclear protein. A, schematic of the strategy employed to generate the 3D7 nPrx-3′ GFP line. hDHFR, human dihydrofolate reductase. B, Southern blots confirming integration of the targeting construct at the endogenous nPrx locus. C, Western blots using the mouse monoclonal anti-nPrx or an anti-GFP antibody confirms that the 3D7nPrx-3′ GFP parasite line expresses the nPrx-GFP chimera. D, epifluorescence microscopy on the 3D7nPrx-3′ GFP line demonstrates that the tagged nPrx localizes to the nucleus in all blood stages. X-over; crossover.
PfnPrx Is a Peroxiredoxin with Unusual Characteristics
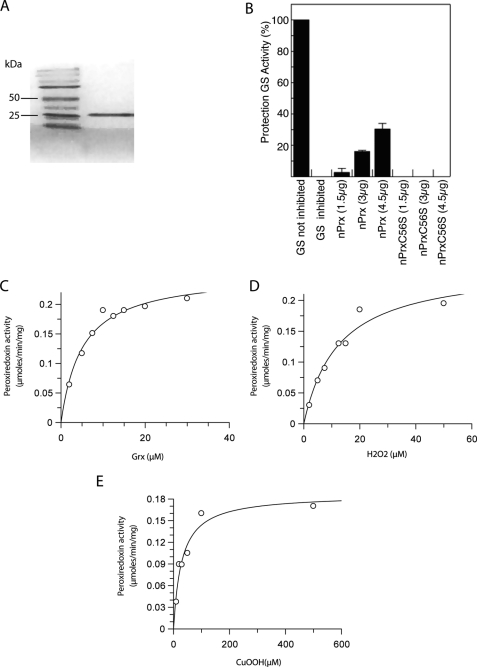
PfnPrx possesses an N-terminal conserved AhpC-TSA domain. To investigate its functionality, the N-terminal portion of the protein (amino acids 1–164) was recombinantly expressed and purified (Fig. 4A), and its potential peroxidase activity was assessed using various enzymatic assay systems. Initially, we determined whether the protein protects glutamine synthetase from inactivation by a thiol-metal catalyzed oxidation system (DTT/Fe3+/O2) (31). 4.5 μg of the recombinant peroxiredoxin-like domain of PfnPrx rescues glutamine synthetase activity by 50% (Fig. 4B), confirming that the AhpC-TSA domain was catalytically active, therefore suggesting that PfnPrx has antioxidant capacity. The specificity of this catalytic activity was further investigated by exchanging the predicted active site cysteine residue at position 56 by serine, which totally abolished the glutamine synthetase protection activity of the recombinant protein (Figs. 1A and 4B). The fact that the recombinant AhpC-TSA domain of PfnPrx showed the ability to protect glutamine synthetase activity is presumably attributable to its capacity to reduce hydrogen peroxide. This suggests that this part of the protein acts as a peroxidase that might potentially protects nuclear components such as DNA from oxidative insults. To identify its natural reducing substrate, the catalytic activity of PfnPrx was analyzed in the presence of P. falciparum thioredoxin 1, glutathione, and glutaredoxin 1. PfnPrx had the ability to reduce hydrogen peroxide, and this activity increased with increasing concentrations of PfTrx1 without reaching saturation at concentrations as high as 100 μm (supplemental Fig. 1), suggesting that is most likely not the favored reductant of the enzyme.
FIGURE 4.
PfnPrx is a peroxiredoxin with unusual characteristics. A, purification of the N-terminal domain of PfnPrx. B, glutamine synthetase (GS) protection assays revealing that PfnPrx is an antioxidant protein and that this activity was dependent on its conserved peroxidatic cysteine. C–E, peroxidase assays showing that glutaredoxin (C) is a potent reductant of PfnPrx and that PfnPrx efficiently reduces hydrogen peroxide (D) and cumene hydroperoxide (E). 2 μm PfnPrx was used for all assays, 20 μm hydrogen peroxide was used for the variable Pf glutaredoxin 1 assay, and 10 μm PfGrx1 was used for the variable peroxide assays. Results are representative of at least three independent experiments.
In addition to the thioredoxin system, P. falciparum possesses a functional glutathione system, and we studied the possibility that it could be involved in the reduction of PfnPrx. No PfnPrx peroxidase activity on hydrogen peroxide was detected when using GSH as a reductant, whereas addition of glutaredoxin 1 (PfGrx1 (15)) to the assay mix supported the reduction of PfnPrx with saturation kinetics (Fig. 4C) and a reasonable reaction rate. At a concentration of 20 μm H2O2, the apparent Km for PfGrx1 was determined to be ∼4 μm, showing that it was an excellent reductant for PfnPrx. When looking at the affinity of PfnPrx for H2O2, an apparent Km of 14 μm was calculated when 10 μm of PfGrx1 was used (Fig. 4D), demonstrating that H2O2 was a good substrate for PfnPrx and corroborating the results obtained with the glutamine synthetase protection assay. In addition, PfnPrx was able to reduce cumene hydroperoxide with an apparent Km of ∼30 μm (Fig. 4E). The specific activity of the recombinant protein was in the range of 6 μmol/min/mg protein, which is low compared with the activity of other Plasmodium peroxiredoxins (16, 30). It cannot be excluded that regulatory or structural features important to define substrate specificity are missing from the recombinant PfnPrx given that only the AhpC-TSA domain of PfnPrx rather than the full-length protein was analyzed.
As PfnPrx is localized to the nucleus, it would be expected that its favored physiological reductant should also localize to this organelle or at least have the ability to translocate to it from the cytoplasm upon oxidative stress. To investigate the cellular localization of PfGrx1, we generated parasites expressing a PfGrx1-GFP fusion. PfGrx1-GFP mostly localizes to the cytoplasm of the parasite though some of protein overlaps with the DAPI-stained nuclear DNA, suggesting that it has the ability to translocate between the two compartments (supplemental Fig. 2). However, we cannot at this stage exclude that the nuclear staining is the result of a mislocalization of the fusion protein due to its overexpression from an episome.
Having demonstrated that PfnPrx had the ability to detoxify peroxides in vitro, we hypothesized that it could potentially be involved in protecting the nucleus of the parasite from oxidative stress. However, it has not been possible to demonstrate a consistent increase in the level of PfnPrx transcript or protein when submitting the parasites to exogenous oxidative stress in vitro using a glucose/glucose oxidase system (data not shown).
PfnPrx Is Potentially Essential in Erythrocytic Stage
To gain further insight into the role of PfnPrx in the blood stage cycle of the malaria parasite, we attempted to generate a knock-out line in both P. falciparum and Plasmodium berghei, a rodent malaria parasite often used as a model for human malaria due to the ease with which knock-out strains can be generated. Despite several attempts, we were unable to generate a nPrx knock-out line in either of the Plasmodium species tested. This was in contrast with the ability to easily tag the PfnPrx with GFP, suggesting that the protein performs an essential/important function in the erythrocytic stage of the malaria parasite (data not shown).
ChIP-seq Analysis Reveals Genome-wide Binding of PfnPrx to Chromatin
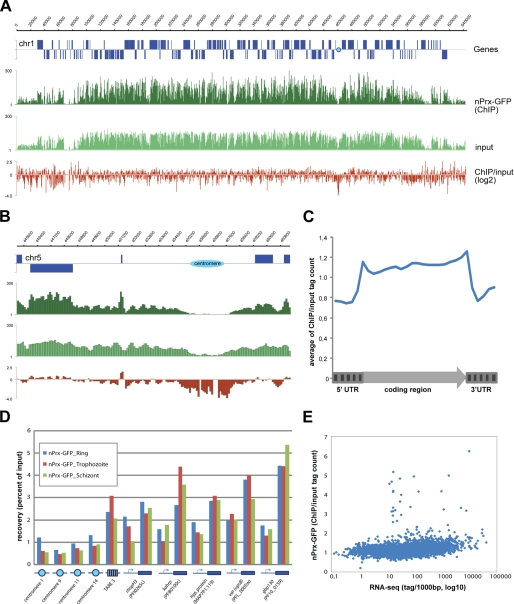
The co-localization of PfnPrx with the DAPI-stained DNA suggested that it was potentially associating with chromatin. To investigate this possibility, we performed a genome-wide ChIP-seq analysis on schizont stage 3D7nPrx-GFP parasites using an anti-GFP antibody. Intriguingly, visual inspection of the data showed nearly even distribution of PfnPrx across the P. falciparum genome (Fig. 5A). An obvious exception was the strong depletion of PfnPrx from the centromeric regions (Fig. 5B). In addition, PfnPrx was found to be slightly enriched in the coding body of genes (Fig. 5B), which was also apparent on the average gene profile (Fig. 5C). Enrichment of PfnPrx in coding regions and its depletion from centromeres were confirmed by ChIP-quantitative PCR and were found to be consistent across all stages of intraerythrocytic development (Fig. 5D). Importantly, recoveries with the GFP antibody for chromatin isolated from nontagged 3D7 parasites were very low for all sites tested, proving the specificity of our assay (data not shown). Finally, we determined whether enrichment of PfnPrx showed a correlation with transcriptional activity. As demonstrated by scatter plot analysis (Fig. 5E), PfnPrx occupancy in coding regions showed limited variation across genes and no or minimal correlation with steady state mRNA levels. Notably, outlier genes with high PfnPrx occupancy tended to be tRNA or rRNA genes. In conclusion, the ChIPseq data demonstrates the intimate association of PfnPrx with the malaria genome.
FIGURE 5.
PfnPrx shows genome wide chromatin association. A and B, broad distribution of PfnPrx (A), its enrichment in coding and depletion at centromeric region (B) demonstrated by coverage plots and ratio track of PfnPrx-GFP ChIP-seq data obtained from schizont stage parasites. C, average gene profile of PfnPrx occupancy. D, quantitative PCR data confirming enrichment of PfnPrx in coding region and its depletion at centromeres at three different stages of intraerythrocytic development. E, scatter plot analysis of correlation between PfnPrx enrichment in coding body of genes and steady state mRNA levels (RNA-seq data of schizont stage parasites from Bartfai et al. (33)).
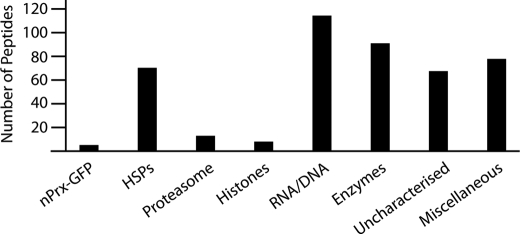
To determine whether the association of PfnPrx with the chromatin was mediated through interactions with other proteins, we performed immunoprecipitation on the 3D7nPrx-GFP line using an anti-GFP antibody. Mass spectrometry analysis revealed, in addition to nPrx-GFP, a high number of different proteins such as core histones, heat shock proteins, proteasome components, and a few transcription factors, but none with very high peptide coverage (Fig. 6). This high variety of proteins was somewhat expected based on the scale of the broad association of nPrx with the genome and the absence of any clear specific interactor suggests that nPrx might interact directly with the chromatin, potentially through its highly positively charged C terminus as has been shown for proteins such as linker histone H1 (38).
FIGURE 6.
PfnPrx-GFP associates with a broad range of proteins. The immunoprecipitated proteins were grouped in categories and plotted according the total number of peptides identified. HSPs, heat shock proteins; RNA/DNA, proteins involved in RNA/DNA metabolism and/or binding. The list of proteins with accession numbers is available in the supplemental Table 1. No GFP peptides were recovered because a P. falciparum-specific database was used for peptide identification.
DISCUSSION
MCP1 was previously described as a protein localizing within the cytoplasm of the merozoite and associating with the moving junction as the parasite invades the host erythrocyte (20, 21). It was noted that this protein contained an oxidoreductase domain, although it had not been demonstrated to be functional. In contrast to the previous observation we show here that MCP1 is exclusively localized within the nucleus of P. falciparum. Moreover, we demonstrate that the protein contains a functional peroxiredoxin domain with unique characteristics. Consequently, we have renamed the protein PfnPrx. The functional properties of PfnPrx as a peroxiredoxin, its subcellular localization within the nucleus, its coverage of a large proportion of P. falciparum chromosomes, and our inability to inactivate its gene in two different species of Plasmodia suggest that it plays an important role in this compartment.
Peroxiredoxins are a family of anti-oxidants that protect the cell from metabolically produced reactive oxygen species. The P. falciparum PfnPrx prefers glutaredoxin over thioredoxin as a reducing substrate and shows saturation kinetics. The protein reduces hydrogen peroxide and cumene hydroperoxide equally well, suggesting that it does not discriminate between anorganic or organic hydroperoxides. Its specificity for glutaredoxin as a reducing substrate is unusual especially because most other known peroxiredoxins or peroxiredoxin-like proteins react with thioredoxin or glutathione. Peroxiredoxins with this substrate specificity have so far only been described in plants, although some of these can also be reduced by thioredoxins (9, 39, 40, and reviewed in Ref. 41)). Intriguingly, PfnPrx was associated strongly with the genome of the parasite, suggesting that the protein has a role in either protecting nuclear components such as DNA from oxidative damage or being involved in maintaining chromatin structure, DNA repair, or mechanisms that affect transcriptional activities. In fact, it has been shown that redox active thiol-containing proteins and peptides are critical for these processes because they can respond to oxidation or reduction processes that allows for the regulation of their activities (42). There are a number of reports that show interaction of human peroxiredoxin 1 with the nucleus. This binding capacity and the downstream effects on transcriptional regulation can be either dependent or independent of the peroxidase activity of the protein, and it has been suggested that apart from acting as an antioxidant the protein can act as a chaperone to stabilize other DNA-binding factors (43–45).
The broad distribution of PfnPrx across the P. falciparum genome suggests that it plays a general role across the chromosomes and it could possibly be associated with nucleosomes. Studies on humans, Drosophila, and worms have shown that nucleosome occupancy increases in coding regions and that this is independent of transcriptional status (46–48). In addition, our previous studies on core histone H3 demonstrated a slight enrichment in the coding body of genes similar to that observed for PfnPrx. Such a similar enrichment profile is consistent with a potential association of PfnPrx with nucleosomes (49). Due to its very highly positively charged C terminus, PfnPrx could potentially interact directly with the phosphate backbone of the DNA, without the need to link through an interacting partner. It is indeed well documented that the lysine-rich C-terminal tails of linker histone H1s bind directly to the linker DNA entering and exiting a nucleosome leading to the formation of higher order folding states of chromatin (50, 51). Although linker histones of multicellular eukaryotes display a tripartite structure made up of a conserved globular domain flanked by two less structured N- and C-terminal domains, their counterparts in several protists and bacteria only contain the lysine-rich C terminus (reviewed in Ref. 52). It is worth mentioning that although H1-like basic proteins have been found in dinoflagellates (53) and ciliates (54), none have been identified for any of the apicomplexans, also members of the alveolates family. It is tempting to speculate that the lysine-rich C terminus of PfnPrx could potentially play a similar role in chromatin structure; however, our attempts to recombinantly express the full-length PfnPrx or its C-terminal tail to investigate a potential association with nucleosomes in vitro have unfortunately been unsuccessful so far. In any case, if PfnPrx is involved in the protection of the genome against oxidative stress, the pairing of a peroxiredoxin domain with a highly basic domain suggests an ingenious means of making sure that the enzyme would always be in a position to quickly get rid of any oxidative insult as soon as it is produced, therefore limiting the chances of DNA damage.
Moreover, we were unable to disrupt the PfnPrx gene either in P. falciparum or P. berghei, suggesting that it plays an essential role for nuclear integrity or function possibly through protecting nuclear DNA and chromatin-binding, thiol-containing proteins from oxidative damage or because it is intimately involved in the regulation of gene expression. In conclusion, we have identified a dedicated nuclear peroxiredoxin with broad substrate specificity that associates with chromatin throughout the genome potentially demonstrating a specific adaptation of the malaria parasite to its hostile environment.
Supplementary Material
Acknowledgments
Human erythrocytes were kindly provided by the Red Cross Blood Bank (Melbourne, Australia). We thank Wieteke Hoeijmakers for critical reading of the manuscript. We thank Tanya De Koning-Ward and Rachel Lundie for the P. berghei KO assays.
This work was supported by the National Health and Medical Research Council of Australia (NHMRC).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
- AhpC
- alkyl hydroperoxide peroxidase C
- TSA
- thiol-specific antioxidant
- IFA
- immunofluorescence assay.
REFERENCES
- 1. Snow R. W., Guerra C. A., Noor A. M., Myint H. Y., Hay S. I. (2005) Nature 434, 214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sachs J., Malaney P. (2002) Nature 415, 680–685 [DOI] [PubMed] [Google Scholar]
- 3. White N. J. (2008) Science 320, 330–334 [DOI] [PubMed] [Google Scholar]
- 4. Becker K., Tilley L., Vennerstrom J. L., Roberts D., Rogerson S., Ginsburg H. (2004) Int. J. Parasitol. 34, 163–189 [DOI] [PubMed] [Google Scholar]
- 5. Hunt N. H., Stocker R. (1990) Blood Cells 16, 499–526 [PubMed] [Google Scholar]
- 6. Müller S. (2004) Mol. Microbiol. 53, 1291–1305 [DOI] [PubMed] [Google Scholar]
- 7. Sztajer H., Gamain B., Aumann K. D., Slomianny C., Becker K., Brigelius-Flohé R., Flohé L. (2001) J. Biol. Chem. 276, 7397–7403 [DOI] [PubMed] [Google Scholar]
- 8. Wood Z. A., Schröder E., Robin Harris J., Poole L. B. (2003) Trends Biochem. Sci. 28, 32–40 [DOI] [PubMed] [Google Scholar]
- 9. Rouhier N., Gelhaye E., Jacquot J. P. (2002) J. Biol. Chem. 277, 13609–13614 [DOI] [PubMed] [Google Scholar]
- 10. Wood Z. A., Poole L. B., Hantgan R. R., Karplus P. A. (2002) Biochemistry 41, 5493–5504 [DOI] [PubMed] [Google Scholar]
- 11. Butterfield L. H., Merino A., Golub S. H., Shau H. (1999) Antioxid. Redox. Signal 1, 385–402 [DOI] [PubMed] [Google Scholar]
- 12. Cox A. G., Winterbourn C. C., Hampton M. B. (2010) Methods Enzymol. 474, 51–66 [DOI] [PubMed] [Google Scholar]
- 13. Zykova T. A., Zhu F., Vakorina T. I., Zhang J., Higgins L., Urusova D. V., Bode A. M., Dong Z. (2010) J. Biol. Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krnajski Z., Gilberger T. W., Walter R. D., Müller S. (2001) Mol. Biochem. Parasitol. 112, 219–228 [DOI] [PubMed] [Google Scholar]
- 15. Rahlfs S., Becker K. (2001) Eur. J. Biochem. 268, 1404–1409 [DOI] [PubMed] [Google Scholar]
- 16. Boucher I. W., McMillan P. J., Gabrielsen M., Akerman S. E., Brannigan J. A., Schnick C., Brzozowski A. M., Wilkinson A. J., Müller S. (2006) Mol. Microbiol. 61, 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kawazu S., Tsuji N., Hatabu T., Kawai S., Matsumoto Y., Kano S. (2000) Mol. Biochem. Parasitol. 109, 165–169 [DOI] [PubMed] [Google Scholar]
- 18. Krnajski Z., Walter R. D., Müller S. (2001) Mol. Biochem. Parasitol. 113, 303–308 [DOI] [PubMed] [Google Scholar]
- 19. Sarma G. N., Nickel C., Rahlfs S., Fischer M., Becker K., Karplus P. A. (2005) J. Mol. Biol. 346, 1021–1034 [DOI] [PubMed] [Google Scholar]
- 20. Klotz F. W., Hadley T. J., Aikawa M., Leech J., Howard R. J., Miller L. H. (1989) Mol. Biochem. Parasitol. 36, 177–185 [DOI] [PubMed] [Google Scholar]
- 21. Hudson-Taylor D. E., Dolan S. A., Klotz F. W., Fujioka H., Aikawa M., Koonin E. V., Miller L. H. (1995) Mol. Microbiol. 15, 463–471 [DOI] [PubMed] [Google Scholar]
- 22. Trager W., Jensen J. B. (1976) Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 23. Lambros C., Vanderberg J. P. (1979) J. Parasitol. 65, 418–420 [PubMed] [Google Scholar]
- 24. Richard D., Kats L. M., Langer C., Black C. G., Mitri K., Boddey J. A., Cowman A. F., Coppel R. L. (2009) PLoS Pathog. 5, e1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richard D., MacRaild C. A., Riglar D. T., Chan J. A., Foley M., Baum J., Ralph S. A., Norton R. S., Cowman A. F. (2010) J. Biol. Chem. 285, 14815–14822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rug M., Wickham M. E., Foley M., Cowman A. F., Tilley L. (2004) Infect. Immun. 72, 6095–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilberger T. W., Thompson J. K., Reed M. B., Good R. T., Cowman A. F. (2003) J. Cell Biol. 162, 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Przyborski J. M., Miller S. K., Pfahler J. M., Henrich P. P., Rohrbach P., Crabb B. S., Lanzer M. (2005) EMBO J. 24, 2306–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gilberger T. W., Walter R. D., Müller S. (1997) J. Biol. Chem. 272, 29584–29589 [DOI] [PubMed] [Google Scholar]
- 30. Akerman S. E., Müller S. (2003) Mol. Biochem. Parasitol. 130, 75–81 [DOI] [PubMed] [Google Scholar]
- 31. Kim K., Kim I. H., Lee K. Y., Rhee S. G., Stadtman E. R. (1988) J. Biol. Chem. 263, 4704–4711 [PubMed] [Google Scholar]
- 32. Akerman S. E., Müller S. (2005) J. Biol. Chem. 280, 564–570 [DOI] [PubMed] [Google Scholar]
- 33. Flueck C., Bartfai R., Volz J., Niederwieser I., Salcedo-Amaya A. M., Alako B. T., Ehlgen F., Ralph S. A., Cowman A. F., Bozdech Z., Stunnenberg H. G., Voss T. S. (2009) PLoS Pathog. 5, e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bártfai R., Hoeijmakers W. A., Salcedo-Amaya A. M., Smits A. H., Janssen-Megens E., Kaan A., Treeck M., Gilberger T. W., Françoijs K. J., Stunnenberg H. G. (2010) PLoS Pathog. 6, e1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bozdech Z., Llinás M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. (2003) PLoS Biol 1, E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rost B., Yachdav G., Liu J. (2004) Nucleic Acids Res. 32, W321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ralph S. A., Scheidig-Benatar C., Scherf A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5414–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Subirana J. A. (1990) Biopolymers 29, 1351–1357 [DOI] [PubMed] [Google Scholar]
- 39. Rouhier N., Gelhaye E., Sautiere P. E., Brun A., Laurent P., Tagu D., Gerard J., de Faÿ E., Meyer Y., Jacquot J. P. (2001) Plant Physiol. 127, 1299–1309 [PMC free article] [PubMed] [Google Scholar]
- 40. Bréhélin C., Meyer E. H., de Souris J. P., Bonnard G., Meyer Y. (2003) Plant Physiol. 132, 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rouhier N., Koh C. S., Gelhaye E., Corbier C., Favier F., Didierjean C., Jacquot J. P. (2008) Biochim. Biophys. Acta 1780, 1249–1260 [DOI] [PubMed] [Google Scholar]
- 42. Go Y. M., Jones D. P. (2010) Antioxid. Redox Signal 13, 489–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang X., He S., Sun J. M., Delcuve G. P., Davie J. R. (2010) Mol. Biol. Cell 21, 2987–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Neumann C. A., Fang Q. (2007) Curr. Opin. Pharmacol. 7, 375–380 [DOI] [PubMed] [Google Scholar]
- 45. Park S. Y., Yu X., Ip C., Mohler J. L., Bogner P. N., Park Y. M. (2007) Cancer Res. 67, 9294–9303 [DOI] [PubMed] [Google Scholar]
- 46. Tilgner H., Nikolaou C., Althammer S., Sammeth M., Beato M., Valcárcel J., Guigó R. (2009) Nat. Struct. Mol. Biol. 16, 996–1001 [DOI] [PubMed] [Google Scholar]
- 47. Schwartz S., Meshorer E., Ast G. (2009) Nat. Struct. Mol. Biol. 16, 990–995 [DOI] [PubMed] [Google Scholar]
- 48. Andersson R., Enroth S., Rada-Iglesias A., Wadelius C., Komorowski J. (2009) Genome Res. 19, 1732–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Salcedo-Amaya A. M., van Driel M. A., Alako B. T., Trelle M. B., van den Elzen A. M., Cohen A. M., Janssen-Megens E. M., van de Vegte-Bolmer M., Selzer R. R., Iniguez A. L., Green R. D., Sauerwein R. W., Jensen O. N., Stunnenberg H. G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9655–9660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Syed S. H., Goutte-Gattat D., Becker N., Meyer S., Shukla M. S., Hayes J. J., Everaers R., Angelov D., Bednar J., Dimitrov S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 9620–9625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hendzel M. J., Lever M. A., Crawford E., Th'ng J. P. (2004) J. Biol. Chem. 279, 20028–20034 [DOI] [PubMed] [Google Scholar]
- 52. Kasinsky H. E., Lewis J. D., Dacks J. B., Ausió J. (2001) FASEB J. 15, 34–42 [DOI] [PubMed] [Google Scholar]
- 53. Vernet G., Sala-Rovira M., Maeder M., Jacques F., Herzog M. (1990) Biochim. Biophys. Acta 1048, 281–289 [DOI] [PubMed] [Google Scholar]
- 54. Hayashi T., Hayashi H., Iwai K. (1987) J Biochem. 102, 369–376 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.