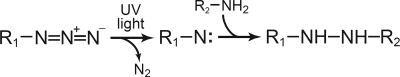Abstract
The mechanism of ATP modulation of E2P dephosphorylation of sarcoplasmic reticulum Ca2+-ATPase wild type and mutant forms was examined in nucleotide binding studies of states analogous to the various intermediates of the dephosphorylation reaction, obtained by binding of metal fluorides, vanadate, or thapsigargin. Wild type Ca2+-ATPase displays an ATP affinity of 4 μm for the E2P ground state analog, 1 μm for the E2P transition state and product state analogs, and 11 μm for the E2 dephosphoenzyme. Hence, ATP binding stabilizes the transition and product states relative to the ground state, thereby explaining the accelerating effect of ATP on dephosphorylation. Replacement of Phe487 (N-domain) with serine, Arg560 (N-domain) with leucine, or Arg174 (A-domain) with alanine or glutamate reduces ATP affinity in all E2/E2P intermediate states. Alanine substitution of Ile188 (A-domain) increases the ATP affinity, although ATP acceleration of dephosphorylation is disrupted, thus indicating that the critical role of Ile188 in ATP modulation is mechanistically based rather than being associated with the binding of nucleotide. Mutants with alanine replacement of Lys205 (A-domain) or Glu439 (N-domain) exhibit an anomalous inhibition by ATP of E2P dephosphorylation, due to ATP binding increasing the stability of the E2P ground state relative to the transition state. The ATP affinity of Ca2E2P, stabilized by inserting four glycines in the A-M1 linker, is similar to that of the E2P ground state, but the Ca2+-free E1 state of this mutant exhibits 3 orders of magnitude reduction of ATP affinity.
Keywords: ATP, ATPases, Calcium ATPase, Enzyme mechanisms, Membrane Proteins, Mutant, Sarcoplasmic Reticulum, Photolabeling, TNP-8N3-ATP, TNP-ATP
Introduction
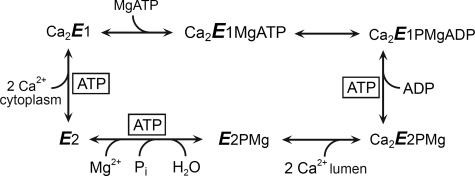
The sarco(endo)plasmic reticulum Ca2+-ATPase (Ca2+-ATPase)3 is a membrane-bound P-type ATPase that translocates Ca2+ from the cytosol to the endoplasmic reticulum, thereby allowing rapid oscillations of Ca2+ during cellular activation events. Insight into the structural organization of the Ca2+-ATPase has come from the elucidation of several crystal structures at atomic resolution, representing the pump in various intermediate states (reviewed in Refs. 1 and 2). The membrane-buried region of the Ca2+-ATPase is made up of 10 membrane spanning helices and is connected to a large cytoplasmic headpiece, which is further separated into three distinct domains, denoted A (“actuator”), P (“phosphorylation”), and N (“nucleotide binding”). Ca2+ transport is achieved by means of a reaction cycle (Scheme 1) involving the formation and decay of a phosphorylated intermediate and extensive protein conformational changes between four major states, E1, E1P, E2P, and E2. The catalytic function in E1 (autokinase activity) and E2P (autophosphatase activity) as well as the movement of Ca2+ ions across the membrane can be understood on the basis of the sequential gathering and displacement of certain conserved amino acid motifs of the N- and A-domains relative to the catalytic site in the P-domain and the coupling of these events to rearrangements of the transmembrane helices containing the Ca2+ sites. In the E1 and E1P states, the highly conserved 181TGES loop of the A-domain is distant from the catalytic center containing nucleotide binding residues and the phosphorylated Asp351 of the P-domain, which in this condition is able to react with ATP/ADP. However, during the Ca2E1P → E2P transition the A domain rotates ∼90° around an axis nearly perpendicular to the membrane, thereby moving the 181TGES loop into close contact with the catalytic site, such that Glu183 can catalyze dephosphorylation of E2P by hydrolysis (3–5). During the dephosphorylation, Glu183 likely coordinates the water molecule attacking the aspartyl phosphoryl bond and withdraws a hydrogen.
SCHEME 1.
Ca2+-ATPase reaction cycle. Major conformational changes and substrate binding and dissociation steps are shown. Boxed ATP indicates steps for which the rate is enhanced by additional binding of ATP or MgATP that is not hydrolyzed (“modulatory ATP”).
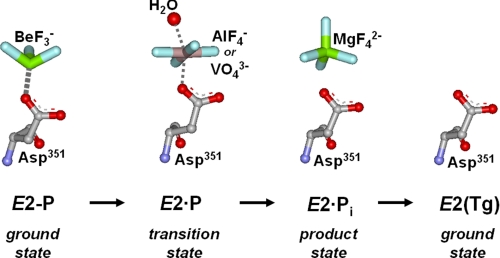
ATP in addition to being the substrate in the phosphorylation of the Ca2E1 state also functions in a non-phosphorylating mode (boxed ATP in Scheme 1), enhancing the rates of the steps involved in phosphoenzyme turnover (Ca2E1P → E2P and E2P → E2) as well as the E2 → Ca2E1 transition of the dephosphoenzyme (6–17). The mechanisms underlying these modulatory effects of ATP remain largely unresolved. A debated issue is whether the modulatory ATP molecule binds at the same site as the phosphorylating ATP or at a distinct, “allosteric” site (14, 18–23). During dephosphorylation the 181TGES loop of the A-domain occupies the position in close contact with the P-domain taken up by part of the ATP and ADP in Ca2E1 and Ca2E1P, however, ATP may still bind to residues of the N- and A-domains under these conditions. Recently, mutagenesis studies have pinpointed several amino acid residues in the N- and A-domains as critical for the modulatory effects of ATP and have provided evidence of an overlap between the catalytic and modulatory ATP binding sites (16, 17, 24), i.e. favoring the existence of a single site that exhibits a high degree of plasticity and flexibility, being reconfigured from slightly different positions of the P-, N-, and A-domains in the conformational states occurring during the transport cycle. The conformational transitions of the cycle likely change the affinity of the site for ATP, thus explaining the enhancing effects of ATP on the kinetics. Hence, the forward flow of a reversible reaction will be enhanced by ATP, if the product state possesses higher affinity for ATP than the ground state (23). Moreover, a relative stabilization of the transition state of a partial reaction of the cycle, leading to acceleration of this reaction, would be achieved in the presence of ATP, if ATP bound with higher affinity to the transition state than to the corresponding ground state. In the present study we have focused on the role of ATP as a modulator of E2P dephosphorylation. We present, for the first time, direct measurements of the ATP affinity of wild type and mutant Ca2+-ATPases stabilized in states analogous to the various intermediate forms occurring during the E2P dephosphorylation reaction sequence. This was achieved by use of the metal-fluoride compounds BeF, AlF, and MgF (Scheme 2) to mimic the phosphoryl group in the ground, transition, and product states of E2P dephosphorylation, respectively (4, 5, 25, 26), and vanadate, which like AlF is thought to mimic the bipyramidal transition state of the phosphoryl group (27, 28). The ATP affinity was also determined for a stable form of the E2 dephosphoenzyme obtained by binding of thapsigargin (29, 30) and for the Ca2E2P phosphoenzyme intermediate that still has Ca2+ bound. The latter was stabilized by inserting four glycines in the A-M1 linker (mutation “4Gi-46/47”) (31, 32). For these stable states the affinity constants for ATP could be determined by studying the competitive inhibition by ATP of TNP-8N3-ATP photolabeling of Lys492, an assay that has proven very powerful for determination of nucleotide affinity of the E1 state of expressed wild type and mutant Ca2+-ATPases (e.g. Refs. 33 and 34), but has not previously been applied to the phosphoenzyme or analog states of intermediates of E2P dephosphorylation. We demonstrate that, for the wild type Ca2+-ATPase, the affinity for modulatory ATP varies during the dephosphorylation sequence, being intermediate in the E2P ground state (with or without bound Ca2+), highest in the E2P transition and product states, and lowest in the E2 dephosphoenzyme. Our analysis of mutant Ca2+-ATPases provides insight into the varying contributions by individual amino acid residues to nucleotide binding during the dephosphorylation reaction sequence and allows distinction between roles in nucleotide binding and in mediating the response to binding that accelerates the dephosphorylation.
SCHEME 2.
Molecular details of the E2P dephosphorylation reaction sequence. Stable analogs of intermediate states of the E2P → E2 reaction sequence can be formed by incubation of the Ca2+-ATPase with BeF (E2P ground state), AlF (E2·P transition state), vanadate (E2·P transition state), MgF (E2·Pi product state), and Tg (E2 ground state) in the absence of Ca2+ (25, 28).
EXPERIMENTAL PROCEDURES
The cDNA encoding the mutant Ca2+-ATPases studied in the present work was the same as that applied in our previous studies (16, 17, 33–35). The cDNA was inserted into the expression vector pMT2 (36). To express wild type and mutant cDNA, COS-1 cells were transfected using the calcium phosphate precipitation method (37). Microsomal vesicles containing either expressed wild type or mutant Ca2+-ATPase were isolated by differential centrifugation (38). SR vesicles isolated from rabbit hind leg muscles (prepared as described in Refs. 39 and 40)) were a gift from Dr. Philippe Champeil (Saclay, France). The concentration of expressed Ca2+-ATPase was determined by an enzyme-linked immunosorbent assay (41) and by measurement of the maximum capacity for phosphorylation with ATP or Pi (“active site concentration” (42)). As previously described, the expression levels of the mutants were similar to that of the wild type (16, 17, 33, 34). The amount of endogenous endoplasmic reticulum Ca2+-ATPase present in the preparation is less than 1% that of the exogenous expressed enzyme, hence labeling corresponding to endogenous Ca2+-ATPase is negligible (cf. Fig. 1 of Ref. 33, compare “wild type” with “control”).
FIGURE 1.
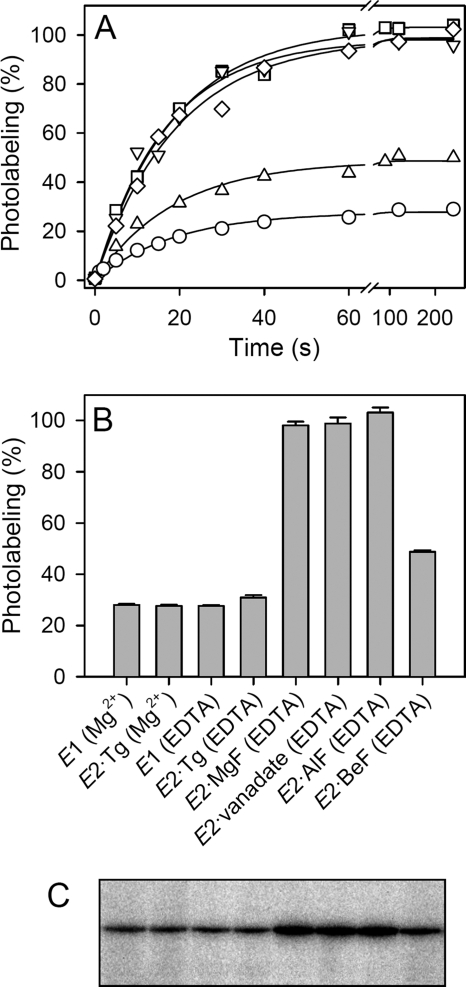
Time dependence of TNP-8N3-ATP photolabeling and photolabeling levels of Ca2+-ATPase in various conformational states. Microsomes containing expressed wild type Ca2+-ATPase were pre-equilibrated with or without metal fluoride or vanadate, and subjected to TNP-8N3-ATP photolabeling for various time intervals, as indicated on the abscissa in panel A, or for 60 s (panel B). The symbols in panel A are as follows: circles, E1 (Mg2+); triangles pointing downward, E2·MgF; diamonds, E2·vanadate; squares, E2·AlF; triangles pointing upward, E2·BeF. In each case, the concentration of TNP-8N3-ATP used was 3× the K0.5 for TNP-8N3-ATP (cf. Fig. 2). The labeling medium contained either MgCl2 (with EGTA) or EDTA as indicated in panel B (for panel A, E1 was studied with Mg2+ present). The labeling level (in %), relative to the average of that obtained with E2·MgF, E2·vanadate, and E2·AlF, is in each panel indicated at the ordinate. Panel C shows a representative gel with lanes corresponding to the columns in the bar chart.
Formation of the complexes of SR or expressed wild type or mutant Ca2+-ATPase in the E2 state with Tg, MgF, AlF, vanadate, and BeF prior to photolabeling was achieved by pre-equilibration of the enzyme for 30 min at 25 °C in 25 mm MOPS/tetramethyl ammonium hydroxide (pH 7.0), 80 mm KCl, 2 mm EGTA, and the concentrations of inhibitors and co-factors indicated as follows: Tg, 1 μm Tg; MgF, 5 mm MgCl2 and 5 mm NaF; vanadate, 0.1 mm orthovanadate and 5 mm MgCl2; AlF, 0.5 mm AlCl3, 2 mm NaF, and 0.2 mm MgCl2; BeF, 0.1 mm BeSO4, 2 mm NaF, and 0.2 mm MgCl2. The enzyme-inhibitor complexes were formed immediately prior to the initiation of the photolabeling experiments and kept on ice throughout (<1 h). The inhibited state of wild type and mutants at the inhibitor concentrations applied, as well as the stability of the enzyme-inhibitor complexes under photolabeling conditions, is validated under supplemental Table S1 and Figs. S1 and S2).
The synthesis of the [γ-32P]TNP-8N3-ATP photolabel, its application as a specific photolabel of the Ca2+-ATPase (Scheme 3), the competitive inhibition by ATP of [γ-32P]TNP-8N3-ATP photolabeling, and the quantification of 32P-labeled bands by electronic autoradiography following SDS-PAGE were carried out using the previously established procedures (20, 33). The medium used for photolabeling contained 25 mm EPPS/tetramethyl ammonium hydroxide (pH 8.5), 2 mm EDTA (to remove Ca2+ and any Mg2+ not tightly bound in the enzyme-inhibitor complex), 17.4% (v/v) glycerol, and [γ-32P]TNP-8N3-ATP without or with ATP at the concentrations indicated in the figures. For photolabeling in the presence of Mg2+, 2 mm EDTA was replaced by 1 mm MgCl2 and 0.5 mm EGTA. The concentration of [γ-32P]TNP-8N3-ATP used in competition experiments with ATP was 3× the K0.5, where K0.5 is the [γ-32P]TNP-8N3-ATP concentration giving half-maximum labeling. For photolabeling the enzyme was diluted 25-fold into ice-cold labeling medium immediately prior to irradiation. The concentration of Ca2+-ATPase in the final photolabeling mixture was typically ∼2 nm. Such low Ca2+-ATPase concentration was critical, because of the high TNP-8N3-ATP affinity of certain mutants (in some cases, K0.5 values as low as 5 nm were obtained, as demonstrated below). Irradiations were performed in a total volume of 75 μl in an ice-cold 500-μl quartz cuvette using the collimated light beam from an LSH102 Arc Light Source (LOT-Oriel Group Europe, Darmstadt, Germany) assembled with a 150 W ozone-free xenon arc lamp, a rear light reflector, an F/1.3 35-mm aperture quartz condenser, and a glass filter with 295-nm wavelength cut-off. The power supply was adjusted to 38 W and the irradiation time was 35 s, unless stated otherwise. The cuvette was placed at a fixed position 5 cm from the tip of the filter holder mounted in front of the condenser. The diameter of the collimated light beam covered the entire reaction volume contained in the cuvette. The irradiation setup was kept fixed for all photolabeling experiments carried out in the present study. The experiment shown in supplemental Fig. S3 confirms that the 35-s irradiation time generally used in the photolabeling experiments did not affect the activity of the enzyme substantially.
SCHEME 3.
TNP-8N3-ATP photolabeling reaction. When the azide of TNP-8N3-ATP is exposed to ultraviolet light, it forms a highly reactive but short-lived nitrene that can initiate reactions with neighboring reactive groups, such as the amino group of the Lys492 side chain, thereby forming a stable covalent bond between the nucleotide and the protein (20, 45, 46). R1 represents the TNP-ATP moiety of TNP-8N3-ATP (see supplemental Fig. S4), and R2-NH2 is the Lys492 side chain of the Ca2+-ATPase.
To form the Ca2E2P state of mutant 4Gi-46/47 (31, 35), phosphorylation was carried out for 10 min at 25 °C in 25 mm MOPS/tetramethyl ammonium hydroxide (pH 7.0), 10 mm MgCl2, 15% DMSO, 38 μm calcium ionophore A23187, 1 mm EGTA, and 0.5 mm Pi, followed by cooling on ice. Immediately prior to photolabeling, 2 μl of the phosphorylated microsomes were supplemented with 2 μl of ice-cold 42 mm CaCl2, to give a final free Ca2+ concentration of 20.5 mm (on both the lumenal and the cytoplasmic sides of the membrane, because of the presence of the calcium ionophore). Then, 3 μl of the phosphorylated and Ca2+-saturated enzyme were diluted 25-fold into the pre-mixed photolabeling medium, followed by irradiation.
Measurements of phosphorylation from [γ-32P]ATP or 32Pi were carried out by acid quenching followed by acid SDS-polyacrylamide gel electrophoresis and quantification of the radioactivity associated with the Ca2+-ATPase band, using the previously established procedures (16, 17, 43). To study the ATP concentration dependence of phosphorylation, microsomes were incubated for 15 s at 0 °C in 40 mm MOPS/Tris (pH 7.0), 80 mm KCl, 5 mm MgCl2, 100 μm CaCl2, and varying concentrations of [γ-32P]ATP. For studies of the ATP dependence of dephosphorylation of E2P, phosphorylation with 0.5 mm 32Pi was carried out for 10 min at 25 °C in 100 mm MES/Tris (pH 6.0), 10 mm MgCl2, 2 mm EGTA, and 30% (v/v) dimethyl sulfoxide. The phosphorylated sample was chilled in ice water, and dephosphorylation was followed at 0 °C by a 19-fold dilution into ice-cold medium containing 50 mm MOPS/Tris (pH 7.0), 2 mm EGTA, 10 mm EDTA, 5 mm H3PO4, and various concentrations of ATP.
The data were analyzed by nonlinear regression using the SigmaPlot program (SPSS, Inc.). The analysis of the TNP-8N3-ATP photolabeling data were based on the hyperbolic function, Y = Ymax × [TNP-8N3-ATP]/(K0.5 + [TNP-8N3-ATP]) + m × [TNP-8N3-ATP], in which Y is the amount of photolabeled Ca2+-ATPase, Ymax is the maximum amount of photolabeled Ca2+-ATPase, K0.5 is the concentration of TNP-8N3-ATP giving half-maximum labeling, and m × [TNP-8N3-ATP] is a linear component, which has been subtracted from the data shown (33). The analysis of the data obtained from ATP inhibition of TNP-8N3-ATP photolabeling was based on the Hill equation modified to describe inhibition, Y = Ymax × (1 − [ATP]n/(K0.5n + [ATP]n)), in which Y and Ymax are defined as above, K0.5 is the concentration of ATP giving half-maximum effect, and n is the Hill coefficient (varying between 0.6 and 1.1 for the present data). The “true” dissociation constant, KD, for ATP binding was calculated from the measured K0.5 values using the validated equation for competitive inhibition (33). The analysis of the ATP dependence of phosphorylation from [γ-32P]ATP was based on the Hill equation, EP = EPmax × [ATP]n/(K0.5n + [ATP]n). For analysis of the modulatory effect of ATP on the rate of E2P dephosphorylation, the ATP concentration dependence of the rate constant was analyzed according to the hyperbolic function, kobs = k0 + (kmax − k0) × [ATP]/(K0.5 + [ATP]), in which kobs is the rate constant observed at the indicated ATP concentration, k0 is the rate constant in the absence of ATP, and kmax is the extrapolated value of the rate constant corresponding to infinite ATP concentration (17). The experiments were conducted at least twice on independent microsomal preparations, and average values are shown.
RESULTS AND DISCUSSION
TNP-8N3-ATP Photolabeling of Wild Type Ca2+-ATPase in E2·Tg, E2·MgF, E2·Vanadate, E2·AlF, and E2·BeF States
To study the interaction of nucleotides with the wild type Ca2+-ATPase in stable analog forms of the intermediate states occurring during E2P dephosphorylation, SR vesicles or microsomes containing expressed enzyme were incubated with saturating concentrations of Tg, MgF, vanadate, AlF, or BeF and subjected to nucleotide binding analysis by TNP-8N3-ATP photolabeling of Lys492 as previously described for the E1 form (33). The formation of the complex with metal fluoride or vanadate took place in the presence of Mg2+, whereas subsequent photolabeling was carried out in medium without free Mg2+ (EDTA added), considering that the substrate that binds to E2P with reasonable affinity and accelerates E2P dephosphorylation is metal-free ATP (10, 14, 15). Enzyme with Tg bound and enzyme in the E1 form was photolabeled either in the absence or presence of Mg2+. In the latter case, EGTA was present to specifically chelate Ca2+, because it was essential to remove Ca2+ to prevent enzyme activation and consequent hydrolysis of the photolabel and ATP. Photolabeling was carried out at a rather high pH of 8.5 to prevent unspecific labeling (33) and to ensure that even in the absence of Ca2+ the enzyme without Tg or metal fluoride bound resides predominantly in the E1 state rather than E2 (44). We were concerned that the enzyme-inhibitor complexes remained stable during photolabeling, and by studying the time course of reactivation following addition of Ca2+, evidence was obtained that all five enzyme-inhibitor complexes were very stable under the photolabeling conditions, despite the high pH of the medium (supplemental Fig. S2).
The time dependence of photolabeling of expressed wild type Ca2+-ATPase pre-equilibrated with or without inhibitor is shown in Fig. 1A. With the current irradiation setup, photolabeling proceeded at a rate of ∼3 min−1 irrespective of the enzyme conformational state/inhibitor bound. The photolabeling rate of ∼3 min−1 is comparable with the rate of photolysis of the azido group in TNP-8N3-ATP of ∼2 min−1, determined using the same irradiation setup, cf. supplemental Fig. S4, implying that for Ca2+-ATPase with or without bound fluoride complex or vanadate the rate-limiting step in the labeling reaction is the formation of the reactive nitrene (cf. Scheme 3). The subsequent chemical reaction between the nitrene of the photoactivated nucleotide and Lys492 of the Ca2+-ATPase (Scheme 3) is likely much faster, given the typical short lifetime and high reactivity of nitrene intermediates (45, 46). Based on the time dependence of TNP-8N3-ATP photolabeling of the Ca2+-ATPase (Fig. 1A) as well as the time dependence of TNP-8N3-ATP photolysis (supplemental Fig. S4), a pre-steady state irradiation time of 35 s was chosen for all subsequent photolabeling experiments.
The maximum levels of photolabeling (corresponding to saturation with photolabel) of the expressed wild type Ca2+-ATPase as well as SR were ∼3.5-fold higher in the E2·vanadate, E2·AlF, and E2·MgF states and ∼1.7-fold higher in the E2·BeF state than in E2·Tg or E1 (Fig. 1). The labeling stoichiometry corresponding to the highest labeling levels indicated as 100% in Fig. 1 can be roughly estimated to be ∼0.7 mol of label incorporated per mol of Ca2+-ATPase present in the microsomal membrane, assuming that no label or protein is lost during gel electrophoresis. The higher maximum levels of photolabeling in the vanadate- and metal fluoride-complexed states as compared with E2·Tg or E1 do not result from additional labeling of other residues than Lys492, because no labeling of the mutant K492L was seen in any of the enzyme states examined (Table 1 and supplemental Fig. S5). Rather, the increased labeling levels may reflect a conformational change resulting in shortening of the interaction distance between Lys492 and the azido group of TNP-8N3-ATP and/or desolvation of the intervening space thereby removing competing water molecules and increasing the nucleophilicity of the amino group, resulting in increased efficiency of the coupling reaction between Lys492 and the reactive nitrene of the photolabel following UV irradiation (cf. Scheme 3). Apart from this clue, little detail is known about the interactions of the photolabel in the various conformational states. Although competition of the photolabel with ATP for binding indicates that there is at least a partial overlap of binding sites, it is also clear from the difference in mutational effects on affinities for photolabel and ATP that the photolabel must bind rather differently from ATP (see below, compare Tables 1 and 2, the difference is particularly striking for mutants F487S and R560L).
TABLE 1.
Affinity for TNP-8N3-ATP of SR and expressed wild type Ca2+-ATPase and mutants in various stabilized states
K0.5 values for photolabeling derived from Figs. 2, 7, and supplemental Fig. S6 are indicated relative (in %) to that of the expressed wild type under the same conditions (for wild type the absolute K0.5 value is shown). The S.E. is indicated with the number of experiments in parentheses.
| EDTAa |
EGTA/Mg2+b | |||||
|---|---|---|---|---|---|---|
| E2 (Tg) | E2·Pi (MgF) | E2·P (vanadate) | E2·P (AlF) | E2−P (BeF) | E1 | |
| Wild type | 100 ± 14 (n = 4) (62 nm) | 100 ± 12 (n = 6) (10.5 nm) | 100 ± 10 (n = 8) (9.6 nm) | 100 ± 9 (n = 9) (11.6 nm) | 100 ± 14 (n = 7) (54 nm) | 100 ± 18 (n = 5) (290 nm) |
| SR | 84 ± 11 (n = 3) | 90 ± 14 (n = 5) | 74 ± 7 (n = 4) | 69 ± 10 (n = 3) | 83 ± 6 (n = 3) | 89 |
| R174A | 331 ± 17 (n = 2) | 269 ± 7 (n = 2) | 251 ± 7 (n = 2) | 255 ± 7 (n = 2) | 795 ± 27 (n = 2) | 73 |
| R174E | 536 ± 43 (n = 2) | 1781 ± 171 (n = 2) | 1735 ± 0 (n = 2) | 878 ± 49 (n = 2) | 1582 ± 340 (n = 2) | 80 |
| I188A | 127 ± 1 (n = 2) | 109 ± 20 (n = 2) | 100 ± 16 (n = 2) | 122 ± 18 (n = 2) | 64 ± 18 (n = 2) | 42 |
| I188F | 87 ± 5 (n = 2) | 154 ± 16 (n = 2) | 76 ± 5 (n = 2) | 69 ± 6 (n = 2) | 19 ± 2 (n = 2) | 68 |
| K205A | 357 ± 2 (n = 2) | 854 ± 196 (n = 2) | 1131 ± 123 (n = 2) | 1031 ± 9 (n = 2) | 567 ± 6 (n = 2) | 56 |
| E439A | 141 ± 6 (n = 2) | 110 ± 6 (n = 2) | 123 ± 11 (n = 2) | 177 ± 11 (n = 3) | 22 ± 3 (n = 2) | 35 |
| F487L | 36 ± 5 (n = 2) | 52 ± 15 (n = 2) | 48 ± 5 n = 2) | 60 ± 0 (n = 2) | 19 ± 0 (n = 2) | 43 |
| F487S | 90 ± 16 (n = 2) | 89 ± 15 (n = 3) | 101 ± 1 (n = 2) | 81 ± 1 (n = 2) | 88 ± 1 (n = 2) | 25 |
| R489L | 965 ± 107 (n = 3) | 116 ± 12 (n = 2) | 108 ± 3 (n = 2) | 120 ± 2 (n = 2) | 702 ± 109 (n = 2) | 85 |
| K492L | No labeling | No labeling | No labeling | No labeling | No labeling | No labeling |
| R560L | 311 ± 0 (n = 2) | 77 ± 19 (n = 2) | 98 ± 16 (n = 3) | 64 ± 8 (n = 2) | 138 ± 5 (n = 2) | 1250 |
| L562F | 136 ± 17 (n = 2) | 100 ± 4 (n = 2) | 81 ± 15 (n = 2) | 110 ± 20 (n = 2) | 52 ± 8 (n = 2) | 25 |
| 4Gi-46/47 | 151 ± 23 (n = 2) | 83 ± 0 (n = 2) | 96 ± 5 (n = 2) | 106 ± 2 (n = 2) | 83 ± 7 (n = 2) | 96 ± 17 (n = 2) |
a EDTA refers to the condition without free Mg2+ present described under “Experimental Procedures.”
TABLE 2.
Affinity for ATP of SR and expressed wild type Ca2+-ATPase and mutants in various stabilized states
KD values derived from competitive inhibition of photolabeling by ATP (Figs. 2, 5, and 7) are indicated relative (in %) to that of the expressed wild type obtained under the same conditions (for wild type the absolute KD value is shown). The S.E. is indicated with the number of experiments in parentheses.
| EDTAa |
EGTA/Mg2+b | |||||
|---|---|---|---|---|---|---|
| E2 (Tg) | E2·Pi (MgF) | E2·P (vanadate) | E2·P (AlF) | E2−P (BeF) | E1 | |
| Wild type | 100 ± 9 (n = 8) (10.8 μm) | 100 ± 19 (n = 5) (0.89 μm) | 100 ± 9 (n = 4) (1.01 μm) | 100 ± 7 (n = 7) (1.33 μm) | 100 ± 7 (n = 6) (4.1 μm) | 100 ± 5 (n = 4) (0.14 μm) |
| SR | 96 ± 8 (n = 5) | 90 ± 7 (n = 3) | 81 ± 6 (n = 3) | 52 ± 6 (n = 4) | 60 ± 6 (n = 3) | 78 |
| R174A | 316 ± 38 (n = 2) | 256 ± 29 (n = 2) | 224 ± 42 (n = 2) | 256 ± 8 (n = 2) | 1070 ± 61 (n = 2) | 182 |
| R174E | 503 ± 4 (n = 2) | 3445 ± 168 (n = 2) | 2579 ± 272 (n = 2) | 1157 ± 15 (n = 2) | 4307 ± 964 (n = 3) | 425 |
| I188A | 75 ± 1 (n = 2) | 43 ± 4 (n = 2) | 47 ± 4 (n = 2) | 56 ± 6 (n = 2) | 31 ± 4 (n = 2) | 45 |
| I188F | 97 ± 17 (n = 3) | 187 ± 12 (n = 2) | 91 ± 2 (n = 2) | 56 ± 0 (n = 2) | 476 ± 68 (n = 2) | 110 |
| K205A | 267 ± 2 (n = 2) | 1147 ± 117 (n = 2) | 1257 ± 10 (n = 2) | 1198 ± 26 (n = 2) | 249 ± 36 (n = 3) | 139 |
| E439A | 78 ± 1 (n = 2) | 195 ± 31 (n = 2) | 237 ± 8 (n = 2) | 631 ± 74 (n = 3) | 32 ± 5 (n = 3) | 235 |
| F487L | 1594 ± 121 (n = 2) | 133 ± 10 (n = 2) | 138 ± 0 (n = 2) | 127 ± 17 (n = 2) | 354 ± 10 (n = 2) | 2,200 |
| F487S | 1219 ± 14 (n = 2) | 1717 ± 450 (n = 4) | 1294 ± 303 (n = 3) | 1104 ± 53 (n = 2) | 2309 ± 229 (n = 3) | >100,000 |
| R489L | 1896 ± 97 (n = 2) | 252 ± 48 (n = 2) | 285 ± 38 (n = 2) | 203 ± 8 (n = 2) | 1168 ± 283 (n = 2) | 1600 |
| K492L | Not feasible | Not feasible | Not feasible | Not feasible | Not feasible | Not feasible |
| R560L | 3555 ± 23 (n = 2) | 2019 ± 419 (n = 2) | 1977 ± 356 (n = 3) | 1172 ± 150 (n = 2) | 3762 ± 369 (n = 3) | >100,000 |
| L562F | 213 ± 36 (n = 3) | 67 ± 3 (n = 2) | 64 ± 12 (n = 2) | 106 ± 1 (n = 2) | 132 ± 18 (n = 2) | 6,900 |
| 4Gi-46/47 | 261 ± 62 (n = 2) | 89 ± 10 (n = 2) | 75 ± 1 (n = 2) | 102 ± 5 (n = 2) | 93 ± 17 (n = 2) | >100,000 (n = 4) |
a EDTA refers to the condition without free Mg2+ present described under “Experimental Procedures.”
Nucleotide Affinity of Wild Type Ca2+-ATPase in E2·Tg, E2·MgF, E2·Vanadate, E2·AlF, and E2·BeF States
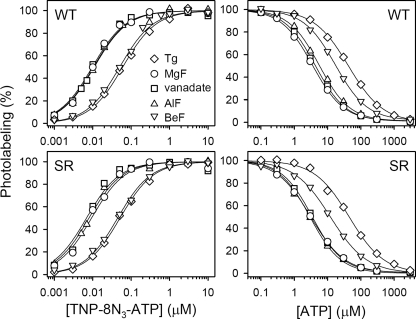
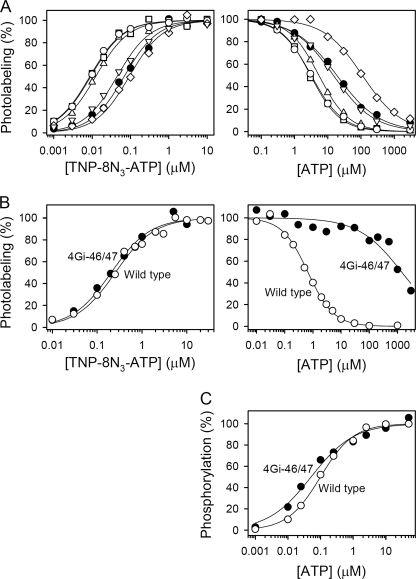
We then proceeded to study the TNP-8N3-ATP concentration dependence of photolabeling of SR and expressed wild type Ca2+-ATPase in the E2·Tg, E2·MgF, E2·vanadate, E2·AlF, and E2·BeF states (Fig. 2, left panels). The TNP-8N3-ATP affinity of the expressed wild type Ca2+-ATPase with thapsigargin bound was 62 nm (Fig. 2, upper left panel, and Table 1).4 A similar affinity of 54 nm (although, as noted above, with a 1.7-fold higher maximum labeling level) was obtained for the E2·BeF state of the expressed wild type. In contrast, the TNP-8N3-ATP affinities of the E2·MgF, E2·vanadate, and E2·AlF states of the expressed wild type were significantly higher, in the 9–12 nm range (Table 1), possibly due to the same conformational change that results in the ∼3.5-fold higher maximal labeling levels described above. In comparison, the affinity for TNP-8N3-ATP of the uncomplexed wild type in the presence of Mg2+ (E1 state) was 290 nm (Table 1).4 The TNP-8N3-ATP affinities of SR with bound Tg, MgF, vanadate, AlF, and BeF were rather similar to those obtained with the expressed wild type (compare Fig. 2, upper left and lower left panels).
FIGURE 2.
TNP-8N3-ATP (left panels) and ATP (right panels) concentration dependences of photolabeling of wild type Ca2+-ATPase stabilized in the intermediate states occurring during E2P dephosphorylation. Expressed wild type Ca2+-ATPase (upper panels) or SR (lower panels) was incubated with Tg, MgF, vanadate, AlF, or BeF as described under “Experimental Procedures,” and subjected to TNP-8N3-ATP photolabeling at the indicated concentrations of TNP-8N3-ATP without ATP (left panels), or at 3× the K0.5 for TNP-8N3-ATP with the indicated concentrations of ATP (right panels). In each case, the maximum level of specific labeling was defined as 100%. Symbols for all panels are indicated in the upper left panel.
Fig. 2, right panels, illustrates the inhibition by ATP of TNP-8N3-ATP photolabeling, showing also that the ATP affinity of the E2·vanadate, E2·AlF, and E2·MgF forms of expressed wild type Ca2+-ATPase or SR Ca2+-ATPase is much higher (KD ∼1 μm) than that of E2·Tg (KD 11 μm), although not nearly as high as the affinity of the E1 state (KD 0.14 μm).4 In E2·BeF, the affinity for ATP was of an intermediate magnitude (KD 4 μm). These data are summarized in Table 2. The higher ATP affinity of the E2P transition state analogs E2·vanadate and E2·AlF, as compared with the E2P ground state analog, E2·BeF, and the E2 dephosphoenzyme, makes it conceivable that stimulation of E2P dephosphorylation by ATP is accomplished by increasing the stability of the transition state of E2P dephosphorylation, thereby lowering the energy barrier for formation of the transition state. The binding of the modulatory ATP may lead to a more compact packing of the A-, P-, and N-domains and to an optimal positioning of Glu183 in the 181TGES motif of the A-domain for coordinating the attacking water molecule during dephosphorylation (3–5) (cf. Fig. 3). In addition, stabilization of the product state E2·Pi, as evidenced by the high ATP affinity of the E2·MgF complex, could be of importance for the modulatory effect on the dephosphorylation.
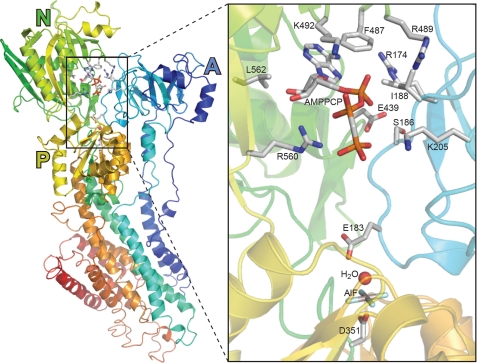
FIGURE 3.
Structural arrangement of the nucleotide binding site in Ca2+-ATPase crystallized in the E2·AlF·AMPPCP state. The Protein Data Bank accession code corresponding to the structure shown is 3B9R (26). Amino acid side chains are shown for residues discussed in the text. Carbon and aluminum atoms are shown in gray, nitrogen in blue, oxygen in red, phosphorous in orange, and fluoride in cyan.
Nucleotide Binding to E2·Tg·AlF
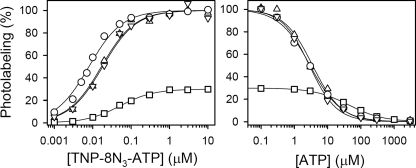
Thapsigargin has been widely applied in crystallization studies of the Ca2+-ATPase either without (30, 47) or with additional inhibitors such as MgF (5), AlF (4, 48), or BeF (48). To address the issue whether thapsigargin binding influences the nucleotide affinity of the metal fluoride complex, we measured TNP-8N3-ATP and ATP binding to enzymes complexed by both Tg and AlF (Fig. 4). SR vesicles were incubated under optimal conditions for forming the E2·AlF complex, followed by supplementation and further incubation with 1 μm Tg. Alternatively, incubation was first carried out with Tg and then with AlF. As seen in Fig. 4, the result was independent of which complex was formed first. The affinity of E2·Tg·AlF for TNP-8N3-ATP was 17–18 nm, i.e. ∼2-fold lower and ∼3-fold higher than the TNP-8N3-ATP affinity of the E2·AlF and E2·Tg complexes, respectively (Fig. 4, left panel). The affinity of E2·Tg·AlF for ATP was on the other hand very similar to that of E2·AlF but 12- to 15-fold higher than that of E2·Tg (Fig. 4, right panel), implying that AlF binding dominates over Tg binding with respect to influencing the conformation of the ATP binding site in the E2·Tg·AlF complex. Also with respect to the maximal labeling levels seen in Fig. 4 did the E2·Tg·AlF complex resemble E2·AlF more than E2·Tg (cf. Fig. 1). Thus, the maximum labeling level of E2·Tg·AlF was ∼3.5-fold higher than that of E2·Tg. It can be concluded that phosphorylation and nucleotide binding sites are fully flexible in the Tg-bound state, readily able to bind the AlF complex, and subsequently be photolabeled by TNP-8N3-ATP with a K0.5 for the photolabel, a K0.5 for the inhibition by ATP of the photolabeling, and a maximal photolabeling level similar to that of the AlF-complexed, but Tg-free, enzyme. Hence, the inhibitory effect on catalysis of thapsigargin binding between transmembrane helixes M3, M5, and M7 (30) is a local effect in the membrane, leaving the cytoplasmic domains free to bind nucleotide and adopt the various conformations characteristic of the transitional states of E2P dephosphorylation.
FIGURE 4.
TNP-8N3-ATP (left panel) and ATP (right panel) concentration dependences of photolabeling of Ca2+-ATPase in the E2·Tg·AlF state. Formation of the E2·Tg·AlF state with SR was accomplished by first forming the E2·Tg state, followed by supplementation with AlCl3, NaF, and MgCl2 to final concentrations of 0.5, 2, and 0.2 mm, respectively (triangles pointing downward), or by first forming the E2·AlF state, followed by supplementation with 1 μm Tg (triangles pointing upward), and further incubation for 30 min at 25 °C. TNP-8N3-ATP photolabeling was then carried out at the indicated concentrations of TNP-8N3-ATP without ATP (left panel) or at 3× the K0.5 for TNP-8N3-ATP with the indicated concentrations of ATP (right panel). For comparison, results with SR in the E2·Tg (squares) and E2·AlF states (circles) (cf. Fig. 2) are included in the panels. In each case, the labeling level (in %) is shown relative to the labeling level of E2·AlF.
ATP Affinity of Mutant Ca2+-ATPases in E2·Tg, E2·MgF, E2·Vanadate, E2·AlF, and E2·BeF States
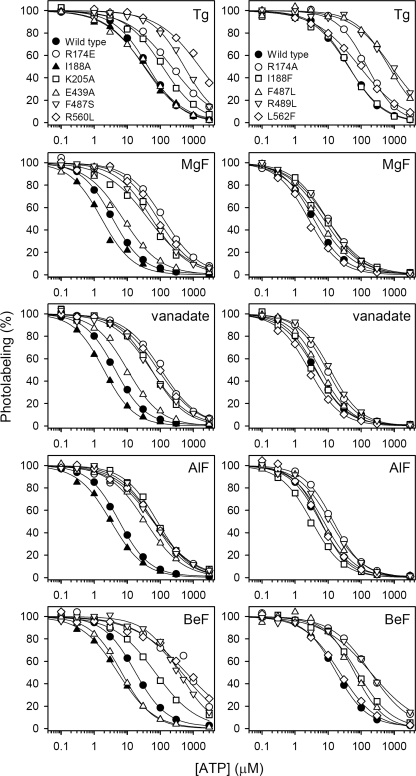
Studies of the ATP dependence of the rate of E2P dephosphorylation in mutants (16, 17, 24) have pinpointed certain residues as critical for ATP modulation of E2P dephosphorylation, including Glu439, Phe487, and Arg560 in the N-domain and Arg174, Ile188, and Lys205 in the A-domain (Fig. 3). To understand how the effect of ATP on E2P dephosphorylation is brought about, a critical question to answer for each of these residues is whether the residue is directly involved in binding of the modulatory nucleotide, or its role is instead associated with mediating the response to binding. By applying the photolabeling assay to determine the ATP affinity of the various states of the E2P dephosphorylation reaction sequence in mutants, it is possible to distinguish between mutational effects on ATP modulation caused by direct interference with ATP binding and effects caused by interference with the consequences of the binding. This analysis was carried out with a series of mutants previously shown to be defective in ATP modulation of E2P dephosphorylation as observed in functional studies (R174A, R174E, I188A, I188F, K205A, E439A, F487S, and R560L, cf. Refs. 16 and 17) and with selected mutants previously found defective in ATP binding at the catalytic site in the E1 conformation (F487S, F487L, R489L, R560L and L562F, cf. Refs. 33 and 34). The binding data are displayed in Fig. 5 for ATP and under supplemental Fig. S6 for TNP-8N3-ATP, and the resulting affinity constants are listed in Tables 1 and 2.
FIGURE 5.
ATP concentration dependences of photolabeling of Ca2+-ATPase mutants stabilized in the intermediate states occurring during E2P dephosphorylation. Expressed Ca2+-ATPase was incubated with Tg, MgF, vanadate, AlF, or BeF as described under “Experimental Procedures,” and subjected to TNP-8N3-ATP photolabeling at 3× the K0.5 for TNP-8N3-ATP (the TNP-8N3-ATP concentration dependences are shown in supplemental Fig. S6) in the presence of the indicated concentrations of ATP. In each case, the maximum level of specific labeling was defined as 100%. The affinity constants determined are listed in Table 2. Symbols for the five panels above each other are indicated in the top panels.
Mutants with alterations to N-domain residues Phe487, Arg489, Arg560, and Leu562 (cf. Fig. 3) were previously shown to be severely defective with respect to ATP binding at the catalytic site in the E1 conformation (33, 34) (Table 2, right column). In functional studies mutations F487S and R560L were, furthermore, found to reduce the apparent affinity for ATP modulation of E2P dephosphorylation as much as >50- and 30-fold, respectively (16), which on the basis of the binding data in Fig. 5 and Table 2 can be ascribed to a deficiency of binding of the modulatory ATP throughout the E2P dephosphorylation reaction sequence. Hence, ATP binding affinity in the five E2/E2P states was reduced 11–23-fold for mutant F487S and 12–38-fold for mutant R560L, relative to wild type. Phe487 and Arg560 are in close proximity to the nucleotide in the E1·AMPPCP, E2·Tg·AMPPCP, E2·MgF·AMPPCP/ATP/ADP, and E2·AlF·AMPPCP crystal structures, with Phe487 apparently interacting with the adenine ring and Arg560 with the ribose and/or the β-phosphate (5, 26, 47–50), and the present data support the notion that the catalytic ATP binding site in the E1 state and the modulatory ATP binding site responsible for stimulation of E2P dephosphorylation are overlapping. Mutation F487L, retaining the bulk and hydrophobicity of the side chain, was, however, much less detrimental to ATP binding than F487S, in the E2P-like analog states stabilized with MgF, vanadate, AlF, and BeF. Hence, a wild type-like affinity for ATP was seen for mutant F487L in E2·MgF, E2·vanadate, and E2·AlF states, and in E2·BeF the ATP affinity was only moderately (3.5-fold) reduced. The effect of the F487L mutation was much more pronounced in the E1 and E2·Tg states (22- and 16-fold reduction of ATP affinity, respectively, cf. Table 2). In most of the crystal structures with bound nucleotide the adenine ring is interposed between Phe487 and Leu562. In the Ca2E1 state, the aromatic ring of the phenylalanine side chain is nearly parallel to the adenine ring, indicating a π-stacking interaction that explains the marked effect of the leucine substitution in this state. In the E2·MgF and E2·AlF crystal structures the phenylalanine and adenine rings are more angled toward each other, suggesting less efficient π-stacking, which might be the reason that despite the lack of aromaticity the leucine is able to substitute quite well for phenylalanine in these states. In the E2·Tg state, however, mutation F487L was just as detrimental to ATP binding as in E1, despite a non-parallel orientation of the phenylalanine ring and the adenine ring in the E2·Tg·AMPPCP crystal structures, thus implying that positioning of the adenine ring in the native enzyme in the E2·Tg state differs somewhat from that seen in the E2·Tg·AMPPCP crystal structures.
Mutation L562F was previously shown to reduce MgATP affinity of E1 69-fold (34) but appears much less distorting in the intermediate E2/E2P states of E2P dephosphorylation, the most marked effect being a 2-fold reduced ATP affinity in E2·Tg (Table 2), which is somewhat surprising, because the Leu562 side chain occupies almost exactly the same position relative to the nucleotide in the Ca2E1·AMPPCP, E2·Tg·AMPPCP, E2·MgF·AMPPCP/ATP/ADP, and E2·AlF·AMPPCP crystal structures, being located 3.2–3.7 Å from the ribose and 3.3–4.9 Å from the adenine ring (26, 47–50). It is possible that because of a higher mobility of the bound ATP in the E2/E2P states, as reflected by the lower ATP affinity of wild type E2/E2P states (KD values in the 1–11 μm range; Table 2) compared with that of E1 (KD = 0.14 μm; Table 2), the bound nucleotide in the E2/E2P states can be correctly positioned by any large hydrophobic side chain replacing Leu562. This would not be feasible in the very tight enzyme-nucleotide complex normally seen for E1, where a phenylalanine side chain cannot be accommodated in place of the leucine without destabilization of the complex. Accordingly, the affinity of the E1 state of L562F for MgATP (∼10 μm) is a bit lower than the affinity of the E2P states of L562F for ATP (0.5–5 μm) (Table 2).
Mutation R489L was equally detrimental to ATP binding in E2·Tg and E2·BeF (19- and 12-fold reduction of affinity, respectively, relative to wild type) as to MgATP binding in E1 (16-fold reduction), whereas the effect was much less pronounced for E2·MgF, E2·vanadate, and E2·AlF (2–3-fold reduced ATP affinities, relative to wild type) (Table 2). This finding provides additional evidence that ATP is bound differently in the E2 dephosphoenzyme and the E2P ground state compared with the E2P transition and product states. The minor effects seen for the E2P transition state and product state analogs accord with the crystal structures of E2·AlF·AMPPCP and E2·MgF·AMPPCP/ATP/ADP, where the distance between the Arg489 side chain guanidinium group and the nucleotide ribose-OH is somewhat larger (4–6 Å) compared with the ∼3 Å seen for the various Ca2E1 crystals. In the E2·Tg·AMPPCP crystal structures the corresponding distance is 5.2 Å, again indicating that details of the positioning of the nucleotide differ somewhat from the native enzyme in the E2·Tg state, where Arg489 according to our result is an important interaction partner. Because the E2·BeF crystal structures (26, 48) do not contain bound nucleotide, there is so far no structural correlation of the marked effect of the R489L mutation on the ATP affinity of E2·BeF.
The A-domain residues Arg174, Ile188, and Lys205 are not involved in nucleotide binding in the E1 conformation, but were in our previous functional studies identified as critical for the ATP-induced acceleration of E2P dephosphorylation (17). The Arg174 side chain is rather close (∼4 Å) to the adenine ring of the nucleotide in the product state (E2·MgF·AMPPCP/ATP/ADP) and transition state (E2·AlF·AMPPCP) (Fig. 3) analog crystal structures (5, 26, 49), whereas in the E2·Tg·AMPPCP crystal structure the bound nucleotide is too far away from Arg174 for any direct interaction (47). Substitution of Arg174 with alanine or glutamate leads to reduced apparent affinity for ATP modulation of E2P dephosphorylation, most markedly for R174E, in which the charge of the side chain is reversed (17). PPi (pyrophosphate) was on the other hand found effective in stimulating dephosphorylation of mutant R174A with an affinity similar to that seen for the wild type, although the affinity of R174E for PPi was reduced (17). Assuming that PPi binds at the same site as modulatory ATP, mimicking the effect of the β- and γ-phosphates of ATP, the functional data would suggest that the role of Arg174 in ATP modulation of E2P dephosphorylation is associated primarily with binding of the adenosine part of the nucleotide and not the mechanism of mediating the effect of binding. The present binding data support this concept by showing that mutation R174A reduces the affinity for ATP as much as 11-fold in the E2P ground state (E2·BeF) and ∼3-fold in the other E2/E2P-like analog states, and that R174E reduces the ATP affinity markedly (12–43-fold) in the four E2P-like analog states (Fig. 5 and Table 2). The more pronounced effect of replacement with glutamate, having a negatively charged side chain, may be a consequence of electrostatic repulsion of the phosphates of the ATP molecule (cf. Fig. 3).
Our previous functional analysis (17), furthermore, showed that stimulation by ATP of E2P dephosphorylation was completely abolished in mutant I188A, whereas I188F displayed a minor, 2-fold reduction of the apparent affinity for the modulation by ATP of E2P dephosphorylation. Because of the 3–5-Å proximity of the Ile188 side chain to the α-phosphate or adenine ring of the nucleotide in E2·MgF·ADP and E2·AlF·AMPPCP crystal structures (Fig. 3), one might have expected Ile188 to be directly involved in ATP binding during E2P dephosphorylation, which might explain the marked effect of mutation I188A on ATP modulation (17). The present binding analysis showed, however, that contrary to the expected lowering of affinity, the I188A mutation actually causes a significant 2–3-fold increase of ATP affinity in the four E2P-like analog states (Table 2, the corresponding KD values are 0.4–1.3 μm), suggesting that in fact the larger side chain of isoleucine present in the wild type is a little disturbing to the binding of the nucleotide, the alanine of the mutant accommodating the nucleotide better. In this light, the complete absence of ATP modulation of dephosphorylation of E2P in I188A (Fig. 5 of Ref. 17) provides a clear indication of a mechanistic role of Ile188 in mediating the stimulating effect of ATP on E2P dephosphorylation. Ile188 is located at the start of the loop containing the TGES motif with Glu183, and a slight clash between Ile188 and the nucleotide might be instrumental in moving Glu183 to the optimal position for catalyzing dephosphorylation. Because of the shorter side chain, alanine would not be able to fulfill this role, whereas the larger phenylalanine would (the efficiency of modulation is actually higher for I188F than for the wild type, see Table 3 in Ref. 17).
Mutants K205A and E439A are both modulated by ATP in a rather anomalous way, displaying inhibition rather than stimulation of E2P dephosphorylation by ATP (16, 17). This is illustrated for mutant E439A in Fig. 6, where more data points have been included than previously (16), showing inhibition between 0.2 and 5 mm ATP with a K0.5 for inhibition of 3.1 mm. In comparison, wild type is stimulated by ATP with a K0.5 of 34 μm. Phosphoenzyme decay curves for mutant E439A at various ATP concentrations are shown under supplemental Fig. S7. Detailed inhibition data for mutant K205A obtained in the same way were previously shown in Fig. 5 of Ref. 17. In the E2·MgF·AMPPCP/ATP/ADP and E2·AlF·AMPPCP (Fig. 3) crystal structures the side chain of Lys205 in the A-domain is close (3–4 Å) to the β- and γ-phosphates of the nucleotide, whereas the side chain of Glu439 in the N-domain is further away from the nucleotide (5–7 Å distant from the adenine ring). Indeed, the role of Glu439 in ATP modulation of E2P dephosphorylation is likely to be of an indirect nature, relating to an interdomain hydrogen bond between Glu439 and A-domain residue Ser186, cf. Fig. 3 (16, 24), rather than to direct interaction with the modulatory nucleotide. The binding data in Fig. 5 provide a tentative explanation of the anomalous inhibitory effect of ATP seen for K205A and E439A. The KD values for ATP binding are for K205A, 10.2 μm in E2·BeF, 15.9 μm in E2·AlF, and 12.7 μm in E2·vanadate; and for E439A, 1.3 μm in E2·BeF, 8.4 μm in E2·AlF, and 2.4 μm in E2·vanadate, which should be compared with the wild type affinity constants of 4.1 μm in E2·BeF, 1.3 μm in E2·AlF, and 1 μm in E2·vanadate. Hence, for both mutants the ATP affinity is higher (i.e. KD lower) in the ground state (E2·BeF) of E2P compared with the transition state (E2·AlF and E2·vanadate). This is contrary to the situation seen with wild type and all the other mutants studied here, where the ATP affinity of E2·BeF is lower than that of E2·AlF and E2·vanadate. Thus, whereas the nucleotide increases stability of the E2P transition state relative to the E2P ground state in the wild type, and consequently stimulates dephosphorylation, the opposite takes place in K205A and E439A, with the nucleotide instead increasing stability of the ground state relative to the transition state, thereby inhibiting dephosphorylation.
FIGURE 6.
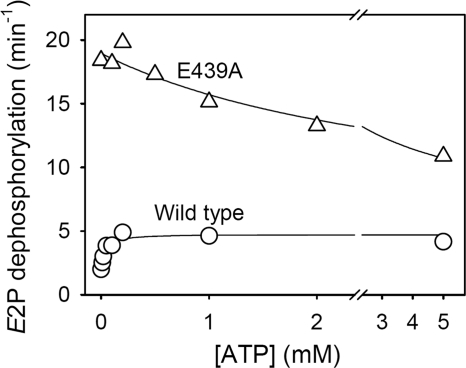
ATP dependence of the rate of dephosphorylation of E2P for mutant E439A. Dephosphorylation of the phosphoenzyme formed in the presence of 32Pi was followed at various ATP concentrations at pH 7.0 in the absence of Mg2+ as described under “Experimental Procedures.” Examples of the decay curves are shown under supplemental Fig. S7. The dephosphorylation rate constants are shown here as a function of the ATP concentration. The parameters derived by fitting a hyperbolic function as described under “Experimental Procedures” are as follows (in each case, the total number of data points included in the fit is indicated in parentheses): wild type, K0.5 = 34 ± 9 μm, k0 = 2.0 min−1, kmax = 4.7 min−1 (n = 22); E439A, K0.5 = 3115 ± 2171 μm, k0 = 19.0 min−1, kmax = 5.6 min−1 (n = 19).
Nucleotide Affinity of the Stable Ca2E2P State of Mutant 4Gi-46/47
For wild type Ca2+-ATPase, the conformational transition of the phosphoenzyme, Ca2E1P → Ca2E2P, is a rate-limiting step of the overall pump cycle, and is succeeded by rapid dissociation of the two Ca2+ ions from lumenally exposed Ca2+ sites to the endoplasmic reticulum lumen, thus forming the Ca2+-free E2P ground state (cf. Scheme 1). The Ca2E2P state is thought to be an unstable and short-lived intermediate that cannot be readily isolated. However, elongation of the A-M1 linker between the A-domain and transmembrane helix M1 by insertion of four glycines between Gly46 and Lys47 (mutant “4Gi-46/47”) has been shown to result in an extremely stable Ca2E2P state (31), in which the A-domain seems to have rotated horizontally from its position in the Ca2E1P state, whereas the inclining (vertical) motion of the top part of transmembrane helix M2 and the A- and P-domains has yet to take place to reach the structure corresponding to the Ca2+-free E2P ground state (32). It is then relevant to ask whether the latter conformational change also affects the nucleotide site, or the modulatory ATP binding site of E2P has already been assembled from the gathering of the A-, P-, and N-domains, before the vertical tilt occurs.
To determine the nucleotide affinity of the stable Ca2E2P state of mutant 4Gi-46/47, phosphorylation of Ca2+-free E2 from the mutant was carried out with inorganic phosphate, followed by supplementation of the microsomes with an excess amount of Ca2+ to saturate the lumenal Ca2+ sites. These experiments were carried out in the presence of the Ca2+ ionophore A23187 to allow Ca2+ access to the lumenal side of the microsomal vesicles. The data under supplemental Fig. S8 confirm the high stability of the phosphoenzyme accumulated with mutant 4Gi-46/47 under the buffer conditions used in the photolabeling assay. The TNP-8N3-ATP and ATP binding data obtained with the Ca2E2P state of mutant 4Gi-46/47 are shown in Fig. 7A. For comparison, we furthermore, measured the TNP-8N3-ATP and ATP affinities of mutant 4Gi-46/47 in the Tg-, MgF-, vanadate-, AlF-, and BeF-complexed states (Fig. 7A), as well as the TNP-8N3-ATP and ATP affinities under E1 conditions in the absence of Ca2+ (Fig. 7B).
FIGURE 7.
Nucleotide binding properties of mutant 4Gi-46/47 in (A) E2·Tg, E2·MgF, E2·vanadate, E2·AlF, E2·BeF, and Ca2E2P states, (B) E1 state, and (C) Ca2E1 state. A, TNP-8N3-ATP (left panels) and ATP (right panels) concentration dependences of photolabeling of mutant 4Gi-46/47 in E2·Tg (diamonds), E2·MgF (open circles), E2·vanadate (squares), E2·AlF (triangles pointing upward), E2·BeF (triangles pointing downward), and Ca2E2P (closed circles) states. Photolabeling was carried out as for Figs. 2, 5, and supplemental Fig. S6. The complexed state of mutant 4Gi-46/47 at the concentrations of metal fluoride, thapsigargin, or vanadate applied is confirmed by the results shown under supplemental Table S1 and Fig. S1. The Ca2E2P state of mutant 4Gi-46/47 was formed by first phosphorylating the enzyme with inorganic phosphate and then adding a high concentration of Ca2+ to saturate the lumenal low affinity Ca2+ sites (see “Experimental Procedures”). The results of supplemental Fig. S8 confirm the high stability of the phosphoenzyme accumulated with mutant 4Gi-46/47 under the conditions used in the photolabeling assay. The nucleotide affinity constants obtained with mutant 4Gi-46/47 in Tg, MgF, vanadate, AlF, and BeF are listed in Tables 1 and 2, and those obtained with the Ca2E2P state are as follows: K0.5 (TNP-8N3-ATP) = 69 ± 3 nm (n = 3); KD(ATP) = 4.6 ± 0.5 μm (n = 3). B, TNP-8N3-ATP (left panels) and ATP (right panels) concentration dependence of photolabeling of wild type and mutant 4Gi-46/47 in the E1 state (Mg2+ present). The affinity constants obtained are listed in Tables 1 and 2, respectively. C, ATP concentration dependence of phosphorylation from [γ-32P]ATP. The maximum phosphorylation level was taken as 100%. The following affinity constants were obtained: wild type, K0.5 = 99 ± 6 nm (n = 5); 4Gi-46/47, K0.5 = 51 ± 8 nm (n = 2).
As shown in Fig. 7A, the Ca2E2P state of mutant 4Gi-46/47 binds TNP-8N3-ATP and ATP with affinities that do not differ significantly from those of the Ca2+-free E2·BeF state of the mutant, implying that any conformational change taking place during the transition to the Ca2+-free E2P ground state does not affect the nucleotide binding site. Furthermore, there was no marked difference between nucleotide affinities of wild type and mutant 4Gi-46/47 in any of the four E2P-like analog states stabilized with MgF, vanadate, AlF, and BeF, suggesting that the mutation does not disturb conformation of the E2P ground state, transition state, or product state appreciably, in accordance with the wild type-like rate of E2P dephosphorylation of the mutant (31).
Under E1 conditions mutant 4Gi-46/47 also displayed an affinity for TNP-8N3-ATP very similar to that of wild type (Fig. 7B, left panel). Surprisingly, however, under these conditions the ATP affinity of mutant 4Gi-46/47 was reduced by more than 3 orders of magnitude, relative to wild type (right panel of Fig. 7B). To assess whether this effect of the 4Gi-46/47 mutation was caused by the absence of Ca2+ in the photolabeling medium (needed to prevent phosphorylation during labeling), we measured ATP dependence of phosphorylation of the Ca2+-saturated E1 from [γ-32P]ATP. As seen in Fig. 7C, the apparent affinity for ATP obtained with 4Gi-46/47 in the phosphorylation assay differed only 2-fold from that of the wild type enzyme. A possible explanation of these findings is that increased flexibility of the lengthened A-M1 linker in the mutant leads to detachment of the A-, P-, and N-domain interactions, resulting in stabilization in the absence of Ca2+ of an open structure similar to that seen in the crystal structure of the nucleotide-free Ca2E1 state (51). In such a state, only the N-domain would be expected to contribute to nucleotide binding, as opposed to tight packing of the nucleotide between the N- and P-domains seen in the Ca2E1·AMPPCP state (50). In accordance with this hypothesis, the ATP affinity of expressed N-domain from Ca2+-ATPase or Na+,K+-ATPase is in the millimolar range (52–54), rather than the typical submicromolar/micromolar affinity range of the intact enzymes. The wild type-like high affinity of mutant 4Gi-46/47 for TNP-8N3-ATP may then suggest that the N-domain generally is the only critical contributor to the binding of the photolabel, thus again illustrating the notion that the photolabel binds in a way rather different from that of ATP, although the binding sites overlap.
Conclusions
By applying the TNP-8N3-ATP Lys492 photolabeling method (20, 33, 34) we have measured nucleotide binding to the various intermediate states occurring during E2P dephosphorylation of wild type and mutant Ca2+-ATPases. The distinct ATP affinities determined for these states in the wild type Ca2+-ATPase suggest a certain degree of flexibility of the modulatory ATP site during E2P dephosphorylation, with a pronounced tightening of the enzyme-nucleotide interaction going from the E2P ground state to the transition state, perpetuation of this tight interaction further into the product state, and then loosening up the site again going into the E2 dephosphoenzyme. Hence, on the basis of the present results the acceleration of dephosphorylation by ATP can be understood in terms of stabilization by ATP binding of the transition and product states in the dephosphorylation reaction. Among the mutations studied here F487S, R560L, and R174A/E interfere with binding of the modulatory nucleotide, whereas I188A interferes mechanistically, possibly disrupting the effect of ATP binding on the optimal positioning of Glu183 for catalysis. The anomalous inhibition of E2P dephosphorylation by ATP seen for mutants K205A and E439A is caused by a reversal of the stabilities of the E2P ground and transition states. The present results fully support a model in which the adenine always slots into the gap between Phe487 and Leu562, whether the ATP is phosphorylating the Ca2E1 form or modulating E2P dephosphorylation, and then the rest of the ATP molecule stretches or folds, as is energetically best depending on domain positions, rather like an anchor and chain.
Supplementary Material
Acknowledgments
We thank Lene Jacobsen and Karin Kracht for expert technical assistance, Dr. Anne Nyholm Holdensen for supplying the cDNA encoding mutant 4Gi-46/47, and Dr. Philippe Champeil (Saclay, France) for generously donating the SR vesicles.
This work was supported in part by grants from the Danish Medical Research Council (to J. P. A.), the Novo Nordisk Foundation (to J. P. A.), and the Danish National Research Foundation (PUMPKIN Centre).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S8.
In our previous studies of TNP-8N3-ATP photolabeling of the Ca2+-ATPase, [γ-32P]TNP-8N3-ATP synthesis and irradiation experiments were performed at the University of Cape Town. In the present study [γ-32P]TNP-8N3-ATP synthesis and irradiation experiments were carried out at Aarhus University. For unidentified reasons, the TNP-8N3-ATP and ATP affinity constants measured in Aarhus with our new photolabeling setup were generally 2–3-fold lower (i.e. higher affinity) than those previously measured in Cape Town. For instance, under E1 conditions in the presence of Mg2+, the affinities of the expressed wild type for TNP-8N3-ATP and ATP measured in Aarhus were 0.29 and 0.14 μm (Tables 1 and 2), respectively, the previously published values being 0.79 and 0.51 μm, respectively. Similarly, in the E2·Tg state, the affinities of the expressed wild type for TNP-8N3-ATP and ATP measured in Aarhus were 62 nm and 10.8 μm (Tables 1 and 2), respectively, the previously published values being 150 nm and 20 μm, respectively (16). We do not presently know the exact reason for the difference.
- Ca2+-ATPase
- Ca2+-transporting adenosine triphosphatase (EC 3.6.3.8)
- AlF
- complex of Al3+ and fluoride
- AMPPCP
- adenosine 5′-(β,γ-methylene)triphosphate
- BeF
- complex of Be2+ and fluoride
- EPPS
- N-2-hydroxyethylpiperazine-N′-3-propanesulfonic acid
- MgF
- complex of Mg2+ and fluoride
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- SR
- sarcoplasmic reticulum vesicles purified from rabbit hind leg muscle
- Tg
- thapsigargin
- TNP-8N3-ATP
- 2′,3′-O-(2,4,6-trinitrophenyl)-8-azidoadenosine 5′-triphosphate
- WT
- wild type Ca2+-ATPase.
REFERENCES
- 1. Toyoshima C. (2009) Biochim. Biophys. Acta 1793, 941–946 [DOI] [PubMed] [Google Scholar]
- 2. Møller J. V., Olesen C., Winther A. M., Nissen P. (2010) Q. Rev. Biophys. 43, 501–566 [DOI] [PubMed] [Google Scholar]
- 3. Clausen J. D., Vilsen B., McIntosh D. B., Einholm A. P., Andersen J. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2776–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olesen C., Sørensen T. L., Nielsen R. C., Møller J. V., Nissen P. (2004) Science 306, 2251–2255 [DOI] [PubMed] [Google Scholar]
- 5. Toyoshima C., Nomura H., Tsuda T. (2004) Nature 432, 361–368 [DOI] [PubMed] [Google Scholar]
- 6. Scofano H. M., Vieyra A., de Meis L. (1979) J. Biol. Chem. 254, 10227–10231 [PubMed] [Google Scholar]
- 7. Wakabayashi S., Shigekawa M. (1990) Biochemistry 29, 7309–7318 [DOI] [PubMed] [Google Scholar]
- 8. McIntosh D. B., Boyer P. D. (1983) Biochemistry 22, 2867–2875 [DOI] [PubMed] [Google Scholar]
- 9. Wakabayashi S., Ogurusu T., Shigekawa M. (1986) J. Biol. Chem. 261, 9762–9769 [PubMed] [Google Scholar]
- 10. Champeil P., Guillain F. (1986) Biochemistry 25, 7623–7633 [DOI] [PubMed] [Google Scholar]
- 11. Bodley A. L., Jencks W. P. (1987) J. Biol. Chem. 262, 13997–14004 [PubMed] [Google Scholar]
- 12. Shigekawa M., Dougherty J. P. (1978) J. Biol. Chem. 253, 1451–1457 [PubMed] [Google Scholar]
- 13. Ariki M., Boyer P. D. (1980) Biochemistry 19, 2001–2004 [DOI] [PubMed] [Google Scholar]
- 14. Champeil P., Riollet S., Orlowski S., Guillain F., Seebregts C. J., McIntosh D. B. (1988) J. Biol. Chem. 263, 12288–12294 [PubMed] [Google Scholar]
- 15. Andersen J. P., Møller J. V. (1985) Biochim. Biophys. Acta 815, 9–15 [DOI] [PubMed] [Google Scholar]
- 16. Clausen J. D., McIntosh D. B., Anthonisen A. N., Woolley D. G., Vilsen B., Andersen J. P. (2007) J. Biol. Chem. 282, 20686–20697 [DOI] [PubMed] [Google Scholar]
- 17. Clausen J. D., McIntosh D. B., Woolley D. G., Andersen J. P. (2008) J. Biol. Chem. 283, 35703–35714 [DOI] [PubMed] [Google Scholar]
- 18. Cable M. B., Feher J. J., Briggs F. N. (1985) Biochemistry 24, 5612–5619 [DOI] [PubMed] [Google Scholar]
- 19. Bishop J. E., Al-Shawi M. K., Inesi G. (1987) J. Biol. Chem. 262, 4658–4663 [PubMed] [Google Scholar]
- 20. Seebregts C. J., McIntosh D. B. (1989) J. Biol. Chem. 264, 2043–2052 [PubMed] [Google Scholar]
- 21. Coll R. J., Murphy A. J. (1991) Biochemistry 30, 1456–1461 [DOI] [PubMed] [Google Scholar]
- 22. Suzuki H., Kubota T., Kubo K., Kanazawa T. (1990) Biochemistry 29, 7040–7045 [DOI] [PubMed] [Google Scholar]
- 23. Reynolds J. A., Johnson E. A., Tanford C. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 3658–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu X., Daiho T., Yamasaki K., Wang G., Danko S., Suzuki H. (2009) J. Biol. Chem. 284, 25190–25198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Danko S., Yamasaki K., Daiho T., Suzuki H. (2004) J. Biol. Chem. 279, 14991–14998 [DOI] [PubMed] [Google Scholar]
- 26. Olesen C., Picard M., Winther A. M., Gyrup C., Morth J. P., Oxvig C., Møller J. V., Nissen P. (2007) Nature 450, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 27. Cantley L. C., Jr., Cantley L. G., Josephson L. (1978) J. Biol. Chem. 253, 7361–7368 [PubMed] [Google Scholar]
- 28. Pick U. (1982) J. Biol. Chem. 257, 6111–6119 [PubMed] [Google Scholar]
- 29. Sagara Y., Wade J. B., Inesi G. (1992) J. Biol. Chem. 267, 1286–1292 [PubMed] [Google Scholar]
- 30. Toyoshima C., Nomura H. (2002) Nature 418, 605–611 [DOI] [PubMed] [Google Scholar]
- 31. Daiho T., Yamasaki K., Danko S., Suzuki H. (2007) J. Biol. Chem. 282, 34429–34447 [DOI] [PubMed] [Google Scholar]
- 32. Daiho T., Danko S., Yamasaki K., Suzuki H. (2010) J. Biol. Chem. 285, 24538–24547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McIntosh D. B., Woolley D. G., Vilsen B., Andersen J. P. (1996) J. Biol. Chem. 271, 25778–25789 [DOI] [PubMed] [Google Scholar]
- 34. Clausen J. D., McIntosh D. B., Vilsen B., Woolley D. G., Andersen J. P. (2003) J. Biol. Chem. 278, 20245–20258 [DOI] [PubMed] [Google Scholar]
- 35. Clausen J. D., Andersen J. P. (2010) J. Biol. Chem. 285, 20780–20792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaufman R. J., Davies M. V., Pathak V. K., Hershey J. W. (1989) Mol. Cell. Biol. 9, 946–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen C., Okayama H. (1987) Mol. Cell. Biol. 7, 2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maruyama K., MacLennan D. H. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 3314–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Champeil P., Büschlen-Boucly S., Bastide F., Gary-Bobo C. (1978) J. Biol. Chem. 253, 1179–1186 [PubMed] [Google Scholar]
- 40. Champeil P., Guillain F., Vénien C., Gingold M. P. (1985) Biochemistry 24, 69–81 [DOI] [PubMed] [Google Scholar]
- 41. Vilsen B., Andersen J. P., MacLennan D. H. (1991) J. Biol. Chem. 266, 16157–16164 [PubMed] [Google Scholar]
- 42. Sorensen T., Vilsen B., Andersen J. P. (1997) J. Biol. Chem. 272, 30244–30253 [DOI] [PubMed] [Google Scholar]
- 43. Vilsen B., Andersen J. P., Clarke D. M., MacLennan D. H. (1989) J. Biol. Chem. 264, 21024–21030 [PubMed] [Google Scholar]
- 44. Forge V., Mintz E., Guillain F. (1993) J. Biol. Chem. 268, 10953–10960 [PubMed] [Google Scholar]
- 45. Chowdhry V., Westheimer F. H. (1979) Annu. Rev. Biochem. 48, 293–325 [DOI] [PubMed] [Google Scholar]
- 46. Kotzyba-Hibert F., Kapfer I., Goeldner M. (1995) Angew. Chem. Int. Ed. Engl. 34, 1296–1312 [Google Scholar]
- 47. Jensen A. M., Sørensen T. L., Olesen C., Møller J. V., Nissen P. (2006) EMBO J. 25, 2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Toyoshima C., Norimatsu Y., Iwasawa S., Tsuda T., Ogawa H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19831–19836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Laursen M., Bublitz M., Moncoq K., Olesen C., Møller J. V., Young H. S., Nissen P., Morth J. P. (2009) J. Biol. Chem. 284, 13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Toyoshima C., Mizutani T. (2004) Nature 430, 529–535 [DOI] [PubMed] [Google Scholar]
- 51. Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000) Nature 405, 647–655 [DOI] [PubMed] [Google Scholar]
- 52. Champeil P., Menguy T., Soulié S., Juul B., de Gracia A. G., Rusconi F., Falson P., Denoroy L., Henao F., le Maire M., Moller J. V. (1998) J. Biol. Chem. 273, 6619–6631 [DOI] [PubMed] [Google Scholar]
- 53. Abu-Abed M., Mal T. K., Kainosho M., MacLennan D. H., Ikura M. (2002) Biochemistry 41, 1156–1164 [DOI] [PubMed] [Google Scholar]
- 54. Hilge M., Siegal G., Vuister G. W., Güntert P., Gloor S. M., Abrahams J. P. (2003) Nat. Struct. Biol. 10, 468–474 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.