Abstract
Although several recent publications have suggested that microRNAs contribute to the pathogenesis of diabetic nephropathy, the role of miRNAs in vivo still remains poorly understood. Using an integrated in vitro and in vivo comparative miRNA expression array, we identified miR-29c as a signature miRNA in the diabetic environment. We validated our profiling array data by examining miR-29c expression in the kidney glomeruli obtained from db/db mice in vivo and in kidney microvascular endothelial cells and podocytes treated with high glucose in vitro. Functionally, we found that miR-29c induces cell apoptosis and increases extracellular matrix protein accumulation. Indeed, forced expression of miR-29c strongly induced podocyte apoptosis. Conversely, knockdown of miR-29c prevented high glucose-induced cell apoptosis. We also identified Sprouty homolog 1 (Spry1) as a direct target of miR-29c with a nearly perfect complementarity between miR-29c and the 3′-untranslated region (UTR) of mouse Spry1. Expression of miR-29c decreased the luciferase activity of Spry1 when co-transfected with the mouse Spry1 3′-UTR reporter construct. Overexpression of miR-29c decreased the levels of Spry1 protein and promoted activation of Rho kinase. Importantly, knockdown of miR-29c by a specific antisense oligonucleotide significantly reduced albuminuria and kidney mesangial matrix accumulation in the db/db mice model in vivo. These findings identify miR-29c as a novel target in diabetic nephropathy and provide new insights into the role of miR-29c in a previously unrecognized signaling cascade involving Spry1 and Rho kinase activation.
Keywords: Cell Death, Diabetes, Kidney, MicroRNA, Rho, Diabetic Nephropathy
Introduction
MicroRNAs (miRNAs)2 comprise a broad class of small non-coding RNAs that negatively regulate gene expression by base-pairing to partially complementary sites in the 3′-untranslated regions (UTR) of specific target mRNAs (1, 2). An emerging body of evidence suggests that miRNAs serve as important therapeutic targets in a wide range of complex human diseases, including cancer and cardiovascular diseases, by targeting multiple transcripts (3–6). Recent studies have also revealed the involvement of miRNAs in diabetic nephropathy (DN) (7–9). However, despite the growing evidence for the regulatory effects of miRNAs in DN, limited information is available on the consequences of modulating miRNAs expression in vivo.
We hypothesized that an unbiased global miRNA expression profiling might reveal novel miRNAs, which may play critical regulatory roles in the pathogenesis of DN. Accordingly, by using an integrated in vitro and in vivo comparative miRNA expression profiling, we identified up-regulated miR-29c as a signature miRNA in the diabetic environment.
Previously published work suggested that down-regulation of miR-29c resulted in cardiac fibrosis (10, 11). In contrast, herein we identified miR-29c as a signature miRNA in the diabetic milieu whose expression was increased in hyperglycemic conditions both in vitro and in vivo. Thus, our objective was to explore the role of increased miR-29c expression in DN.
We found that miR-29c targets Sprouty homolog 1 (Spry1) in hyperglycemic conditions. Spry1 is a cytoplasmic protein that plays a critical role in kidney development (12–14) and is primarily known to inhibit the Ras/MEK/ERK pathway (15, 16). Spry1 and its related proteins have also been implicated as negative regulators of RhoA and its downstream effector Rho kinase through the non-canonical Wnt signaling (17–19). Importantly, Rho kinase plays a key role in DN (20–23). Published studies from our laboratory and others have shown that pharmacological inhibition of Rho kinase in experimental models of diabetes results in a significant reduction in albuminuria and accumulation of glomerular matrix accumulation (22, 23). At the cellular level, Rho kinase has been implicated in cell proliferation, fibrosis, and apoptosis via multiple signaling pathways (24–26). However, the mechanisms that regulate Rho kinase activation in DN are not fully understood.
Here, we report the identification of miR-29c as a signature miRNA in DN. Together with functional studies, our results establish the role of miR-29c as a key miRNA governing kidney remodeling through a coordinated coupling of Spry1 with Rho kinase activation.
EXPERIMENTAL PROCEDURES
Cell Lines
Conditionally immortalized mouse podocytes were kindly provided by Dr. Jochen Reiser (University of Miami, Miami, FL) and cultured as reported previously (27). Briefly, podocytes were cultured on BD BioCoat Collagen I plates (BD Biosciences) at 33 °C in the presence of 20 units/ml mouse recombinant IFN-γ (Invitrogen) to enhance expression of a thermosensitive T antigen. To induce differentiation, podocytes were maintained at 37 °C without IFN-γ for 10–12 days. A conditionally immortalized renal microvascular endothelial cell line was a kind gift of Dr. Robert Langley (M.D. Anderson Cancer Center, University of Texas, Houston, TX) (28). Mouse mesangial cells (CRL-1927) were purchased from ATCC and cultured as reported previously (22).
miRNA Extraction and Microarray Analysis
miRNAs were extracted using the miRNeasy mini kit (Qiagen) according to the manufacturer's instructions. The miRNA microarray was performed using μParaflo microfluidic chips (LC Sciences, Houston, TX). Data were analyzed by normalizing the signals using a LOWESS filter (locally weighted regression) (29). For two-color experiments, the ratio of the two sets of detected signals (log2-transformed, balanced) and p values of the Student's t test were calculated. Those with p < 0.05 were considered as differentially expressed miRNAs.
Real-time RT-PCR and Northern Blot for miRNAs
miRCURY LNA universal RT microRNA PCR system (Exiqon, Woburn, MA) was used in conjunction with qPCR and SYBR Green supermix (Bio-Rad) for quantification of miRNA transcripts according to the manufacturer's instructions. U6 snRNA was used as an internal control with the following primers: 5′-CGCTTCGGCAGCACATATAC-3′ (forward) and 5′-TTCACGAATTTGCGTGTCAT-3′ (reverse). The reactions were incubated in a 96-well plate at 95 °C for 3 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 1 min. Individual samples were run in triplicate, and each experiment was repeated at least three times. Relative gene expression was calculated using the 2−ΔΔCT method (30). Northern blots were carried out using [γ-32P]ATP (PerkinElmer Life Sciences) end-labeled miRNA locked nucleic acid (LNA) probes (Exiqon) (31).
Computational Targeted Gene Predictions of miR-29c
The full-length mRNA of mouse Spry1 (NM_011896) was obtained from the National Center for Biotechnology Information (NCBI) database. The miRNA sequence database (miRBase) was obtained from the University of Manchester. Three separate algorithms (miRanda, TargetScan, and PicTar) were used to assess potential targets sites for miR-29c. The RNA Hybrid program (32) was used to predict the secondary structure of the RNA/miRNA duplex.
Plasmids, Mutagenesis, Transfection, and Luciferase Reporter Assays
The 3′-UTR of the mouse Spry1 gene (NM_011896) was amplified from podocyte genomic DNA by PCR using the HotMaster Taq DNA polymerase (5 PRIME), with the following primers: 5′-GTCTCGAGCGGTGTTGGTCTTCACATCAGA-3′ (forward) and 5′-GAGAATTCAGACATGAGTACATTTCAACAGTC-3′ (reverse). The 1048-bp PCR product was cloned between the XhoI and EcoRI site of the luciferase reporter vector 3.1-luc, kindly provided by Dr. Ralph Nicholas (Dartmouth Medical School, Hanover, NH) (33), to generate 3.1-luc-Spry1. Putative miR-29c binding site UGGUGCU (nucleotides 773–779) was mutated into GAUGUGC by oligonucleotide-directed PCR. The open reading frame (without the 3′-UTR) of mouse Spry1 gene was amplified from podocyte genomic DNA by PCR with the following primers: 5′-GTGAATTCGATTCCCCAAGTCAGCATGGCGCCAC-3′ (forward) and 5′-ACGGATCCTCATGACAGTTTGCCCTGAGCTTGA-3′ (reverse). The 942-bp PCR product was cloned between the EcoRI and SalI site of a modified FLAG-tagged mammalian expression vector pRK5 (34) to generate FLAG-Spry1. Mouse miR-29c precursor was amplified from mouse podocyte genomic DNA by PCR using AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen), with the following primers: 5′-GACTCGAGGACTGAGATCCATGGAGCACC-3′ (forward) and 5′-GAGAATTCGACTTGAAGTTAGGAACTGGATC-3′ (reverse). The 310-bp PCR product was cloned between the XhoI and EcoRI site of lentiviral vector pLB2 CAG P2Gm (Addgene, Cambridge, MA) to generate pLB2-CAG-miR29c. All constructs were verified by sequencing. The pEGP-miR-29c was obtained from Cell Biolabs (San Diego, CA). The luciferase reporter vector pGL4.10 [luc2] was from Promega.
Pre-miRNA precursor and anti-miR miRNA molecules were purchased from Ambion (Austin, TX). These include: pre-miR negative control 1 (AM17110); pre-miR-29c (AM17100); anti-miRNA inhibitor negative control 1 (AM17010); and anti-miR-29c (AM10518). Spry1 siRNA was purchased from Santa Cruz Biotechnology. miRNA mimics, miRNA inhibitors, and siRNAs were transfected into podocytes using Lipofectamine 2000 (Invitrogen) at a final concentration of 30 nm.
For experiments using 3.1-luc luciferase reporter constructs in vitro, 2.0 × 105 podocytes were plated onto 10-cm plates and well differentiated for 7–10 days. 4.0 μg of 3.1-luc empty vector or 3.1-luc-Spry1 construct, 200 ng of pSV-β-gal control vector (Promega), and 30 nm pre-miRNA mimics were transfected using Lipofectamine 2000 (Invitrogen). Following 48 h of transfections, luciferase activity was measured using a Dual-Glo luciferase reporter assay kit (Promega) on a FLUOstar Omega luminometer (BMG Labtech, Cary, NC) as reported previously (35), using β-gal as internal control.
Lentivirus-mediated miR-29c Expression in Podocytes and Mesangial Cells
Lentiviral miR-29c and control lentivirus were prepared from lentiviral construct pLB2-CAG-miR29c or empty pLB2-CAG-P2Gm vector by the Gene Vector Core Laboratory at the Baylor College of Medicine. Lentiviral constructs were then used to infect undifferentiated podocytes with 0.8 mg/ml Polybrene (Sigma) and were subsequently selected with 1 μg/ml puromycin (Sigma) in the presence of 20 units/ml IFN-γ at 33 °C. Mouse mesangial cells were also infected with the same lentiviral constructs and selected with 1 μg/ml puromycin at 37 °C. Overexpression of miR-29c in the stable cells was confirmed by real-time qPCR analysis of miR-29c.
RT-qPCR
Total RNA was extracted using TRIzol Reagent (Invitrogen). First-strand cDNAs were generated using iScript cDNA Synthesis Kit (Bio-Rad). SYBR Green-based qPCR on a DNA Engine Opticon (Bio-Rad) were used to analyze the relative expression levels of the following genes with the following primer sets: fibronectin (Fn1), 5′-GCGGTTGTCTGACGCTGGCT-3′ (forward) and 5′-TGGGTTCAGCAGCCCCAGGT-3′ (reverse); GAPDH, 5′-CCTTCATTGACCTCAACTAC-3′ (forward) and 5′-GGAAGGCCATGCCAGTGAGC-3′ (reverse).
Immunofluorescence Microscopy
Immunofluorescent staining was performed as described previously (9, 36). Briefly, expression of miR-29c and endogenous Spry1 in the stable Lenti-miR-29c-infected podocytes were detected using GFP and anti-Spry1 antibodies (Santa Cruz Biotechnology). Coverslips were imaged on an Applied Precision SoftWoRx Image using a deconvolution restoration microscope. For immunostaining, rabbit anti-Spry1 and anti-fibronectin antibodies (Santa Cruz Biotechnology) were used. Sections were incubated at 4 °C overnight. Sections were imaged on a Zeiss LSM 510 inverted laser scanning microscope.
Animal Studies
All animal studies were conducted according to the “Principles of Laboratory Animal Care” (National Institutes of Health publication number 85023, revised 1985) and the guidelines of the Institutional Animal Care and Use Committee (IACUC) of the Baylor College of Medicine. The diabetic db/db mice and their control littermates db/m mice were obtained from The Jackson Laboratory (Bar Harbor, ME). All animals were maintained on a normal chow diet and housed in a room with a 12:12-h light/dark cycle and an ambient temperature of 22 °C. Kidney glomeruli were isolated by perfusion using Dynabeads (Invitrogen) as described previously (37). For biochemical and histological analyses, mice were housed in individual metabolic cages for collection of urine. Urinary albumin and creatinine were measured using the Albuwell M and Creatinine Companion assay kits (Exocell, Philadelphia, PA), respectively. Blood glucose was measured after a 12-h fast using the OneTouch UltraSmart blood glucose meter (Lifescan, Milpitas, CA). For morphometric studies, the kidneys were fixed in 10% neutral buffered formalin and subsequently embedded in paraffin. The 4-μm sections of paraffin-embedded tissues were stained with periodic acid-Schiff. Light microscopic views of 50 consecutive glomerular cross-sections per mouse were scanned into a computer. Mesangial matrix was quantified in a blinded fashion using an image analysis system (MetaMorph version 6.1; Universal Imaging). The mesangial matrix index was calculated as the ratio of mesangial area to glomerular area × 100 (percentage of area) as described previously (22).
miR-29c ASO Microinjection in Vivo
Chemically modified antisense RNA oligonucleotides (ASOs) complementary to the mature miR-29c sequence were used to inhibit miR-29c activity in vivo as suggested previously (10, 38, 39). The miR-29c ASO sequence was: 5′-AUAACCGAUUUCAAAUGGUGCUA-3′; the miR-29c mismatch control sequence was: 5′-AUAUCCCAUAUCUAAUCGAGCUA-3′. All oligonucleotides were 2′-O-methyl-modified and purified through HPLC (Sigma-Genosys), and all oligonucleotides were free of endotoxin for in vivo delivery. miR-29c ASO or miR-29c mismatch control oligonucleotides were injected at 80 μg/g of body weight intraperitoneally into 8-week-old db/db mice (n = 5) every other week (10, 40, 41). Body weight and blood glucose were monitored, and 24-h urine samples were collected every week. Mice were sacrificed after 12 weeks of treatment.
Assessment of Apoptosis and Mesangial Matrix Expansion
TUNEL assays were carried out using the ApopTag fluorescein in situ apoptosis detection kit (Millipore) as described previously (42). Caspase-3 activation assay was performed using the Caspase-3 assay kit (Millipore) in accordance with the manufacturer's instructions. Analysis of apoptosis in live cells by flow cytometry was performed as described previously (43). Briefly, transfected podocytes were serum-starved overnight before treating with normal or high glucose for 36 h. Cells were then resuspended in 0.5 ml of FACS binding buffer (Hanks' balanced salt solution containing 5 mm Ca2+ and Mg2+ with 1% BSA) and counterstained with Sytox Red and annexin V-phycoerythrin (Invitrogen) for 15 min at room temperature. At last, samples were run on a CantoII flow cytometer (BD Biosciences).
Rho Kinase Activation Assay
Rho kinase activity was measured as described previously (22). Briefly, kidney cortex or podocytes were lysed in radioimmune precipitation buffer (Sigma). To assess Rho kinase activity, 500 ng of myosin phosphatase target subunit 1 (MYPT1) (654-880, Millipore) and Tris/ATP (final concentration: 1 mm ATP, 50 mm Tris-HCl, 0.1 mm EGTA, 0.1 m DTT, 10 mm MgCl2) were added to the lysate. The lysates were incubated for 30 min at 30 °C. The reaction was stopped by adding 2× Laemmli sample buffer (Bio-Rad) and by subsequent boiling for 5 min. Phosphorylation of MYPT1 by Rho kinase was detected by running the samples on an SDS-PAGE gel and by subsequent immunoblotting with an anti-phospho-MYPT1 (Thr-850, Millipore). As a loading control, internal β-actin was detected by immunoblotting with an anti-β-actin antibody (Sigma). Appropriate DyLight Fluor-conjugated secondary antibodies (Thermo Fisher Scientific) were chosen for signal detection on an Odyssey infrared imager (LI-COR Biosciences, Lincoln, NE).
Statistical Analysis
All data are shown as mean ± S.E. Statistical significance was assessed by performing analysis of variance followed by the Tukey-Kramer post hoc analysis for multiple comparisons using an α value of 0.05 in GraphPad Prism software (San Diego, CA). Western blot signals were quantitated using software National Institutes of Health ImageJ version 1.42q.
RESULTS
Comparative miRNA Profiling Reveals Up-regulation of miR-29c in Hyperglycemic Conditions Both in Vitro and in Vivo
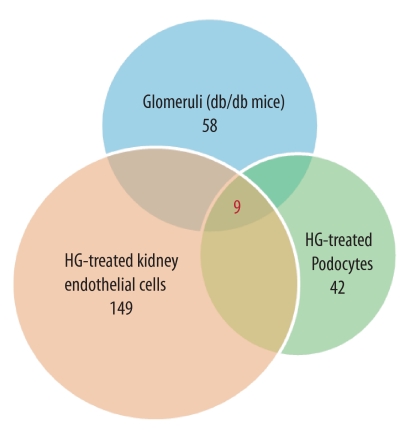
We have previously described an unbiased approach to identify regulatory miRNAs in DN by examining the expression profiles of miRNAs in samples obtained from glomeruli from db/db mice, an established mouse model of type 2 diabetes, and in kidney cell lines exposed to hyperglycemic conditions (9). Here, by employing an integrated in vitro and in vivo approach, we analyzed the expression of miRNAs at several time points (6, 12, and 24 h) following stimulation with high glucose (25 mm) in cultured podocytes and kidney microvascular endothelial cells as well as in glomeruli from diabetic db/db mice at 24 weeks of age when the animals develop biochemical and histological changes consistent with DN. We found that nine miRNAs were differentially up-regulated in our experimental conditions both in vitro and in vivo (Fig. 1, Table 1 and supplemental Table 1). Among these up-regulated miRNAs, we focused on miR-29c, which was differentially expressed in all samples with a significant fold of induction in our experimental models both in vitro and in vivo.
FIGURE 1.
miRNA expression profiling under hyperglycemic conditions in vitro and in vivo. Comparative microarray analysis indicated that nine miRNAs were differentially up-regulated within all samples. Specifically, 58 miRNAs were up-regulated in db/db glomeruli, 149 miRNAs were up-regulated in high glucose (HG)-treated kidney microvascular endothelial cells, and 42 miRNAs were up-regulated in podocytes treated with HG (25 mm) for 24 h as compared with normal glucose conditions.
TABLE 1.
Identification of nine miRNAs that are differentially up-regulated in db/db glomeruli and high glucose-treated kidney microvascular endothelial cells and cultured podocytes
The relative expression of the up-regulated miRNAs is shown. All data are expressed as the mean signal intensity. Log2 represents mean signal intensity of Group2/Group1. miRNAs are listed in the order of their log2 differences based on values obtained from kidney glomeruli (from high to low).
| miRNA ID | Kidney glomeruli |
Mouse kidney microvascular endothelial cells |
Mouse cultured podocytes |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group1 (db/m) | Group2 (db/db) | Log2 | p | Group1 (NG) | Group2 (HG) | Log2 | p | Group1 (NG) | Group2 (HG) | Log2 | p | |
| mmu-miR-192 | 361 | 1,902 | 2.40 | 0.002 | 90 | 190 | 2.40 | 0.004 | 133 | 492 | 1.88 | 0.050 |
| mmu-miR-29c | 395 | 1,693 | 2.10 | 0.002 | 67 | 693 | 3.37 | 0.005 | 292 | 417 | 0.51 | 0.049 |
| mmu-miR-26b | 168 | 387 | 1.21 | 0.005 | 1,132 | 1,914 | 0.76 | 0.003 | 4,826 | 8,336 | 0.79 | 0.042 |
| mmu-miR-1196 | 189 | 318 | 0.75 | 0.016 | 162 | 411 | 1.35 | 0.001 | 282 | 793 | 1.49 | 0.000 |
| mmu-miR-1195 | 165 | 266 | 0.69 | 0.004 | 286 | 741 | 1.38 | 0.006 | 580 | 750 | 0.37 | 0.032 |
| mmu-miR-200b | 142 | 217 | 0.61 | 0.015 | 95 | 503 | 2.40 | 0.005 | 1,871 | 3,756 | 1.01 | 0.007 |
| mmu-let-7g | 1,235 | 1,890 | 0.61 | 0.007 | 2,162 | 3,269 | 0.60 | 0.010 | 935 | 2,071 | 1.15 | 0.023 |
| mmu-miR-200a | 182 | 265 | 0.54 | 0.016 | 212 | 547 | 1.37 | 0.027 | 322 | 990 | 1.62 | 0.017 |
| mmu-miR-705 | 557 | 716 | 0.36 | 0.013 | 243 | 336 | 0.47 | 0.006 | 11,928 | 20,979 | 0.81 | 0.001 |
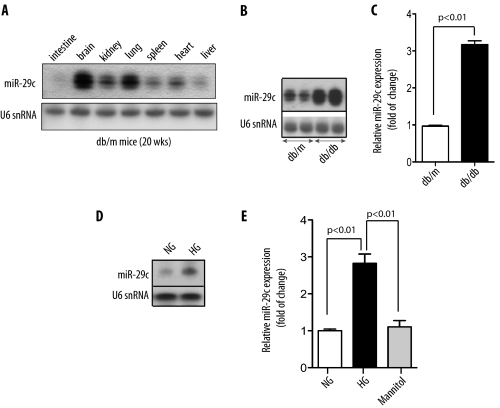
Northern blot analysis of multiple mouse tissues from control db/m mice for miR-29c indicated that the expression of miR-29c was highest in the brain and lung with significant expression patterns in the kidney and the heart (Fig. 2A). Next, we validated our preliminary profiling data on miR-29c in the db/db model of DN by Northern blot. miR-29c levels were significantly increased in the kidney glomeruli from db/db diabetic mice as compared with their littermates db/m mice (Fig. 2B). Northern blot analysis also indicated increased miR-29c expression in several other tissues in db/db mice (supplemental Fig. S1). Similarly, in the streptozotocin-induced murine model of DN, miR-29c expression in the kidney was also significantly increased (supplemental Fig. S2). Using qPCR analysis, we confirmed that the expression level of miR-29c was about 3-fold higher in the kidney cortices of db/db mice as compared with that in db/m control mice (Fig. 2C).
FIGURE 2.
Up-regulation of miR-29c expression in vivo and in vitro. A, expression of miR-29c in different tissues of a control db/m mouse as detected by Northern blot analysis. U6 snRNA served as a loading control. B, Northern blot analysis shows representative results from glomerular miR-29c expression in control db/m and diabetic db/db mice. C, real-time qPCR analysis shows glomerular miR-29c expression in control db/m and diabetic db/db mice. Measured transcript levels were normalized to U6 snRNA expression. Data are shown as mean ± S.E. (n = 5). D, Northern blot analysis shows representative results of miR-29c expression in cultured podocytes treated with normal glucose (NG, 5 mm) or HG (25 mm) for 24 h. E, real-time qPCR analysis shows miR-29c expression in podocytes treated with high glucose (25 mm) for 24 h as compared with podocytes treated with either normal glucose (5 mm) or mannitol (25 mm) for 24 h. Measured transcript levels were normalized to U6 snRNA expression. Data are shown as mean ± S.E. (n = 3).
We also validated the expression levels of miR-29c in kidney podocytes and kidney microvascular endothelial cells stimulated with high glucose (25 mm). Northern blot analysis indicated that miR-29c was significantly increased following 24 h of exposure to high glucose (Fig. 2D). Similarly, real-time qPCR analysis demonstrated that high glucose treatment led to ∼3-fold increase in miR-29c expression in podocytes and kidney microvascular endothelial cells, whereas mannitol had no effect on miR-29c expression (Fig. 2E and supplemental Fig. S3). Taken together, these findings validated our initial observations confirming that miR-29c is up-regulated in response to hyperglycemic conditions both in vitro and in vivo.
Spry1 Is the Target of miR-29c under Hyperglycemic Conditions
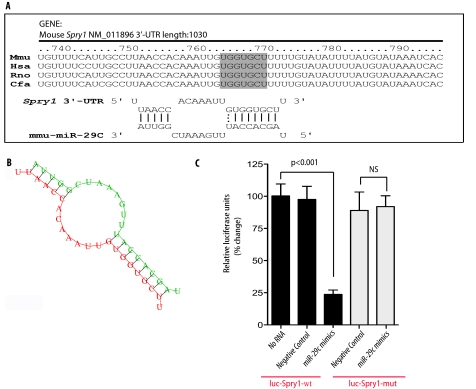
We next set out to identify the potential target(s) of miR-29c under hyperglycemic conditions. There were at least six previously reported targets for the miR-29 family members, which include: DNA methyltransferase 3 (44), Mcl-1 (45, 46), collagen (10, 47, 48), phosphatidylinositol 3-kinase (49), CDK6 (50), and YY1 (51). By using TargetScan v5.1 and PicTar target prediction algorithms, we identified Sprouty homolog 1 (Spry1) as another potential target of miR-29, which has not been previously experimentally validated and was known to play a critical role in the kidney development and remodeling (12–14). We found that there was a highly conserved binding site for miR-29c in the 3′-UTR region of Spry1 in several species (Fig. 3A, upper panel). Fig. 3A also shows the alignment between miR-29c and 3′-UTR of mouse Spry1. The 3′-UTR of the mouse Spry1 gene contains a 7-mer (UGGUGCU), which is perfectly complementary to the seed region of miR-29c. The minimum free energy predicted for hybridization with the Spry1 3′-UTR and miR-29c at this site was ΔG = −21.6 kcal mol−1, consistent with an authentic miRNA targeting (52). As predicted by RNA Hybrid analysis, miR-29c and its binding site in Spry1 could potentially form a very stable secondary structure (Fig. 3B). Thus, we hypothesized that Spry1 might serve as a target for miR-29c in the diabetic milieu.
FIGURE 3.
miR-29c is predicted to target Spry1 gene. A (upper panel), a predicted miR-29c target site resides at nucleotides 773–779 (shown in the gray box) of the mouse Spry1 3′-UTR and is highly conserved in several species. Lower panel, sequence alignment of miR-29c with the mouse Spry1 3′-UTR. Mmu, Mus musculus; Hsa, Homo sapiens; Rno, Rattus norvegicus; and Cfa, Canis lupus familiaris. B, miR-29c can potentially form a strong secondary structure with the target sequence of 3′-UTR of Spry1 (predicted by the RNA Hybrid program). C, differentiated podocytes were co-transfected with plasmid 3.1-luc-Spry1 wild type 3′-UTR (luc-Spry1-wt) or miR-29c mutant 3′-UTR (luc-Spry1-mut) and the indicated miRNA mimics. Luciferase activities were normalized to β-gal activities. Results were obtained from three independent experiments. Data are shown as mean ± S.E.
To address whether binding of miR-29c to Spry1 mRNA leads to its translational suppression, we cloned 3′-UTR of the mouse Spry1 gene into luciferase reporter vector 3.1-luc (33). We also generated a mutant where the putative miR-29c binding site UGGUGCU was mutated into GAUGUGC. Transient co-transfection of miR-29c mimics with luciferase expression plasmids resulted in a significant repression of luciferase reporter gene in podocytes, whereas transfection of cells with miRNA control mimics (negative control) did not have any effect on the expression of luciferase (Fig. 3C). However, mutations within the seed sequence binding site of Spry1 abrogated the effect of miR-29c mimics (Fig. 3C). These results suggest that miR-29c directly targets and inhibits Spry1 by binding to the 3′-UTR binding site of Spry1.
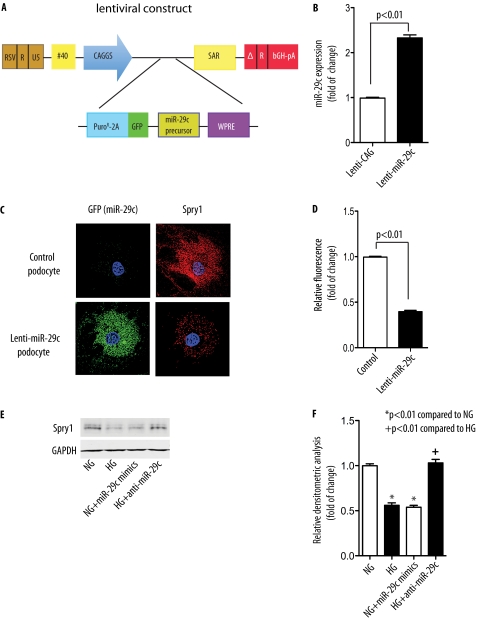
To further provide evidence that Spry1 is a target of miR-29c, we generated a stable podocyte cell line overexpressing miR-29c (Lenti-miR-29c) by taking advantage of a recently described lentivirus system (53). To this end, mouse primary miR-29c transcript was subcloned into the 3′-UTR of a dual selection cassette that consisted of a puromycin-N-acetyl-transferase gene fused to a segment of the foot-and-mouth disease virus 2A peptide followed by GFP. A woodchuck hepatitis post-transcriptional regulatory element (WPRE) was also incorporated for optimal gene expression (54). Expression of the whole Puro-2A-GFP-miR-29c cassette is driven in this system by the CAGGS (CMV enhancer-chicken β-actin-globin intron promoter) (Fig. 4A). Using RT-PCR analysis, we confirmed that the expression of miR-29c in this stable podocyte cell line was ∼2.3-fold higher than that in the control podocytes infected with the lentiviral vector alone (Lenti-CAG) (Fig. 4B). Using immunofluorescence microscopy, we addressed whether miR-29c overexpression had an inhibitory effect on endogenous Spry1 protein expression. As shown in Fig. 4C, we observed significantly reduced fluorescent immunoreactivity of Spry1 in the stable podocyte cell line infected with lentiviral miR-29c as compared with control podocytes (Fig. 4, C and D). This result suggests that miR-29c down-regulates Spry1 expression in podocytes.
FIGURE 4.
miR-29c targets Spry1. A, schematic diagram of lentiviral miR-29c construct. CAGGS, CMV enhancer-chicken β-actin-globin intron promoter; WPRE, woodchuck hepatitis post-transcriptional regulatory element. B, real-time qPCR analysis of miR-29c expression in stable podocytes infected with empty vector lentivirus (Lenti-CAG) or miR-29c lentivirus (Lenti-miR-29c). Measured transcript levels were normalized to U6 snRNA expression. Data are shown as mean ± S.E. (n = 3). C, expression of Spry1 (red) in control podocytes or stable podocytes infected with miR-29c lentivirus (Lenti-miR-29c) harboring GFP marker (green) was assessed by deconvolution microscopy. Original magnification, ×400. D, quantitative analysis based on fluorescence intensity of Spry1 (n = 3). E, podocytes were transfected with miR-29c mimics or inhibitor as indicated and treated with HG (25 mm) for 24 h as compared with normal glucose. Spry1 expression in total cell lysates was analyzed by immunoblot. GAPDH served as a loading control. F, densitometric analysis of Spry1 expression in E (n = 3).
We next used Lenti-miR-29c-infected podocytes to address whether miR-29c overexpression down-regulates Spry1 protein expression by using Western blot analysis. We found that high glucose treatment (25 mm) significantly decreased Spry1 protein levels in Lenti-CAG-infected podocytes, whereas overexpression of miR-29c with Lenti-miR-29c reduced Spry1 protein in both normal glucose-treated and high glucose-treated podocytes (supplemental Fig. S4). To further examine the effect of miR-29c on Spry1, we next transfected podocytes with miR-29c mimics or inhibitors and then treated podocytes with high glucose (25 mm for 24 h). Spry1 immunoblot of the whole cell lysate confirmed that high glucose treatment, as well as miR-29c mimics, significantly down-regulated Spry1 expression (Fig. 4, E and F). Importantly, high glucose-induced down-regulation of Spry1 was reversed when podocytes were co-transfected with a miR-29c inhibitor (Fig. 4, E and F). Taken together, these findings indicate that high glucose down-regulates Spry1 protein expression through a miR-29c-regulated pathway.
miR-29c Promotes Cell Apoptosis and Increased Fibronectin Synthesis by Targeting Spry1
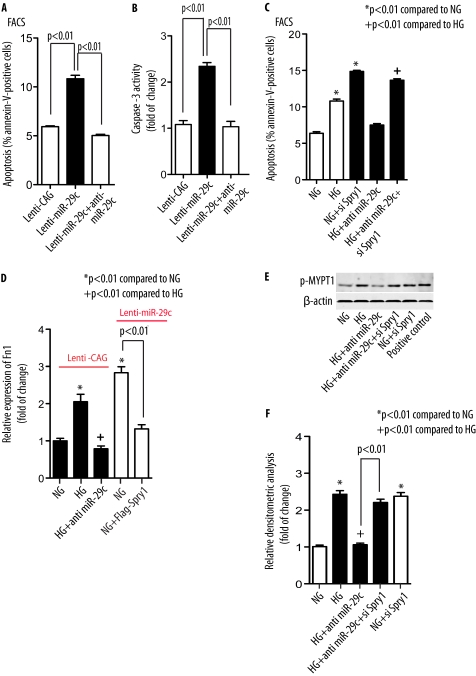
Because DN is characterized by increased frequency of apoptosis in podocytes (55, 56), we asked whether miR-29c might be an active component of the apoptotic pathway in podocytes. Stable transfection of miR-29c with Lenti-miR-29c in podocytes resulted in more than 2-fold increase in apoptosis as assayed by flow cytometry (Fig. 5A and supplemental Fig. S5A). However, the percentage of apoptotic cells induced by Lenti-miR-29c decreased to almost basal levels when cells were transfected with a miR-29c inhibitor, indicating a potent pro-apoptotic effect of miR-29c in podocytes. Consistent with these findings, the increase in caspase-3 activity in Lenti-miR-29c-infected podocytes was blocked when podocytes were transfected with a miR-29c inhibitor (Fig. 5B).
FIGURE 5.
miR-29c promotes cell apoptosis, promotes accumulation of fibronectin, and activates Rho kinase by targeting Spry1. Lenti-CAG and A and B, Lenti-miR-29c podocytes were transfected with miR-29c inhibitor as indicated under NG (5 mm) conditions. Cell apoptosis was analyzed by FACS (A) and caspase-3 activity assay (B). Results were obtained from three independent experiments. Data are shown as mean ± S.E. C, control podocytes were transfected with Spry1 siRNA (siSpry1, 30 nm) and/or miR-29c inhibitor as indicated and treated with HG (25 mm) for 36 h. Apoptosis in the cells was analyzed by FACS. Results were obtained from three independent experiments. Data are shown as mean ± S.E. D, mesangial cells stably infected with empty vector lentivirus (Lenti-CAG) or miR-29c lentivirus (Lenti-miR-29c) were transfected with miR-29c inhibitor or FLAG-Spry1 as indicated and treated with HG (25 mm) for 48 h as compared with NG. Expression of fibronectin (Fn1) in the cells was analyzed by real-time qPCR normalized to GAPDH. Data are shown as mean ± S.E. (n = 3). E, podocytes were transfected with Spry1 siRNA (si Spry1, 30 nm) and/or miR-29c inhibitor as indicated and treated with HG (25 mm) for 36 h as compared with NG. Rho kinase activity in cell lysates was measured as described under “Experimental Procedures.” β-Actin served as a loading control. F, densitometric analysis of Rho kinase activity in E (n = 3).
Because our findings indicated that Spry1 was a target of miR-29c, we also examined the involvement of Spry1 in miR-29c-mediated high glucose-induced cell apoptosis. We hypothesized that Spry1 might mediate the pro-apoptotic effect of miR-29c in the diabetic environment. Both hyperglycemia (25 mm for 36 h) and knocking down of Spry1 by using Spry1 siRNA increased apoptosis in podocytes, whereas a specific inhibitor of miR-29c significantly reduced high glucose-induced apoptosis (Fig. 5C and supplemental Figs. S5B and S6). Importantly, when high glucose-treated podocytes were transfected with both miR-29c inhibitor and Spry1 siRNA, the effect of miR-29c inhibitor on apoptosis was largely reversed (Fig. 5C), highlighting the role of Spry1 in high glucose-induced podocyte apoptosis.
DN is also characterized by excessive accumulation of extracellular matrix proteins in the renal glomerulus. Several studies have previously found altered synthesis of various mesangial extracellular matrix proteins, especially of fibronectin, type-I, -III, and -IV collagens, in DN (57, 58). Thus, we next asked whether the miR-29c/Spry1 pathway may be involved in the increased synthesis of fibronectin in mesangial cells, a key feature of DN. High glucose increased expression of fibronectin as determined by qPCR in mesangial cells (Fig. 5D). However, the effect of high glucose was reversed with miR-29c inhibition. Interestingly, in stable Lenti-miR-29c-infected mesangial cells, mRNA expression of fibronectin was also significantly increased under normal glucose conditions (Fig. 5D), suggesting that miR-29c overexpression increases fibronectin expression. In contrast, the effect of miR-29c was largely blocked when Lenti-miR-29c mesangial cells were co-transfected with FLAG-Spry1 plasmid (Fig. 5D). Taken together, our findings indicate that miR-29c functionally promotes cell apoptosis and increased fibronectin synthesis via a Spry1-dependent pathway.
miR-29c Activates Rho Kinase by Inhibiting Spry1
We and others have previously shown that RhoA and its downstream effector Rho kinase play critical roles in mediating several key pathways critical in the pathogenesis of DN (22, 23, 59, 60). Rho kinase is also known to enhance cell apoptosis (61) and fibronectin matrix assembly (62). Because previous studies have indicated that Spry1 and its related proteins can inhibit RhoA and Rho kinase activation (19), we further characterized the effect of miR-29c and Spry1 on the level of Rho kinase activation. Rho kinase activity was quantified by the extent of MYPT1 phosphorylation with Western blot as described previously (63). We observed a 2-fold increase in Rho kinase activity following exposure of podocytes to high glucose treatment (25 mm for 24 h) (Fig. 5E). However, miR-29c inhibition abrogated high glucose-induced Rho kinase activation. This suggested that high glucose-induced Rho kinase activation might be mediated through up-regulation of miR-29c. Importantly, knockdown of Spry1 rescued the effect of anti-miR-29c on Rho kinase activation (Fig. 5E). To confirm the effect of Spry1 on Rho kinase activity, we also tested the consequence of Spry1 knockdown by using Spry1 siRNA on Rho kinase activation. Under normal glucose conditions, Spry1 knockdown also resulted in ∼2-fold increase in Rho kinase activity (Fig. 5, E and F).
In Vivo Knockdown of miR-29c Ameliorates Progression of DN
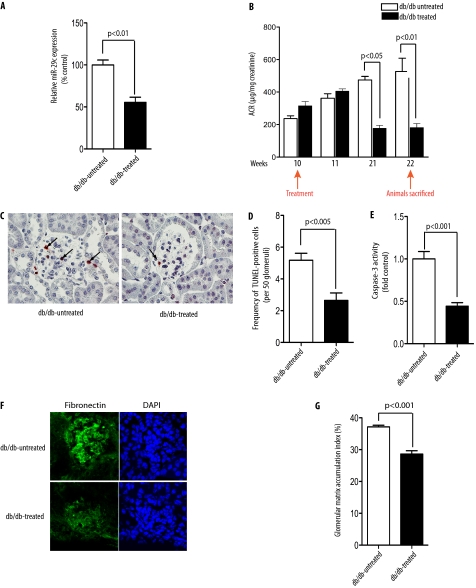
Based on our data in vitro, we hypothesized that inhibition of miR-29c in vivo might ameliorate progression of DN. To test this hypothesis, we used a chemically modified ASO (miR-29c ASO) to knock down miR-29c expression. ASOs have been recently used in several published studies as a tool to knock down the functional role of miRNAs in vivo (10, 39, 40, 64). Thus, to assess the effect of miR-29c in DN in vivo, we injected miR-29c ASO into 8–10-week-old db/db mice intraperitoneally (10, 40, 64). The control db/db diabetic animals were treated either with miR-29c mismatch RNA oligonucleotides or with PBS. Mice were treated every 2 weeks for 12 weeks and were sacrificed at 22 weeks of age. As shown in Fig. 6A, miR-29c expression in the kidney cortices of miR-29c ASO-treated mice was knocked down by ∼50% after 12 weeks (Fig. 6A). Real-time qPCR analysis also indicated efficient knockdown of miR-29c expression in different tissues (supplemental Fig. S7). Importantly, we found that although albuminuria in the control db/db group increased, administration of miR-29c ASO led to a marked improvement in albuminuria (Fig. 6B). These results indicate that the in vivo knockdown of miR-29c ameliorates the progression of DN.
FIGURE 6.
In vivo inhibition of miR-29c ameliorates progression of DN. A, miR-29c expression level was significantly down-regulated in the kidney cortices of miR-29c ASO-treated db/db mice as compared with control db/db mice. Representative RT-qPCR resulting from three different experiments is shown (n = 3). Expression levels were normalized to U6 snRNA expression. Data are shown as mean ± S.E. B, injection of miR-29c ASO led to improved proteinuria in db/db mice. Urine albumin and creatinine from miR-29c ASO-treated and control db/db mice (n = 5/group) were measured at the indicated weeks of age, and the relative albumin/creatinine ratio (ACR) was calculated. C, representative TUNEL assay to detect the apoptosis in miR-29c ASO-treated db/db mice as compared with control db/db mice. Apoptotic cells in the glomeruli are indicated by arrows. D, quantitative analysis of apoptosis in miR-29c ASO-treated mice as compared with control as shown in C (n = 3). E, caspase-3 activity in the kidney cortex lysates of miR-29c ASO-treated db/db mice as compared with control mice. Data are shown as mean ± S.E. (n = 5). F, representative fibronectin immunostaining from kidney cortex of miR-29c ASO-treated db/db mice as compared with control mice. G, the mesangial matrix index of miR-29c ASO-treated db/db mice was significantly decreased as compared with control mice.
To correlate the observed biochemical findings with histological changes in miR-29c ASO-treated mice, we assessed the effect of miR-29c inhibition on glomerular apoptosis, fibronectin immunostaining, and mesangial matrix accumulation. TUNEL assays of db/db mice kidney cortices showed that the miR-29c ASO-treated group had fewer apoptotic cells in the glomeruli as compared with the untreated group (Fig. 6, C and D). Consistent with these data, caspase-3 activity in the kidney cortices from the miR-29c ASO-treated group was significantly lower as compared with the untreated control group (Fig. 6E). Furthermore, miR-29c ASO-treated db/db mice had significantly reduced fibronectin immunostaining, and decreased glomerular matrix accumulation (Fig. 6, F and G).
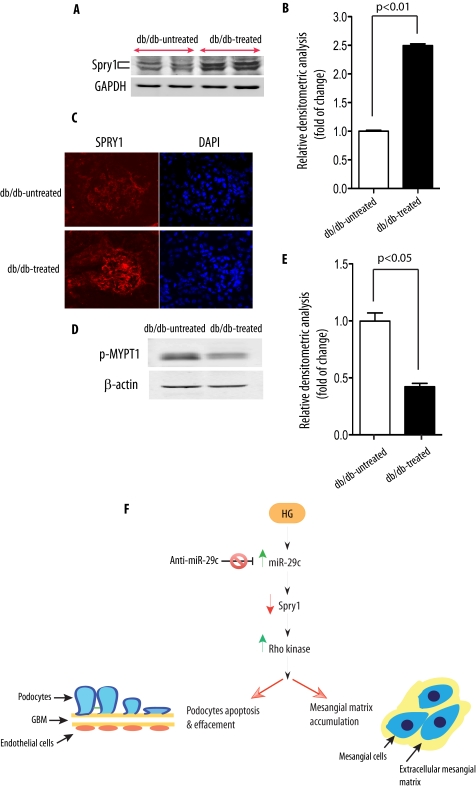
To evaluate the in vivo relevance of miR-29c knockdown on Spry1 expression in the kidneys of db/db mice, we examined Spry1 protein levels in the kidney cortices of miR-29c ASO-treated mice by using Western blot analysis. Importantly, Western blotting showed the up-regulation of Spry1 in the kidney cortices of ASO-treated db/db mice (Fig. 7, A and B). To further validate our findings, we performed immunofluorescent staining for Spry1 protein comparing kidneys from miR-29c ASO-treated db/db mice with the untreated db/db mice. Spry1 protein expression was detected in the glomeruli by immunofluorescence staining and was up-regulated in the majority of glomeruli in miR-29c ASO-treated db/db mice (Fig. 7C). Furthermore, consistent with our in vitro studies, Rho kinase activity was significantly reduced by miR-29c ASO treatment (Fig. 7, D and E). These results demonstrate the therapeutic efficacy of miR-29c ASO in an experimental model of DN in vivo and suggest that the knockdown of miR-29c in vivo leads to increased Spry1 protein level and decreased Rho kinase activity, consistent with our in vitro findings.
FIGURE 7.
In vivo inhibition of miR-29c reduces Rho kinase activation through Spry1. A, Spry1 expression in the kidney cortex extracts of miR-29c ASO-treated db/db mice as compared with control mice was analyzed by Western blot. GAPDH served as a loading control. B, densitometric analysis of Spry1 expression in A (n = 3). C, representative Spry1 staining from kidney cortex of miR-29c ASO-treated db/db mice as compared with control mice. D, Rho kinase activity in the kidney cortex extracts of miR-29c ASO-treated db/db mice as compared with control mice. Representative results from three different experiments are shown. β-Actin served as a loading control. E, densitometric analysis of Rho kinase activity in D (n = 3). F, proposed mechanism for the putative effects of high glucose on the miR-29c-mediated signaling pathway leading to diabetic nephropathy. GBM, glomerular basement membrane.
DISCUSSION
Our findings identify miR-29c as a signature miRNA in the diabetic milieu. Our results also indicate that miR-29c directly targets Spry1 and promotes Rho kinase activation, leading to enhanced cell apoptosis and increased fibronectin synthesis. Importantly, the in vivo inhibition of miR-29c ameliorated progression of DN in an established experimental model of diabetes in vivo.
An emerging body of evidence suggests that miRNAs serve as important therapeutic targets in a wide range of complex human diseases by targeting multiple transcripts (65, 66). We argued that to identify key regulatory miRNAs in DN, it would be necessary to examine miRNA expression patterns in an unbiased manner. Accordingly, we examined expression profiles of miRNAs in samples obtained from glomeruli from db/db mice and in kidney cell lines exposed to hyperglycemic conditions. Our integrated in vitro and in vivo experimental design identified up-regulation of miR-29c as a signature miRNA in hyperglycemic conditions. Several other miRNAs were also up-regulated in our comparative miRNA arrays (Table 1), including miR-192, which was highly expressed in all samples and had been previously demonstrated to be significantly increased in glomeruli isolated from diabetic mice, leading to TGF-β-mediated collagen expression in mesangial cells by down-regulating ZEB2 expression (7).
Another major finding of this study is the identification of Spry1 as a novel target of miR-29c in hyperglycemic conditions. Spry1 has been recently found to serve as a target for miR-21 in myocardiac fibroblast (67). It is becoming increasingly clear that most miRNAs are promiscuous and target multiple genes (65, 68). The miR-29 family of miRNAs is a good example because they exert their effects through modulating multiple targets. Indeed, DNA methyltransferase 3, collagen, Mcl-1, phosphatidylinositol 3-kinase, CDK6, and YY1 have all been previously validated as direct targets of miR-29c (10, 44–51). Our findings demonstrate that Spry1 is a bona fide target of miR-29c under hyperglycemic conditions. We demonstrate that miR-29c directly interacts with 3′-UTR of Spry1 and represses Spry1 expression. Consistent with this notion, an inverse correlation between miR-29c and Spry1 expression was detected in podocytes.
Functionally, our data indicate that miR-29c not only exhibits a potent pro-apoptotic effect, in agreement with other recent reports (45, 46, 49, 69), but also promotes fibronectin synthesis in hyperglycemic conditions. Indeed, overexpression of miR-29c induced apoptosis and increased fibronectin synthesis in podocytes. Importantly, knockdown or overexpression of Spry1 expression modulated the effects of miR-29c on apoptosis and fibronectin expression, indicating the critical role of Spry1 as a target of miR-29c in both high glucose-induced cell apoptosis and fibronectin synthesis in podocytes. It is also important to emphasize that several recent studies have implicated the potential role of miRNAs and Spry1 as regulators of extracellular matrix proteins. For instance, miR-377 was shown to play a pivotal role in increased fibronectin protein production in mesangial cells (8), and a recent report by Sabatel et al. (70) provided evidence that Spry1 may serve as an endogenous angiogenesis inhibitor, whose silencing can modulate the interaction between cells and extracellular matrix proteins. Interestingly, although the role of Spry1 on fibronectin synthesis has recently been described (70), the effect of Spry1 on cell apoptosis is less understood with a number of published studies suggesting that Spry1 can increase or decrease apoptosis at the cellular level, depending on the cellular context (12, 67, 71, 72).
Our study also revealed the role of miR-29c as a regulator of the Rho kinase pathway. The effect of miR-29c on Rho kinase activation is of significant importance because recent studies have suggested that RhoA and Rho kinase are up-regulated in DN, and pharmacological inhibition of Rho kinase results in amelioration of proteinuria (22, 23, 62). Furthermore, several studies have reported that the RhoA/Rho kinase pathway mediates cell apoptosis and promotes fibronectin assembly at the cellular level (61, 62). Our data indicate that Rho kinase is downstream of the high glucose-induced miR-29c/Spry1 pathway because inhibition of Spry1 was able to increase Rho kinase activity. Although Spry1 and its related proteins have previously been implicated as negative regulators of RhoA and its downstream effector Rho kinase (17–19), our findings provide the first evidence for a direct interaction between Spry1 and Rho kinase under hyperglycemic environments. Furthermore, our findings suggest a regulatory role for miR-29c on Rho kinase activation and provide an underlying mechanism for the aberrant expression of Rho kinase in DN.
In summary, we propose that miR-29c is a central component of high glucose-induced signaling. Our findings suggest that up-regulation of miR-29c promotes the progression of DN through a Spry1/Rho kinase pathway (Fig. 7F). Our study also underscores the effectiveness of in vivo inhibition of key miRNAs in complex disease such as DN. These findings provide new insights into the role of miR-29c in the diabetic milieu and open a window of opportunity for a novel therapeutic intervention.
Supplementary Material
Acknowledgments
We are grateful to Drs. R. Nicholas, R. Langley, and J. Reiser for kind gifts of constructs or cell lines. We thank the Gene Vector Core Laboratory and Sequencing Core at the Baylor College of Medicine.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1DK067604 and RO1DK078900 through the NIDDK (to F. R. D.). This work was also partly supported by the Diabetes and Endocrinology Research Center (Grant DK079638) at the Baylor College of Medicine.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Table S1.
- miRNA
- microRNA
- miR-29c
- microRNA-29c
- snRNA
- small nuclear RNA
- Spry1
- Sprouty homolog 1
- DN
- diabetic nephropathy
- ASO
- antisense oligonucleotide
- RT-qPCR
- real-time quantitative polymerase chain reaction
- MYPT1
- myosin phosphatase target subunit 1
- NG
- normal glucose
- HG
- high glucose.
REFERENCES
- 1. Ambros V. (2004) Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 2. Farh K. K., Grimson A., Jan C., Lewis B. P., Johnston W. K., Lim L. P., Burge C. B., Bartel D. P. (2005) Science 310, 1817–1821 [DOI] [PubMed] [Google Scholar]
- 3. Croce C. M., Calin G. A. (2005) Cell 122, 6–7 [DOI] [PubMed] [Google Scholar]
- 4. Lee Y. S., Dutta A. (2009) Annu. Rev. Pathol. 4, 199–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Urbich C., Kuehbacher A., Dimmeler S. (2008) Cardiovasc. Res. 79, 581–588 [DOI] [PubMed] [Google Scholar]
- 6. Kartha R. V., Subramanian S. (2010) J. Cardiovasc. Transl. Res. 3, 256–270 [DOI] [PubMed] [Google Scholar]
- 7. Kato M., Zhang J., Wang M., Lanting L., Yuan H., Rossi J. J., Natarajan R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3432–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Q., Wang Y., Minto A. W., Wang J., Shi Q., Li X., Quigg R. J. (2008) FASEB J. 22, 4126–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Long J., Wang Y., Wang W., Chang B. H., Danesh F. R. (2010) J. Biol. Chem. 285, 23457–23465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Rooij E., Sutherland L. B., Thatcher J. E., DiMaio J. M., Naseem R. H., Marshall W. S., Hill J. A., Olson E. N. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13027–13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Rooij E., Olson E. N. (2009) Circ. Res. 104, 138–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gross I., Morrison D. J., Hyink D. P., Georgas K., English M. A., Mericskay M., Hosono S., Sassoon D., Wilson P. D., Little M., Licht J. D. (2003) J. Biol. Chem. 278, 41420–41430 [DOI] [PubMed] [Google Scholar]
- 13. Basson M. A., Akbulut S., Watson-Johnson J., Simon R., Carroll T. J., Shakya R., Gross I., Martin G. R., Lufkin T., McMahon A. P., Wilson P. D., Costantini F. D., Mason I. J., Licht J. D. (2005) Dev. Cell 8, 229–239 [DOI] [PubMed] [Google Scholar]
- 14. Basson M. A., Watson-Johnson J., Shakya R., Akbulut S., Hyink D., Costantini F. D., Wilson P. D., Mason I. J., Licht J. D. (2006) Dev. Biol. 299, 466–477 [DOI] [PubMed] [Google Scholar]
- 15. Kim H. J., Bar-Sagi D. (2004) Nat. Rev. Mol. Cell Biol. 5, 441–450 [DOI] [PubMed] [Google Scholar]
- 16. Mason J. M., Morrison D. J., Basson M. A., Licht J. D. (2006) Trends Cell Biol. 16, 45–54 [DOI] [PubMed] [Google Scholar]
- 17. Wang Y. (2009) Mol. Cancer Ther. 8, 2103–2109 [DOI] [PubMed] [Google Scholar]
- 18. Wang Y., Janicki P., Köster I., Berger C. D., Wenzl C., Grosshans J., Steinbeisser H. (2008) Genes Dev. 22, 878–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miyoshi K., Wakioka T., Nishinakamura H., Kamio M., Yang L., Inoue M., Hasegawa M., Yonemitsu Y., Komiya S., Yoshimura A. (2004) Oncogene 23, 5567–5576 [DOI] [PubMed] [Google Scholar]
- 20. Gojo A., Utsunomiya K., Taniguchi K., Yokota T., Ishizawa S., Kanazawa Y., Kurata H., Tajima N. (2007) Eur. J. Pharmacol. 568, 242–247 [DOI] [PubMed] [Google Scholar]
- 21. Kikuchi Y., Yamada M., Imakiire T., Kushiyama T., Higashi K., Hyodo N., Yamamoto K., Oda T., Suzuki S., Miura S. (2007) J. Endocrinol. 192, 595–603 [DOI] [PubMed] [Google Scholar]
- 22. Kolavennu V., Zeng L., Peng H., Wang Y., Danesh F. R. (2008) Diabetes 57, 714–723 [DOI] [PubMed] [Google Scholar]
- 23. Peng F., Wu D., Gao B., Ingram A. J., Zhang B., Chorneyko K., McKenzie R., Krepinsky J. C. (2008) Diabetes 57, 1683–1692 [DOI] [PubMed] [Google Scholar]
- 24. Etienne-Manneville S., Hall A. (2002) Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 25. Loirand G., Guérin P., Pacaud P. (2006) Circ. Res. 98, 322–334 [DOI] [PubMed] [Google Scholar]
- 26. Narumiya S. (1996) J. Biochem. 120, 215–228 [DOI] [PubMed] [Google Scholar]
- 27. Mundel P., Reiser J., Zúñiga Mejía Borja A., Pavenstädt H., Davidson G. R., Kriz W., Zeller R. (1997) Exp. Cell Res. 236, 248–258 [DOI] [PubMed] [Google Scholar]
- 28. Langley R. R., Ramirez K. M., Tsan R. Z., Van Arsdall M., Nilsson M. B., Fidler I. J. (2003) Cancer Res. 63, 2971–2976 [PubMed] [Google Scholar]
- 29. Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 30. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 31. Várallyay E., Burgyán J., Havelda Z. (2008) Nat. Protoc. 3, 190–196 [DOI] [PubMed] [Google Scholar]
- 32. Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. (2004) RNA 10, 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Du M., Roy K. M., Zhong L., Shen Z., Meyers H. E., Nichols R. C. (2006) FEBS J. 273, 732–745 [DOI] [PubMed] [Google Scholar]
- 34. Lin X., Duan X., Liang Y. Y., Su Y., Wrighton K. H., Long J., Hu M., Davis C. M., Wang J., Brunicardi F. C., Shi Y., Chen Y. G., Meng A., Feng X. H. (2006) Cell 125, 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Long J., Matsuura I., He D., Wang G., Shuai K., Liu F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9791–9796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zeng L., Xu H., Chew T. L., Eng E., Sadeghi M. M., Adler S., Kanwar Y. S., Danesh F. R. (2005) FASEB J. 19, 1845–1847 [DOI] [PubMed] [Google Scholar]
- 37. Takemoto M., Asker N., Gerhardt H., Lundkvist A., Johansson B. R., Saito Y., Betsholtz C. (2002) Am. J. Pathol. 161, 799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horwich M. D., Zamore P. D. (2008) Nat. Protoc. 3, 1537–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krützfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., Stoffel M. (2005) Nature 438, 685–689 [DOI] [PubMed] [Google Scholar]
- 40. Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., Subramaniam A., Propp S., Lollo B. A., Freier S., Bennett C. F., Bhanot S., Monia B. P. (2006) Cell Metab. 3, 87–98 [DOI] [PubMed] [Google Scholar]
- 41. Esau C. C. (2008) Methods 44, 55–60 [DOI] [PubMed] [Google Scholar]
- 42. Isermann B., Vinnikov I. A., Madhusudhan T., Herzog S., Kashif M., Blautzik J., Corat M. A., Zeier M., Blessing E., Oh J., Gerlitz B., Berg D. T., Grinnell B. W., Chavakis T., Esmon C. T., Weiler H., Bierhaus A., Nawroth P. P. (2007) Nat. Med. 13, 1349–1358 [DOI] [PubMed] [Google Scholar]
- 43. Mukhopadhyay P., Rajesh M., Haskó G., Hawkins B. J., Madesh M., Pacher P. (2007) Nat. Protoc. 2, 2295–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E., Liu S., Alder H., Costinean S., Fernandez-Cymering C., Volinia S., Guler G., Morrison C. D., Chan K. K., Marcucci G., Calin G. A., Huebner K., Croce C. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15805–15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mott J. L., Kobayashi S., Bronk S. F., Gores G. J. (2007) Oncogene 26, 6133–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garzon R., Heaphy C. E., Havelange V., Fabbri M., Volinia S., Tsao T., Zanesi N., Kornblau S. M., Marcucci G., Calin G. A., Andreeff M., Croce C. M. (2009) Blood 114, 5331–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Z., Hassan M. Q., Jafferji M., Aqeilan R. I., Garzon R., Croce C. M., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2009) J. Biol. Chem. 284, 15676–15684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sengupta S., den Boon J. A., Chen I. H., Newton M. A., Stanhope S. A., Cheng Y. J., Chen C. J., Hildesheim A., Sugden B., Ahlquist P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5874–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Park S. Y., Lee J. H., Ha M., Nam J. W., Kim V. N. (2009) Nat. Struct. Mol. Biol. 16, 23–29 [DOI] [PubMed] [Google Scholar]
- 50. Zhao J. J., Lin J., Lwin T., Yang H., Guo J., Kong W., Dessureault S., Moscinski L. C., Rezania D., Dalton W. S., Sotomayor E., Tao J., Cheng J. Q. (2010) Blood 115, 2630–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang H., Garzon R., Sun H., Ladner K. J., Singh R., Dahlman J., Cheng A., Hall B. M., Qualman S. J., Chandler D. S., Croce C. M., Guttridge D. C. (2008) Cancer Cell 14, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Doench J. G., Sharp P. A. (2004) Genes Dev. 18, 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stern P., Astrof S., Erkeland S. J., Schustak J., Sharp P. A., Hynes R. O. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13895–13900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Donello J. E., Loeb J. E., Hope T. J. (1998) J. Virol. 72, 5085–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lewko B., Stepinski J. (2009) J. Cell Physiol. 221, 288–295 [DOI] [PubMed] [Google Scholar]
- 56. Susztak K., Raff A. C., Schiffer M., Böttinger E. P. (2006) Diabetes 55, 225–233 [PubMed] [Google Scholar]
- 57. Brownlee M. (2001) Nature 414, 813–820 [DOI] [PubMed] [Google Scholar]
- 58. Wolf G., Ziyadeh F. N. (1999) Kidney Int. 56, 393–405 [DOI] [PubMed] [Google Scholar]
- 59. Danesh F. R., Sadeghi M. M., Amro N., Philips C., Zeng L., Lin S., Sahai A., Kanwar Y. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8301–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zeng L., Xu H., Chew T. L., Chisholm R., Sadeghi M. M., Kanwar Y. S., Danesh F. R. (2004) J. Am. Soc. Nephrol. 15, 1711–1720 [DOI] [PubMed] [Google Scholar]
- 61. Del Re D. P., Miyamoto S., Brown J. H. (2007) J. Biol. Chem. 282, 8069–8078 [DOI] [PubMed] [Google Scholar]
- 62. Yoneda A., Ushakov D., Multhaupt H. A., Couchman J. R. (2007) Mol. Biol. Cell 18, 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hartshorne D. J., Hirano K. (1999) Mol. Cell Biochem. 190, 79–84 [PubMed] [Google Scholar]
- 64. Davis S., Propp S., Freier S. M., Jones L. E., Serra M. J., Kinberger G., Bhat B., Swayze E. E., Bennett C. F., Esau C. (2009) Nucleic Acids Res. 37, 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Garofalo M., Croce C. M. (2011) Annu. Rev. Pharmacol. Toxicol. 51, 25–43 [DOI] [PubMed] [Google Scholar]
- 66. Nagpal J. K., Rani R., Trink B., Saini K. S. (2010) Curr. Mol. Med. 10, 503–510 [DOI] [PubMed] [Google Scholar]
- 67. Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., Galuppo P., Just S., Rottbauer W., Frantz S., Castoldi M., Soutschek J., Koteliansky V., Rosenwald A., Basson M. A., Licht J. D., Pena J. T., Rouhanifard S. H., Muckenthaler M. U., Tuschl T., Martin G. R., Bauersachs J., Engelhardt S. (2008) Nature 456, 980–984 [DOI] [PubMed] [Google Scholar]
- 68. Inui M., Martello G., Piccolo S. (2010) Nat. Rev. Mol. Cell Biol. 11, 252–263 [DOI] [PubMed] [Google Scholar]
- 69. Xiong Y., Fang J. H., Yun J. P., Yang J., Zhang Y., Jia W. H., Zhuang S. M. (2010) Hepatology 51, 836–845 [DOI] [PubMed] [Google Scholar]
- 70. Sabatel C., Cornet A. M., Tabruyn S. P., Malvaux L., Castermans K., Martial J. A., Struman I. (2010) Mol. Cancer 9, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gross I., Bassit B., Benezra M., Licht J. D. (2001) J. Biol. Chem. 276, 46460–46468 [DOI] [PubMed] [Google Scholar]
- 72. Impagnatiello M. A., Weitzer S., Gannon G., Compagni A., Cotten M., Christofori G. (2001) J. Cell Biol. 152, 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.