Abstract
In late mitosis and G1, Mcm2-7 are assembled onto replication origins to license them for initiation in the upcoming S phase. After initiation, Mcm2-7 provide helicase activity to unwind DNA at the replication fork. Here we examine the structure of Mcm2-7 on chromatin in Xenopus egg extracts. We show that prior to replication initiation, Mcm2-7 is present at licensed replication origins in a complex with a molecular mass close to double that of the Mcm2-7 hexamer. This complex has approximately stoichiometric quantities of the 6 Mcm2-7 proteins and we conclude that it consists of a double heterohexamer. This provides a configuration potentially capable of initiating a pair of bidirectional replication forks in S phase. We also show that after initiation, Mcm2-7 associate with Cdc45 and GINS to form a relatively stable CMG (Cdc45-MCM-GINS) complex. The CMG proteins also associate less strongly with other replication proteins, consistent with the idea that a single CMG complex forms the core of the replisome.
Keywords: Cell Cycle, DNA Helicase, DNA-Protein Interaction, DNA Replication, Xenopus
Introduction
In eukaryotes, precise duplication of the genome during S phase is achieved through the initiation of replication forks at numerous origins distributed throughout the DNA. The central component of the replication fork is the replicative helicase that unwinds the template DNA and coordinates the replication of leading and lagging strands. In eukaryotes, the major replicative helicase activity is provided by the 6 Mcm2-7 proteins (1–4), which are activated as a helicase by association with Cdc45 and the GINS complex (5, 6).
To ensure that no segment of DNA replicates more than once in a single cell cycle, the replication process is divided into two non-overlapping stages (7–10). In the first stage, which occurs in late mitosis and early G1 phase, origins of replication are “licensed” for use in the upcoming S phase by loading Mcm2-7 (without associated Cdc45 or GINS) to form the pre-replicative complex (pre-RC). To prevent DNA from replicating more than once in a single cell cycle it is essential that once S phase starts, no further Mcm2-7 can be loaded onto DNA. This ensures that replicated replication origins cannot reload Mcm2-7 and therefore cannot replicate a second time during a single S phase (7–10).
The licensing reaction has been reconstituted in vitro using proteins from the amphibian Xenopus laevis (11) and from the yeast Saccharomyces cerevisiae (12, 13). First the origin recognition complex (ORC)4 binds to DNA at the origin, and DNA-bound ORC then recruits the Cdt1- and Cdc6-licensing proteins. Finally, in an ATP-consuming reaction ORC, Cdc6 and Cdt1 cooperate to load Mcm2-7 onto DNA. Electron microscopy of the reconstituted reaction in S. cerevisiae shows that the DNA apparently takes a path directly through the MCM loaded onto DNA (12, 13). This is consistent with previous structural studies of archaeal MCM proteins showing that they form a double hexameric complex with a positively charged central channel capable of encircling double-stranded DNA (14–17). The size of the S. cerevisiae complexes on DNA suggest that they form back-to-back double hexamers (12, 13).
This idea that the licensing of origins involves the clamping of a symmetrical double hexamer around DNA is biologically plausible. First, it is important that the association of Mcm2-7 with DNA at licensed origins is very stable, because it must persist from G1 until the completion of DNA replication which may take >10 h in some cell types. The clamping of the ring-shaped Mcm2-7 complex around DNA can potentially provide an association that would be stable over these long periods. Secondly, replication forks are initiated bidirectionally from replication origins, so it is important that origins contain back-to-back Mcm2-7 hexamers that can support this bidirectional movement.
In this report, we provide evidence that the licensing of replication origins in Xenopus egg extracts corresponds to the loading of double Mcm2-7 hexamers onto DNA prior to the onset of S phase. The conservation of this mechanism between yeasts and frogs suggests that it is likely to be a conserved mechanism used by all eukaryotes. We also provide evidence that during S phase in the Xenopus system, Mcm2-7 associate with Cdc45 and GINS to form CMG complexes only when replication forks have been initiated.
EXPERIMENTAL PROCEDURES
Xenopus Egg Extract
Metaphase-arrested Xenopus laevis egg extracts were prepared as previously described (18). Extracts were supplemented with 6.25 μg/ml cycloheximide, 0.625 mm phosphocreatine, and 0.375 μg/ml creatine phosphokinase and released into interphase by 15 min of incubation with 0.3 mm CaCl2. Demembranated Xenopus sperm nuclei were added to final concentrations of 15 ng/μl for S-phase samples, 20 ng/μl for CDK- and Cdc7-blocked samples, and 7 ng/μl for Mcm3-depleted extract. Replication reactions were carried out at 23 °C. Immunodepletion of interphase extract with Mcm3 rabbit antibody serum was performed as described previously (18). DNA synthesis was assessed in extract supplemented with α-[32P]dATP by TCA precipitation as described (18).
Releasing Proteins from Chromatin
Reactions (2 ml for AKTA column sample, 0.48 ml for glycerol gradient sample or 0.32 ml for SMART column sample) were diluted with 10–20 volumes of ice-cold ANIB buffer (50 mm HEPES-KOH pH7.6, 50 mm potassium acetate, 25 mm sodium glycerophosphate pH 7.6, 10 mm magnesium acetate, 0.1% Triton X-100, 2.5 mm MgATP, 0.1 mm sodium vanadate, 0.1 μm microcystin, 1 μg/ml each of leupeptin, pepstatin, and aprotonin, 0.5 mm spermidine, 0.15 mm spermine, 0.1 mm PMSF, 1× Halt protease inhibitor mixture (Thermo Scientific)), underlaid with ANIB buffer containing 30% sucrose and spun 10 min, 2,500 × g, 4 °C. The top of the cushion was washed three times with ANIB buffer and the rest of the cushion removed from above the pellet. Chromatin was resuspended in one-fourth original extract volume in ANIB + 30% sucrose (or for glycerol gradients, ANIB). Benzonase (Novagen) or DNase I (Roche) was added to 2 units/μl or 3 units/μl final concentration respectively and incubated for 10 min, room temperature. Samples were sonicated (Bioruptor, Diagenode) for 5 min (10 s sonication, 45 s break, medium setting), centrifuged 5 min, 20,000 × g, 4 °C, and the supernatant taken as solubilized chromatin. 5 μl of control samples were taken at every stage and analyzed by immunoblotting.
For samples at higher salt concentrations, chromatin was isolated using ANIB buffer with 50 mm potassium acetate as above but cushion buffer contained no detergent. After resuspension, digestion with Benzonase and sonication, the potassium acetate concentration was adjusted as desired and chromatin incubated for further 5 min at room temperature before the final spin.
Gel Filtration and Glycerol Gradients
Chromatin samples for gel filtration or glycerol gradient were prepared as described above; extract samples were prepared by adding 4 volumes of ANIB buffer +10% sucrose and centrifuging 10 min, 20,000 × g, 4 °C to remove insoluble material.
Analytical gel filtration was performed using Superose 6 PC 3.2/30 column and SMARTTM system (Amersham Biosciences, GE Healthcare) with 50 μl of sample. Large-scale gel filtration for immunoprecipitation was performed using a Superose 6 10/300 GL column and AKTA system (GE Healthcare) and a 500-μl sample. Columns were run in ANIB plus 30% sucrose and indicated salt concentration. 30 fractions of 50 μl (SMART) or 500 μl (AKTA) were collected, and aliquots of the first 24 fractions analyzed. Columns were calibrated using a kit for molecular masses 29,000–700,000 (Sigma, MWGF1000).
4 ml of 20–40% glycerol gradients in ANIB buffer (containing appropriate salt concentrations) were prepared in 4-ml thin wall tubes (Beckman Coulter, 328874). 100 μl of released chromatin protein sample was loaded and samples spun in SW60 Ti rotor (Beckman Coulter) 16 h, 45,000 rpm, 4 °C. A separate glycerol gradient with size marker proteins (Sigma, MWGF1000) was included in every run. Spun gradients were divided into 200-μl fractions.
Molecular masses were calculated according to Siegel and Monty (19) using the values: thyroglobulin tetramer (1338 kDa, 107 Å), thyroglobulin dimer (669 kDa, 85 Å, 19.5 S), apoferritin (443 kDa, 67 Å, 17.6 S), β-amylase (200 kDa, 54 Å, 8.9 S), BSA (66 kDa, 35.5 Å, 4.3 S), carbonic anhydrase (29 kDa, 24.3 Å, 3.2 S).
DNA Isolation
For DNA isolation, chromatin samples were prepared from 120 μl of extract. Digestion with Benzonase or DNase I was stopped by addition of EDTA to 65 mm and proteins digested by addition of proteinase K to 2 μg/ml and SDS to 1% and 30 min of incubation at 37 °C. DNA was phenol-chloroform extracted and ethanol precipitated.
Recombinant Proteins, Reagents, and Antibodies
p27KIP1 was a gift from Gaganmeet Chadha (Dundee University). PHA-767491 (20) was synthesized at Dundee University. Mcm2 and Mcm3 affinity-purified antibodies were previously described (21), as were Mcm4, Mcm5, Mcm6, and Mcm7 antibodies (22). Psf2, Sld5, Tipin and Cdc45 antibodies were raised in sheep against full-length His6-tagged X. laevis recombinant proteins expressed and purified from Escherichia coli (RosettaTM(DE3)pLysS, Novagen) using Ni2+-NTA affinity chromatography. His6-tagged C-terminal fragments of Ctf4/And-1 (828–1127 aa) and Mcm10 (278–860 aa) were purified in the same way. Cdc45, Psf2, Ctf4, Mcm10, and Tipin antibodies were affinity purified. Antibody specificity is shown in supplemental Fig. S5. Antibodies were used for immunoblotting at 1:1000 dilutions.
Immunoprecipitations and Mass Spectrometry
Mcm3 and Cdc45 immunoprecipitations were performed using affinity-purified antibodies or control sheep IgG (Sigma, S2763) covalently coupled to Dynabeads M-270 Epoxy (Invitrogen) according to the manufacturer's instructions.
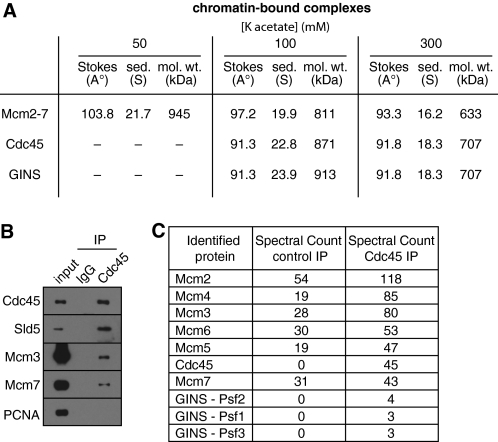
For Mcm3 immunoprecipitation, gel filtration fractions were combined and incubated for 2 h with 350 μl of Mcm3 or IgG beads. After 5 × 5 min washes with ANIB, beads were boiled in 70 μl of LDS sample buffer (Invitrogen) and 40-μl IP samples were run on a 4–12% gradient NuPAGE gel (Invitrogen). The gel was stained with Cypro Ruby Protein Gel Stain (Molecular probes, Invitrogen) or SimplyBlue SafeStain (Invitrogen). Mcm3 and IgG lanes were cut into 40 equal slices for analysis by mass spectrometry. For Cdc45 and Ctf4 immunoprecipitation, chromatin was isolated in mid-S phase from a 4-ml reaction; 500 μl of antibody-coupled beads were used and 40-μl IP samples run on 10% NuPAGE gel (Invitrogen) in MOPS buffer for 2 cm, lanes cut into 10 slices for analysis by mass spectrometry. Mass spectrometry of immunoprecipitates was as described (23). Briefly, samples were reduced with dithiothreitol, alkylated with iodoacetamide and digested with trypsin. Peptide solutions were analyzed using nano LC-MS/MS on an LTQ Orbitrap XL (ThermoFisher, San Jose, CA). Total spectral count for a protein includes repeated identification of the same sequence or any versions with modifications or different charge states. Soluble chromatin fractionated by gel filtration was precipitated with methanol/chloroform, prepared for mass spectrometry and protein abundance was estimated from total ion current as described (21, 24).
RESULTS
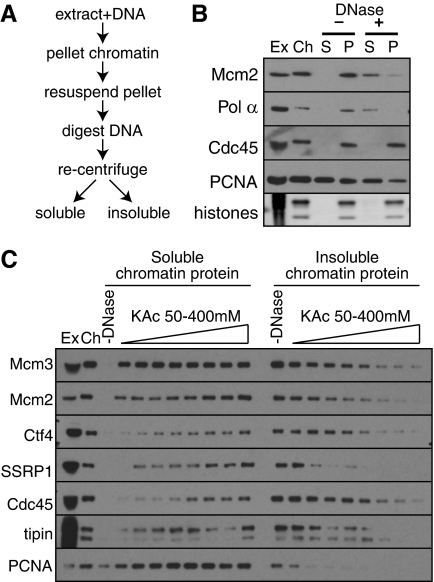
We first devised a method for isolating intact protein complexes from chromatin assembled in the Xenopus cell-free system (Fig. 1A). Demembranated sperm nuclei were incubated in egg extract as appropriate, the extract was diluted, and chromatin was centrifuged into a sucrose cushion. The DNA was extensively degraded by a combination of sonication and DNase treatment, until no large DNA molecules were visible by agarose gel electrophoresis (supplemental Fig. S1). Insoluble material was then removed by centrifugation, and DNase-solubilized chromatin subjected to further analysis. When low salt buffers (50 mm potassium acetate) were used for chromatin resuspension, >80% of Mcm2-7 proteins loaded onto the DNA was released by DNase treatment (Fig. 1B). Certain replication fork proteins, such as DNA polymerase α were also released from chromatin by this protocol, but many others, such as Cdc45, remained in the insoluble pellet. We next investigated the effect of salt concentration on the solubility of replication fork proteins (Fig. 1C). This showed that a wide range of replication fork proteins remained insoluble when isolated in 50 mm potassium acetate, but became soluble at 300–400 mm. Because most of the chromatin-bound Mcm2-7 license dormant replication origins that remain inactive during normal S phases (25–27) this result is consistent with the idea that Mcm2-7 on licensed origins are soluble in low salt, but become insoluble when they associate with replication fork proteins in the active replisome.
FIGURE 1.
Protein release from replicating X. laevis chromatin. A, schematic representation of chromatin protein preparation procedure. B and C, chromatin was isolated from egg extract in the middle of S-phase (when replisome proteins peak on chromatin) in the presence of 50 mm potassium acetate, and DNA was digested by Benzonase and sonication. Ex, 0.5 μl of egg extract; Ch, chromatin after first centrifugation; other lanes correspond to material isolated from 5 μl of egg extract. B, chromatin was maintained in 50 mm potassium acetate throughout the procedure. P, pellet after DNA digestion; S, supernatant after DNA digestion. C, after DNA digestion, the salt concentration was adjusted to the 50, 100, 150, 200, 250, 300, 350, or 400 mm potassium acetate before samples were separated by centrifugation into soluble and insoluble (pellet) fractions. Fractions were separated by SDS-PAGE and immunoblotted with antibodies to the indicated proteins.
Licensed Origins Contain Double Hexamers of Mcm2-7
To analyze the status of Mcm2-7 on licensed chromatin prior to S phase onset, we prevented replication initiation by adding the CDK inhibitor p27KIP1 to the extract. Because Mcm2-7 hexamers are very sensitive to disruption by salt (22, 28), we performed the chromatin manipulations in 50 mm potassium acetate. As expected, KIP1 blocked the assembly of replication fork proteins Cdc45, PCNA, and Psf2 onto the chromatin (Fig. 2A). We examined the structure of chromatin-bound Mcm2-7 by a combination of gel filtration (Fig. 2B) and glycerol gradient sedimentation (Fig. 2C). Gel filtration overestimates the size of elongated molecules, while glycerol gradient sedimentation underestimates the size of elongated molecules, but combining results from both techniques allows the calculation of molecular masses that are not biased by shape (19). The results (Fig. 2D) suggest that chromatin-bound Mcm2-7 on licensed origins has a molecular mass of ∼945 kDa. This value is significantly higher than free Mcm2-7 complex in the extract, which has a calculated molecular mass of ∼530 kDa and consists of a single heterohexamer containing one copy each of Mcm2, Mcm3, Mcm4, Mcm5, Mcm6, and Mcm7 (22). Because chromatin-bound Mcm2-7 has a molecular mass almost double that of free hexameric Mcm2-7, this is consistent with origin licensing representing the assembly of double hexamers onto replication origins (7, 12, 13).
FIGURE 2.
Mcm2-7 form large complexes on chromatin before replication initiation. Chromatin was isolated from egg extract plus or minus 100 nm p27KIP1 and chromatin proteins released in 50 mm potassium acetate. A, Ex, 0.5 μl egg extract; Ch, chromatin after first centrifugation; other lanes correspond to insoluble pellet (P), and soluble chromatin (S) from 5 μl extract. B and C, top panels: chromatin was isolated from extracts treated with p27KIP1 and soluble proteins released in 50 mm potassium acetate were subjected to gel filtration (B) or glycerol gradient fractionation (C) and immunoblotted with antibodies to Mcm2-7. Bottom panels: extract incubated with 100 nm p27KIP1 was subjected to gel filtration (B) or glycerol gradient fractionation (C) in buffer containing 50 mm potassium acetate and immunoblotted with antibodies to Mcm2-7. D, data from B and C were used to determine the molecular weight of Mcm2-7 complexes in whole extract and in chromatin. E, chromatin in mid S phase was isolated from extracts without added p27KIP1 and soluble proteins separated by gel filtration as for B.
The bulk of chromatin-bound Mcm2-7 still migrated as an apparent double-hexamer when KIP1 was omitted from the reaction and chromatin was isolated from mid-S phase (Fig. 2E). This unchanged behavior is likely explained because most of the Mcm2-7 license dormant origins that do not normally initiate replication forks (25), and because replication forks are only poorly soluble under the low salt conditions used (Fig. 1C).
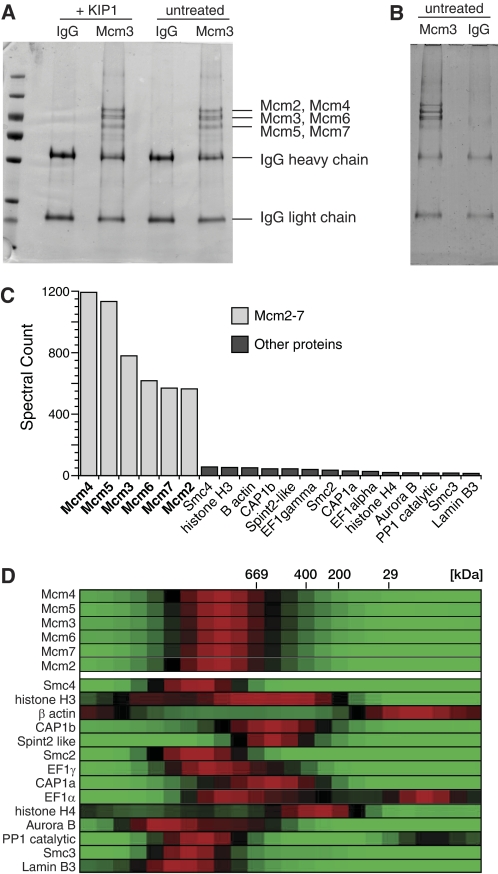
To determine whether the large chromatin-bound Mcm2-7 complex contains proteins other than Mcm2-7, chromatin solubilized in 50 mm potassium acetate was separated by gel filtration; fractions containing the large Mcm2-7 complex were pooled (supplemental Fig. S2) and immunoprecipitated with anti-Mcm3 antibodies. The resulting Mcm3 immunoprecipitates were separated by SDS-PAGE and stained for total protein. The major proteins in the immunoprecipitate migrated as expected of the 6 Mcm2-7 proteins (Fig. 3A). Although the 6 proteins were not fully resolved, doublets corresponding to Mcm2+Mcm4, Mcm3+6, and Mcm5+7 (22, 29) were present in approximately stoichiometric amounts, as expected of a double heterohexamer. Apart from IgG used for the immunoprecipitation, no proteins other than Mcm2-7 could be seen at levels high enough to be stoichiometric with Mcm2-7.
FIGURE 3.
Mcm2-7 complexes immunoprecipitated from chromatin assembled in egg extract with blocked CDK activity look the same as Mcm2-7 at unfired origins in S-phase. Chromatin was isolated from egg extract plus or minus 100 nm p27KIP1 and chromatin proteins were released in 50 mm potassium acetate. Soluble protein was then separated by gel filtration. A and B, high molecular weight fractions containing Mcm2-7 were pooled and immunoprecipitated with either non-immune or anti-Mcm3 antibodies. Immunoprecipitates were separated by SDS-PAGE and gels were stained with colloidal Coomassie stain. C, gel shown in B was sliced into 40 segments, and each segment was subjected to mass spectrometry. The spectral count for each identified protein was summed over all 40 gel slices. The top 20 identified proteins specific for the Mcm3 immunoprecipitate are shown, along with its spectral count. D, each of the gel filtration fractions were analyzed by mass spectrometry, and the abundance of proteins estimated by total ion current. For the 20 proteins shown in C, the relative protein abundance across the gel filtration column is shown as a heat map. Red, peak abundance; black, half-peak abundance; green, no protein detected.
To gain a more quantitative assessment of proteins present in the large Mcm2-7 complex, gel filtration fractions were immunoprecipitated with Mcm3 antibodies, and the proteins separated by SDS-PAGE (Fig. 3B). The gel lanes were then divided into 40 equal slices, and proteins within each slice were analyzed by mass spectrometry. The top 20 proteins specific for the Mcm3 immunoprecipitation are shown in Fig. 3C, with the spectral count (total number of peptide identifications) for each protein shown as the height of the bar. This shows that peptides associated with Mcm2-7 were detected >10 times more often than peptides from any of the other detected proteins, suggesting that Mcm2-7 were the most abundant proteins in the sample. To determine the possible co-elution of these other proteins with Mcm2-7, the gel filtration fractions of solubilized chromatin were individually analyzed by LC-MS/MS mass spectrometry. The relative abundance of the top 20 proteins from Fig. 3C was determined by comparing the total ion current of peptides across the gel filtration fractions (21). For each protein, the peak abundance of the protein was normalized to 1, and the results are shown in Fig. 3D as a heat map. Consistent with the immunoblotting results, all 6 Mcm2-7 polypeptides peak with an apparent molecular mass of ∼1150 kDa. None of the 16 other proteins peaked at the same position as Mcm2-7, suggesting that none of them quantitatively associate with Mcm2-7. Taken together, all these results strongly suggest that the increased molecular mass of Mcm2-7 when loaded onto chromatin is a consequence of formation of a double Mcm2-7 hexamer. Consistent with this interpretation, cross-linking experiments showed that new interactions between Mcm2-7 proteins after they have been loaded onto chromatin (data not shown).
Effect of Cdc7
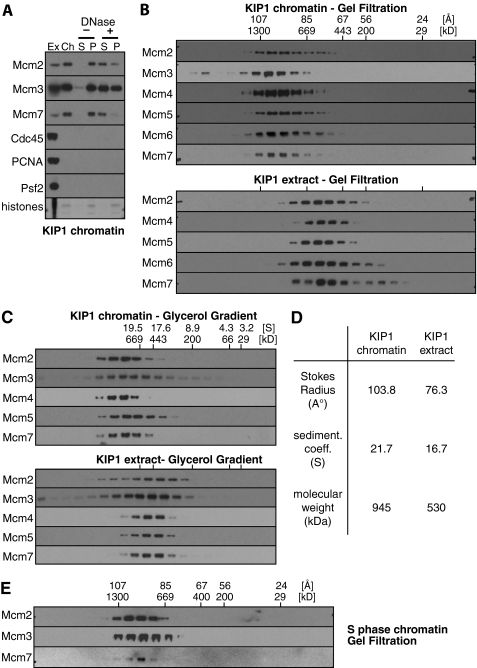
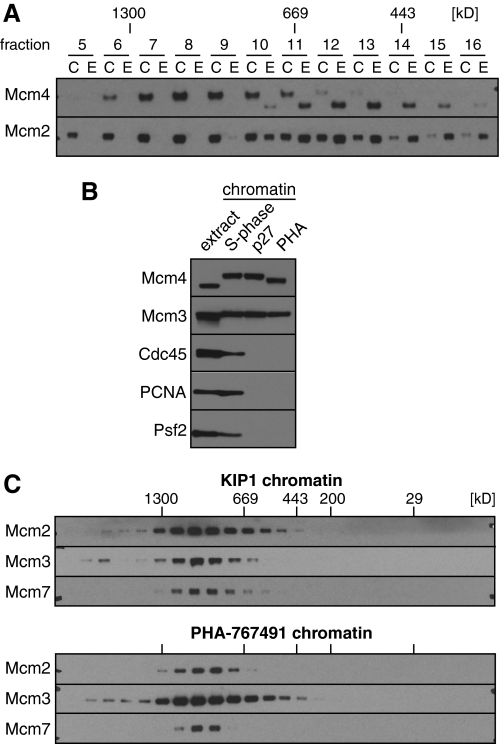
During S phase, a proportion of licensed origins initiate pairs of replication forks that progress bidirectionally away from the origin. Replication initiation requires the activity of two protein kinases: S phase inducing CDKs and Cdc7. Mcm2-7 are the major substrates of Cdc7. It has recently been suggested that phosphorylation of Mcm2-7 by Cdc7 promotes separation of the double hexamers into single hexamers (13). In Xenopus egg extracts, phosphorylation of Mcm2-7 by Cdc7 kinase occurs shortly after licensing and is independent of S phase CDK activity (30, 31). Cdc7 causes a hyperphosphorylation of Xenopus Mcm4 that can be detected as a mobility shift of Mcm4 on SDS gels (32). The hyperphosphorylation of Mcm4 in KIP1-treated extract can be clearly seen when gel filtration fractions of chromatin and whole extract were run next to each other (Fig. 4A). Virtually all chromatin-bound Mcm4 was hyperphosphorylated (Fig. 4B) and the hyperphosphorylated Mcm4 on chromatin migrated on gel filtration with the double hexamer (Fig. 4A). These results suggest that Cdc7 phosphorylation of Mcm2-7 does not significantly break up the double hexamer.
FIGURE 4.
Large Mcm2-7 complexes form independently of Cdc7 kinase activity and are substrates of Cdc7 kinase. A, chromatin was isolated from egg extract containing 100 nm p27KIP1 and chromatin proteins released in 50 mm potassium acetate. Soluble protein was then separated by gel filtration. In parallel, whole extract was separated by gel filtration under identical conditions. Aliquots of chromatin C and extract E fractions were separated by SDS-PAGE and immunoblotted for Mcm2 and Mcm4. B, soluble chromatin proteins were isolated in 50 mm potassium acetate from extract optionally supplemented with 100 nm p27KIP1 or 100 μm PHA-767491, separated by SDS-PAGE and immunoblotted for the indicated proteins. An aliquot of untreated extract is also shown. C, chromatin from extract treated with p27KIP1 or PHA-767491 was prepared as in B and separated by gel filtration. Fractions were separated by SDS-PAGE and immunoblotted for the indicated proteins.
A small molecule inhibitor of Cdc7 kinase activity, PHA-767491, has been described (20). PHA-767491 can inhibit DNA replication and Mcm4 hyperphosphorylation in Xenopus egg extracts and this effect appears to be due to specific inhibition of Cdc7 kinase activity in the extract.5 Fig. 4B shows that like KIP1, PHA-767491 completely blocked the initiation of replication forks, as evidenced by the absence of the fork proteins Cdc45, PCNA, and Psf2 on chromatin. PHA-767491 also substantially reduced the hyperphosphorylation of Mcm4. Fig. 4C shows that the size of the Mcm2-7 complex on chromatin assembled in the presence of PHA-767491 was very similar to the complex formed in the presence of KIP1. Taken together, these results suggest that Cdc7 phosphorylation of Mcm2-7 does not significantly affect stability of the double hexamer.
Active Mcm2-7 at Replication Forks
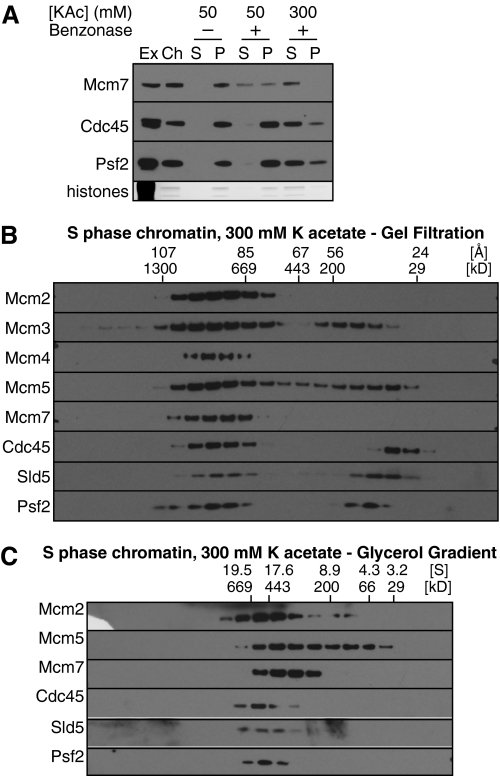
We next investigated what happens to Mcm2-7 once they have initiated replication forks following combined CDK and Cdc7 activity. Analysis of Mcm2-7 in Drosophila cells has shown that when replication forks initiate, Mcm2-7 associate with Cdc45 and the GINS complex to form the CMG (Cdc45-MCM-GINS) helicase (5, 6). Fig. 1A shows that in Xenopus extracts, potassium acetate concentrations of ∼300 mm were required in order to efficiently release replication fork proteins in a soluble form following DNase digestion. At 300 mm, Cdc45 and the Psf2 component of the GINS complex were substantially solubilized by DNase treatment (Fig. 5A). However, as the potassium acetate concentration was increased, the size of the released Mcm2-7 complexes decreased, as evidenced by changes to migration on gel filtration (Fig. 5B) and glycerol gradients (Fig. 5C). In particular, Mcm3 and Mcm5, the subunits most easily detached from the Mcm2-7 hexamer (22, 28), started to migrate on gel filtration as an isolated dimer (Fig. 5, B and C). The same size decrease of the double hexameric Mcm2-7 was observed with chromatin assembled in p27KIP1 treated extract and solubilized at 300 mm potassium acetate (data not shown). This indicates that the inactive form of Mcm2-7 is unstable at higher salt concentrations.
FIGURE 5.
Mid-S phase chromatin. A, chromatin was isolated from egg extract in the middle of S-phase (when replisome proteins peak on chromatin), and proteins were optionally released from chromatin with benzonase in the presence of 50 or 300 mm potassium acetate as indicated. Ex, 0.5 μl of egg extract; Ch, chromatin after first centrifugation; other lanes correspond to insoluble pellet (P) and soluble chromatin (S) from a 5-μl extract after a second centrifugation. B and C, protein complexes released from mid-S phase chromatin by benzonase and solubilized in 300 mm potassium acetate were separated by gel filtration (B) or glycerol gradient (C). Fractions were separated by SDS-PAGE and immunoblotted with antibodies to the indicated proteins.
In whole extract, Cdc45 behaved on gel filtration and glycerol gradients as an isolated monomer of ∼76 kDa (predicted molecular mass 65 kDa), while the GINS subunits Psf1, -2, and -3 and Sld5 behaved as a single isolated hetero-tetramer of ∼86 kDa (predicted molecular mass, 94 kDa) (supplemental Fig. S3). Fig. 6A shows the calculated molecular masses of chromatin-bound Cdc45 and GINS at 100 and 300 mm potassium acetate. At 300 mm potassium acetate, most of the Cdc45 and GINS forms a complex with a calculated molecular mass of ∼707 kDa (Fig. 6A), slightly larger than the main Mcm2-7 peak of ∼633 kDa. Although the Mcm2-7 peak does not precisely correspond to the Cdc45 and GINS peaks, there is still substantial overlap between the profiles (Fig. 5, B and C). This is consistent with the Cdc45 and GINS peak being the CMG complex, which has a predicted molecular mass of 706 kDa. The bulk of Mcm2-7 is likely to come from unfired origins to which Cdc45 and GINS would not be substantially recruited, explaining why Mcm2-7 peak at a slightly lower mass, as double hexamers are unstable at this salt concentration.
FIGURE 6.
Xenopus CMG complex. Chromatin was isolated from egg extract in the middle of S-phase (when replisome proteins peak on chromatin), and proteins were released from chromatin in presence of 50, 100, or 300 mm potassium acetate. A, soluble chromatin protein complexes were separated by gel filtration and glycerol gradient and CMG components (Mcm2-7, Cdc45, and GINS) were followed through fractionation by immunoblotting. The data were used (19) to determine the molecular weight of the complexes. B and C, mid-S phase chromatin solubilized in 300 mm potassium acetate was immunoprecipitated with non-immune or anti-Cdc45 antibodies. Proteins present in the immunoprecipitates were analyzed by immunoblotting (B) or by mass spectrometry (C).
To provide evidence that the ∼700 kDa complexes containing Cdc45 and GINS represent a discrete CMG complex, we immunoprecipitated Cdc45 from chromatin solubilized in 300 mm potassium acetate and analyzed co-precipitating proteins by immunoblotting (Fig. 6B) and mass spectrometry (Fig. 6C). Fig. 6B revealed that Cdc45 efficiently co-precipitated the Sld5 component of the GINS complex, and less efficiently co-precipitated Mcm3 and -7. Mass spectrometry also showed significant enrichment of Mcm2-7 and the Psf1-3 components of the GINS complex in the Cdc45 precipitation (Fig. 6C). These results are consistent with the bulk of Cdc45 being in the form of the CMG complex at 300 mm potassium acetate.
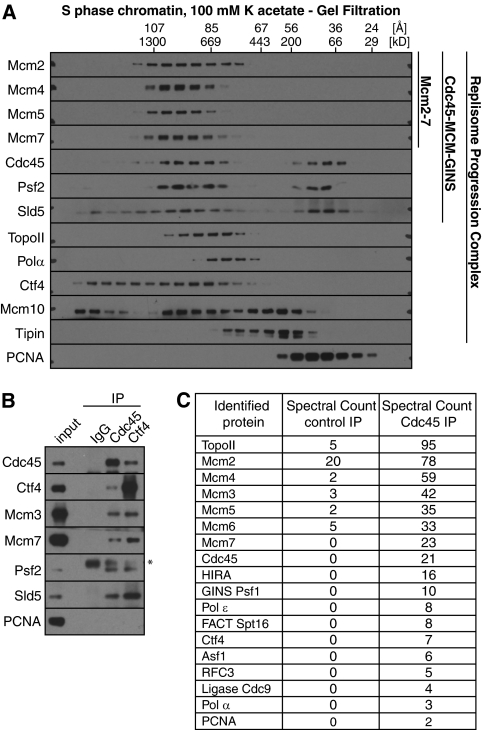
The CMG helicase makes up part of a larger Replisome Progression Complex (RPC), which contains a range of other proteins including the Tipin-Timeless (Tof1-Csm3) complex that allows replication forks to pause at protein-DNA barriers, the histone chaperone FACT and Ctf4 (23, 33–35). To probe for the existence of a Xenopus RPC, we examined complexes solubilized from replicating chromatin in 100 mm potassium acetate. At this salt concentration, only a small fraction of the replisome proteins were solubilized (Fig. 1C), but Mcm2-7 remained together in a high molecular mass complex (Fig. 7A and supplemental Fig. S4) with an estimated molecular mass of ∼811 kDa (Fig. 6A). In 100 mm K acetate, GINS and Cdc45 migrated differently from the bulk of Mcm2-7, particularly on glycerol gradients (supplemental Fig. S4), behaving as though they were part of larger complexes with molecular masses of ∼817 and ∼913 kDa, respectively. This is consistent with the presence in these complexes of other components of the replisome progression complex. Immunoprecipitation of Cdc45 suggested that there was still significant physical association of Cdc45 with Mcm2-7 and GINS in these complexes (Fig. 7B). A proportion of the RPC component Ctf4 was also found co-migrating with Mcm2-7 (Fig. 7A) and co-precipitated with Mcm2-7, Cdc45, and GINS (Fig. 7B). Mass spectrometry of Cdc45 immunoprecipitates from replicating chromatin solubilized in 100 mm potassium acetate also revealed the presence of Ctf4 plus other replication fork proteins, including part of the FACT complex (Fig. 7C). These experiments show that a range of other replisome proteins physically associate with Mcm2-7, Cdc45, and GINS on replicating chromatin.
FIGURE 7.
Behavior of other replisome proteins. Chromatin was isolated from egg extract in the middle of S-phase (when replisome proteins peak on chromatin) and proteins were released from chromatin in 100 mm potassium acetate. A, soluble chromatin protein complexes were separated by gel filtration and immunoblotted for a range of replisome proteins. B, mid-S phase chromatin solubilized in 100 mm potassium acetate was immunoprecipitated with non-immune, anti-Cdc45 or anti-Ctf4 antibodies. Proteins present in the immunoprecipitates were analyzed by immunoblotting. C, mid-S phase chromatin solubilized in 100 mm potassium acetate was immunoprecipitated with non-immune and anti-Cdc45 antibodies, and the precipitates were analyzed by mass spectrometry.
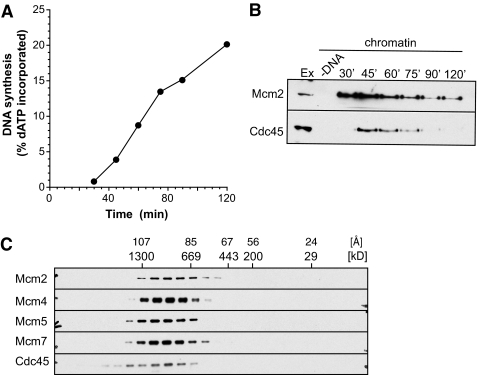
To increase the proportion of Mcm2-7 complexes that undergo initiation and to minimize the number of unfired dormant origins, we lowered total Mcm2-7 levels in the extract ∼90% by immunodepletion. Unfortunately the immunodepletion also reduced overall replication rates so that origin firing was less synchronous than in undepleted extract (Fig. 8, A and B). Despite this, the size of chromatin-bound Mcm2-7 in depleted extract (Fig. 8C; mean peak size ∼90 Å) was slightly smaller than the Mcm2-7 peak seen in undepleted extract (Fig. 7A; mean peak size 97 Å). The reduced size of Mcm2-7 in the depleted extract more closely matched the ∼91 Å) size of Cdc45 and the GINS complex (Figs. 6A, 7A, and 8C). These results are consistent with the idea that reducing the number of dormant origins caused a greater proportion of Mcm2-7 complexes to be assembled into active replisomes, thereby reducing the proportion of double-hexameric Mcm2-7 present at unfired origins.
FIGURE 8.
CMG complex at minimally licensed chromatin. Mcm3 depleted extract was supplemented with 10% of normal extract. A, DNA synthesis was assayed by α-[32P]dATP incorporation. B, chromatin was isolated at the indicated times and immunoblotted for Mcm2 and Cdc45 proteins. C, chromatin was isolated at 45 min and proteins were released from chromatin in 100 mm potassium acetate. Soluble chromatin protein complexes were separated by gel filtration and immunoblotted for the indicated proteins.
DISCUSSION
We have examined the physical composition of chromatin-bound complexes of Mcm2-7 in Xenopus egg extracts. We provide evidence for the existence of at least two different complexes. Prior to replication fork initiation we show that Mcm2-7 bind to chromatin in a large complex that appears to be an Mcm2-7 double hexamer. During S phase, chromatin-bound Mcm2-7 associate with Cdc45 and GINS, to form a complex that appears to represent the Xenopus CMG complex, as well as associating with a range of other replisome proteins.
Licensed Origins Contain Double Hexamers of Mcm2-7
Two recent reports have described the reconstitution of origin licensing using ORC, Cdc6, Cdt1, and Mcm2-7 from budding yeast (12, 13). Electron microscopy showed that when Mcm2-7 was loaded onto DNA, it encircled DNA as a head-to-head double hexamer. Gel filtration analysis showed that this complex eluted on gel filtration with approximately double the size of the free hexamer (12). Previous work has shown that an analogous licensing reaction can be reconstituted using Xenopus sperm and nucleoplasmin, ORC, Cdc6, Cdt1, and Mcm2-7 purified from Xenopus eggs (11). Nucleoplasmin is required to decondense the sperm chromatin and allow ORC to bind DNA (36); the subsequent steps in Xenopus appear very similar to those in budding yeast (11).
Here we have assessed the physical state of Mcm2-7 loaded onto sperm chromatin in Xenopus egg extracts. We show, using gel filtration and glycerol gradient, that once loaded onto chromatin as a consequence of origin licensing, Mcm2-7 form a complex with a molecular mass approximately equal to that of an Mcm2-7 double hexamer. No other proteins are present in the complex at stoichiometric quantities. In contrast, previous work has shown that the bulk of free Mcm2-7 proteins in Xenopus egg extract form a heterohexamer consisting of one each of the 6 proteins Mcm2-7 proteins (22). The relative ratios of the 6 Mcm2-7 proteins on licensed chromatin is approximately stoichiometric. This is consistent with previous data showing that Mcm2-7 proteins are loaded on to chromatin in a similar proportion to the ratios in the free heterohexamer (29). We conclude that in Xenopus egg extract, when Mcm2-7 are loaded onto chromatin they predominantly form double heterohexamers, consistent with the loading of Mcm2-7 in the reconstituted yeast system. The conservation of this mechanism between yeasts and frogs suggests that it is likely to be a conserved mechanism used by all eukaryotes. It makes good sense if the licensing of replication origins represents the loading of two anti-parallel double hexamers, as this is the configuration required to initiate a pair of bidirectional replication forks in S phase.
It is unclear precisely how the double hexamer is assembled on DNA from free hexamers. Several possibilities have been discussed (7, 12, 13). One possibility is that a single molecule of chromatin-bound ORC could cooperatively bind and load the double hexamer. Another possibility is that a pair of ORC molecules could be co-ordinated at each origin in an anti-parallel fashion, each of which loads one hexamer which combine to form the double hexamer. In support of this latter model, archaeal replication origins typically have ORC binding sites arranged in an anti-parallel fashion around the origin (37). Another possibility is that Mcm2-7 are initially loaded as hexamers, which are unstable, but can subsequently become stabilized if they encounter another hexamer to allow a double hexamer to form. This idea is less appealing because there would potentially be a lot of unproductive loading and unloading of hexamers that fail to form double hexamers. There is also some evidence in Xenopus that two molecules of Cdc6 can act together at origins, consistent with either of the first two models (29, 38).
CMG and RPC Complexes in S Phase
Following activation of Cdc7 and S phase CDKs, a proportion of Mcm2-7 at licensed replication origins initiate a pair of bidirectional replication forks. This is accompanied by the association of other replisome (replication fork) proteins with Mcm2-7 and its activation as a helicase. However, the majority of origins remain dormant during S phase, unless replication fork progression is inhibited (25–27, 39). We show here that Mcm2-7 at unreplicated origins remain as double hexamers during S phase. We also show that prior to replication fork initiation, Cdc7 phosphorylation of Mcm4 does not affect its ability to form double hexamers.
Biochemical analysis in Drosophila has provided evidence that for Mcm2-7 to be active as a helicase, it must be associated with Cdc45 and the four GINS proteins Psf1, -2, -3, and Sld5, thereby forming the CMG complex (5, 6). Although a discrete CMG complex has not previously been isolated from other organisms, there is evidence that a similar complex is a conserved feature of the eukaryotic replisome. A group of interacting replisome proteins, the Replisome Progression Complex, has been purified from budding yeast, which contains Mcm2-7, Cdc45, GINS and several other replisome proteins (23, 33, 34). Similarly, in a range of other organisms, physical interactions between Cdc45, Mcm2-7 and GINS have been reported at replication forks (40–46).
We provide here evidence for the existence of a discrete CMG subcomplex that forms part of the replisome in Xenopus egg extracts. When replication forks were fully solubilized in 300 mm potassium acetate, Mcm2-7 proteins behaved as a heterogeneous set of complexes. A subset of the Mcm2-7 proteins co-migrated with Cdc45 and GINS proteins with a molecular mass of 707 kDa, as expected of the Xenopus CMG complex. Immunoprecipitation and mass spectrometry demonstrated that Cdc45 and GINS were major components of this 707 kDa complex. This suggests that the Xenopus CMG complex is a particularly stable subcomponent of the replisome. At lower salt concentrations (100 mm potassium acetate), interactions between the CMG proteins and a range of other replisome proteins were seen, consistent with the existence of a more extensive RPC-like complex.
A more detailed analysis of replisome complexes in Xenopus is complicated by the fact that many replisome proteins, such as Cdc45, remain insoluble in low salt conditions, while other protein complexes, such as Mcm2-7 tend to break up in higher salt. For this reason we have not been able to precisely determine the steps by which the double hexamer of Mcm2-7 is converted into an active helicase. Because Mcm2-7 tend to break up in the salt concentrations necessary to solubilize replisome proteins, we cannot determine whether the two Mcm2-7 hexamers separate at replication initiation, or whether the two opposing replisomes remain associated to extrude rabbit ears of replicated DNA behind the replisome (47–50). At present all we can conclude is that at the lowest salt concentration where we can solubilize replisome proteins, CMG proteins migrate on gel filtration columns at a size significantly smaller than would be expected of a dimeric double replisome. Recent results obtained by stretching DNA that is replicating in Xenopus egg extracts showed that no physical association is required between sister replisomes in order for normal fork progression to occur (51). Further developments of the technique we have used here may be able to address this issue and determine the precise sequence events occurring as Mcm2-7 double hexamers are converted into active replisomes.
Supplementary Material
Acknowledgments
We thank Karim Labib for helpful comments on the manuscript. We thank Gaganmeet Chadha, Grace Cochrane, Michael Munsen, and Alan Score for help purifying and characterizing the antibodies.
This work was supported in part by CRUK (Grants C303/A3135, C303/A8102, C303/A7399, and C303/A5434, BBSRC (Grant BB/H013024/1), and the Wellcome Trust (Grants 08136/Z/06/Z and 083524/Z/07/Z).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
W. T. Poh and J. J. Blow, unpublished data.
- ORC
- origin recognition complex
- aa
- amino acids
- CDK
- cyclin-dependent kinase.
REFERENCES
- 1. Labib K., Diffley J. F. (2001) Curr. Opin. Genet. Dev. 11, 64–70 [DOI] [PubMed] [Google Scholar]
- 2. Forsburg S. L. (2004) Microbiol. Mol. Biol. Rev. 68, 109–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takahashi T. S., Wigley D. B., Walter J. C. (2005) Trends Biochem. Sci. 30, 437–444 [DOI] [PubMed] [Google Scholar]
- 4. Bochman M. L., Schwacha A. (2008) Mol. Cell 31, 287–293 [DOI] [PubMed] [Google Scholar]
- 5. Ilves I., Petojevic T., Pesavento J. J., Botchan M. R. (2010) Mol. Cell 37, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moyer S. E., Lewis P. W., Botchan M. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blow J. J., Dutta A. (2005) Nat. Rev. Mol. Cell Biol. 6, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Machida Y. J., Hamlin J. L., Dutta A. (2005) Cell 123, 13–24 [DOI] [PubMed] [Google Scholar]
- 9. DePamphilis M. L., Blow J. J., Ghosh S., Saha T., Noguchi K., Vassilev A. (2006) Curr. Opin Cell Biol. 18, 231–239 [DOI] [PubMed] [Google Scholar]
- 10. Arias E. E., Walter J. C. (2007) Genes Dev. 21, 497–518 [DOI] [PubMed] [Google Scholar]
- 11. Gillespie P. J., Li A., Blow J. J. (2001) BMC Biochem. 2, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Evrin C., Clarke P., Zech J., Lurz R., Sun J., Uhle S., Li H., Stillman B., Speck C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20240–20245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Remus D., Beuron F., Tolun G., Griffith J. D., Morris E. P., Diffley J. F. (2009) Cell 139, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chong J. P., Hayashi M. K., Simon M. N., Xu R. M., Stillman B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shechter D. F., Ying C. Y., Gautier J. (2000) J. Biol. Chem. 275, 15049–15059 [DOI] [PubMed] [Google Scholar]
- 16. Fletcher R. J., Bishop B. E., Leon R. P., Sclafani R. A., Ogata C. M., Chen X. S. (2003) Nature Struct. Biol. 10, 160–167 [DOI] [PubMed] [Google Scholar]
- 17. Costa A., Pape T., van Heel M., Brick P., Patwardhan A., Onesti S. (2006) J. Struct. Biol. 156, 210–219 [DOI] [PubMed] [Google Scholar]
- 18. Chong J. P., Thömmes P., Rowles A., Mahbubani H. M., Blow J. J. (1997) Methods Enzymol. 283, 549–564 [DOI] [PubMed] [Google Scholar]
- 19. Siegel L. M., Monty K. J. (1966) Biochim. Biophys. Acta 112, 346–362 [DOI] [PubMed] [Google Scholar]
- 20. Montagnoli A., Valsasina B., Croci V., Menichincheri M., Rainoldi S., Marchesi V., Tibolla M., Tenca P., Brotherton D., Albanese C., Patton V., Alzani R., Ciavolella A., Sola F., Molinari A., Volpi D., Avanzi N., Fiorentini F., Cattoni M., Healy S., Ballinari D., Pesenti E., Isacchi A., Moll J., Bensimon A., Vanotti E., Santocanale C. (2008) Nat. Chem. Biol. 4, 357–365 [DOI] [PubMed] [Google Scholar]
- 21. Khoudoli G. A., Gillespie P. J., Stewart G., Andersen J. S., Swedlow J. R., Blow J. J. (2008) Curr. Biol. 18, 838–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prokhorova T. A., Blow J. J. (2000) J. Biol. Chem. 275, 2491–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 24. Gillespie P. J., Khoudoli G. A., Stewart G., Swedlow J. R., Blow J. J. (2007) Curr. Biol. 17, 1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woodward A. M., Göhler T., Luciani M. G., Oehlmann M., Ge X., Gartner A., Jackson D. A., Blow J. J. (2006) J. Cell Biol. 173, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ge X. Q., Jackson D. A., Blow J. J. (2007) Genes Dev. 21, 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ibarra A., Schwob E., Méndez J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thömmes P., Kubota Y., Takisawa H., Blow J. J. (1997) EMBO J. 16, 3312–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oehlmann M., Score A. J., Blow J. J. (2004) J. Cell Biol. 165, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jares P., Blow J. J. (2000) Genes Dev. 14, 1528–1540 [PMC free article] [PubMed] [Google Scholar]
- 31. Walter J. C. (2000) J. Biol. Chem. 275, 39773–39778 [DOI] [PubMed] [Google Scholar]
- 32. Pereverzeva I., Whitmire E., Khan B., Coué M. (2000) Mol. Cell. Biol. 20, 3667–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R. C., Edmondson R. D., Calzada A., Labib K. (2009) EMBO J. 28, 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morohashi H., Maculins T., Labib K. (2009) Curr. Biol. 19, 1943–1949 [DOI] [PubMed] [Google Scholar]
- 35. Tanaka H., Kubota Y., Tsujimura T., Kumano M., Masai H., Takisawa H. (2009) Genes Cells 14, 949–963 [DOI] [PubMed] [Google Scholar]
- 36. Gillespie P. J., Blow J. J. (2000) Nucleic Acids Res. 28, 472–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wigley D. B. (2009) Curr. Opin Struct. Biol. 19, 72–78 [DOI] [PubMed] [Google Scholar]
- 38. Frolova N. S., Schek N., Tikhmyanova N., Coleman T. R. (2002) Mol. Biol. Cell 13, 1298–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blow J. J., Ge X. Q. (2009) EMBO Rep. 10, 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mimura S., Masuda T., Matsui T., Takisawa H. (2000) Genes Cells 5, 439–452 [DOI] [PubMed] [Google Scholar]
- 41. Kubota Y., Takase Y., Komori Y., Hashimoto Y., Arata T., Kamimura Y., Araki H., Takisawa H. (2003) Genes Dev. 17, 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marinsek N., Barry E. R., Makarova K. S., Dionne I., Koonin E. V., Bell S. D. (2006) EMBO Rep. 7, 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pacek M., Tutter A. V., Kubota Y., Takisawa H., Walter J. C. (2006) Mol. Cell 21, 581–587 [DOI] [PubMed] [Google Scholar]
- 44. Bauerschmidt C., Pollok S., Kremmer E., Nasheuer H. P., Grosse F. (2007) Genes Cells 12, 745–758 [DOI] [PubMed] [Google Scholar]
- 45. Im J. S., Ki S. H., Farina A., Jung D. S., Hurwitz J., Lee J. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15628–15632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aparicio T., Guillou E., Coloma J., Montoya G., Méndez J. (2009) Nucleic Acids Res. 37, 2087–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dingman C. W. (1974) J. Theor. Biol. 43, 187–195 [DOI] [PubMed] [Google Scholar]
- 48. Falaschi A. (2000) Trends Genet. 16, 88–92 [DOI] [PubMed] [Google Scholar]
- 49. Kitamura E., Blow J. J., Tanaka T. U. (2006) Cell 125, 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ligasová A., Raska I., Koberna K. (2009) J. Struct. Biol. 165, 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yardimci H., Loveland A. B., Habuchi S., van Oijen A. M., Walter J. C. (2010) Mol. Cell 40, 834–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.