Abstract
The final stage of cytokinesis is abscission, the cutting of the narrow membrane bridge connecting two daughter cells. The endosomal sorting complex required for transport (ESCRT) machinery is required for cytokinesis, and ESCRT-III has membrane scission activity in vitro, but the role of ESCRTs in abscission has been undefined. Here, we use structured illumination microscopy and time-lapse imaging to dissect the behavior of ESCRTs during abscission. Our data reveal that the ESCRT-I subunit tumor-susceptibility gene 101 (TSG101) and the ESCRT-III subunit charged multivesicular body protein 4b (CHMP4B) are sequentially recruited to the center of the intercellular bridge, forming a series of cortical rings. Late in cytokinesis, however, CHMP4B is acutely recruited to the narrow constriction site where abscission occurs. The ESCRT disassembly factor vacuolar protein sorting 4 (VPS4) follows CHMP4B to this site, and cell separation occurs immediately. That arrival of ESCRT-III and VPS4 correlates both spatially and temporally with the abscission event suggests a direct role for these proteins in cytokinetic membrane abscission.
Keywords: superresolution imaging, cell division, centrosomal protein of 55 kDa, mitotic kinesin-like protein 1, Madin–Darby canine kidney cells
Membrane severing of the thin, microtubule-enriched intercellular bridge connecting the two daughter cells at the end of cytokinesis, termed “abscission,” is thought to be regulated at the midbody, a highly dense structure at the center of the intercellular bridge (1–5). Despite the abundance of factors localized to the midbody and known to be necessary for abscission (1–6), the mechanism for mediating the final scission event remains unclear. One family of factors that has been suggested to be involved is the endosomal sorting complex required for transport (ESCRT) machinery (3, 7). Comprising several components, including ESCRT-I, ESCRT-II, ESCRT-III, and vacuolar protein sorting 4 (VPS4), ESCRTs are sequentially recruited to membranes to mediate fission events (8) and have membrane scission activity in vitro (9, 10). During cytokinesis, ESCRT components (including ESCRT-I, ESCRT-III, and VPS4) localize to the midbody (11–13). ESCRT-I localization is mediated by GPPX3Y motifs of the ESCRT-I subunit tumor-susceptibility gene 101 (TSG101) and the ESCRT-related protein apoptosis-linked gene 2-interacting protein X (ALIX) that interact with a noncanonical coiled-coil region of centrosomal protein of 55 kDa (CEP55) (14). CEP55 translocates to the midbody following cyclin-dependent kinase 1 (Cdk1) and Polo-like kinase 1 (Plk1) phosphorylation (15), where it binds to the central spindlin subunit mitotic kinesin-like protein 1 (MKLP1). The ESCRT-III subunit charged multivesicular body protein 1b (CHMP1B) binds to the microtubule-severing enzyme spastin, which is essential for cytokinesis (16, 17). Cell depletion of any of these ESCRT proteins leads to cytokinetic failure, as evidenced by an increase in multinucleated cells (11–13).
These characteristics make ESCRTs attractive candidates for mediating neck cleavage during cytokinetic abscission. However, other properties of cytokinesis and ESCRTs are not consistent with their having a role in abscission (7). The membrane-cutting event during abscission, for example, is thought to occur at a narrow constriction site along the intercellular bridge that is distant from the midbody (3, 5) and where ESCRTs previously have not been localized. In addition, ESCRTs are known to mediate fission of small vesicles (<50 nm diameter) or viruses (<100 nm) (7), making it unclear how they could mediate large-scale scission of membranes >1 μm in diameter (3, 5), as exist within the intercellular bridge during cytokinetic abscission. Finally, cytokinetic failure in the absence of ESCRTs could be an indirect effect of ESCRTs’ having multiple functions in cell division, including roles in maintaining the integrity of centrosomes (18) and the midbody (13).
One way to assess the role of ESCRTs in cytokinesis is to characterize their organization and dynamics during cytokinesis directly using imaging techniques. To do so, we used high-resolution structured illumination microscopy (SIM) and high-sensitivity temporal imaging of fluorescently labeled ESCRT proteins to dissect the spatiotemporal dynamics of ESCRTs at the intercellular bridge during cytokinesis. Our results suggest ESCRTs have a direct role in membrane abscission during cytokinesis. We find that ESCRT proteins assemble into a series of ring-like structures at the midbody center. Immediately before the abscission event, ESCRT-III and VPS4 redistribute to a nearby constriction site where abscission occurs. The tightly correlated recruitment of ESCRT-III/VPS4 to the constriction site and timing of abscission reveal an intimate function for ESCRT machinery in the final events of cytokinetic membrane abscission.
Results
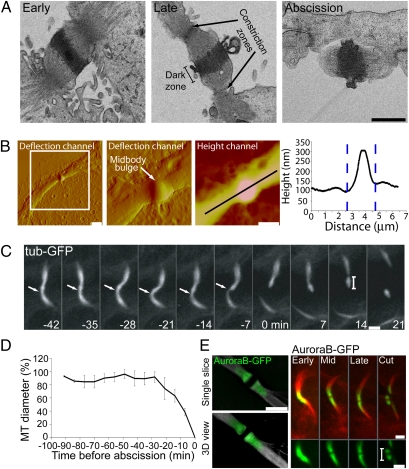
To gain insight into the precise site and timing of cytokinesis, we examined the overall anatomical characteristics of the intercellular bridge found during cytokinesis. EM images of cells undergoing cytokinesis reveal a narrow intercellular bridge with tightly compressed microtubule bundles (Fig. 1A). In the center is a thickened “dark” zone, 0.6 μm wide and with maximum diameter of 1.0 μm (excluding membrane outgrowths). Microtubule bundles extending from either side of the dark zone converge in two narrow membrane-constriction zones that are about 2 μm apart. Imaging using atomic force microscopy (AFM) yields similar horizontal dimensions for the intercellular bridge, confirming the EM data. In addition, AFM reveals a height profile along the intercellular bridge, with the midbody center bulging out about 150–200 nm above the narrow constriction zones found on either side of the midbody (Fig. 1B).
Fig. 1.
Anatomical characteristics of the intercellular bridge during cytokinesis. (A) EM sections of the intercellular bridge from MDCK cells at early to late stages in cytokinesis (left to right) shows that the midbody dark zone (1 μm in diameter and 0.6 μm in width) is separated by constriction zones (arrows) that are 2 μm apart and has a diameter of 0.1–0.5 μm. (Scale bar: 1 μm.) (B) AFM of the intercellular bridge from MDCK cells in cytokinesis. Shown are deflection channels of the same midbody in different scales and a height channel of the zoomed in image. A height profile along the intercellular bridge indicates two constriction zones that are 2 μm apart and are 150–200 nm below the highest point of the midbody. (Scale bars: Left, 5 μm; Right. 2 μm.) (C) Live MDCK cells expressing α-tubulin–GFP during cytokinesis. The length of the midbody remnant (see bar) as measured by the fluorescent intensity profile is 2.18 ± 0.35 μm (n = 17) (Scale bar: 2 μm). (D) The relative change in microtubule diameter over time during cytokinesis at the position indicated by the arrow. Microtubule diameter was normalized to the maximal diameter measured for each cell. Abscission time (time 0) was defined as the time of the first breakage of the microtubule bridge. n = 17. (E) (Left) SIM images of the intercellular bridge of MDCK cells expressing AuroraB-GFP. (Right) Live-cell imaging of cells expressing AuroraB-GFP (green) together with α-tubulin–mCherry (red) during cytokinesis. n = 4. (Scale bars: 2 μm.)
Completion of cytokinesis requires both membrane and microtubule severing. Indeed, microtubule severing is known to correlate tightly with abscission time (19). To test whether the two narrow constriction zones seen by EM and AFM represent sites of cutting during abscission, we imaged dividing Madin–Darby canine kidney (MDCK) cells expressing GFP-tagged α-tubulin. The entire process of cytokinesis in MDCK cells takes about 110 ± 30 min (n = 50). Abscission occurs during the last ~20 min and is characterized by acute narrowing of one side of the midbody bulge followed by microtubule breakage and membrane cutting (arrows in Fig. 1C and in Fig S1; see also Fig. 1D). Narrowing and scission of the other side of the midbody bulge then occurs, leaving a midbody remnant ~2 μm in length (Fig. 1C and Fig. S1). All cells exhibited this abscission phenotype, involving two distinct cuts, separated in time, that occur on either side of the midbody bulge to produce a midbody remnant dissociated from both daughter cells.
To examine whether the remnant includes portions of the intercellular bridge flanking the midbody dark zone, we imaged the microtubule-binding protein AuroraB-GFP during cytokinesis. AuroraB-GFP is excluded from the central region of the midbody representing the dark zone (Fig. 1E Left). Notably, the protein is included in the midbody remnant, rimming either side of the dark zone at a distance of ~1 μm (Fig. 1E Right). This observation suggests that abscission occurs at a similar distance on either side of the midbody dark zone. This distance is consistent with cutting occurring at the two constriction zones seen by EM and AFM. These zones would be cleaved sequentially, giving rise to a 2-μm midbody remnant.
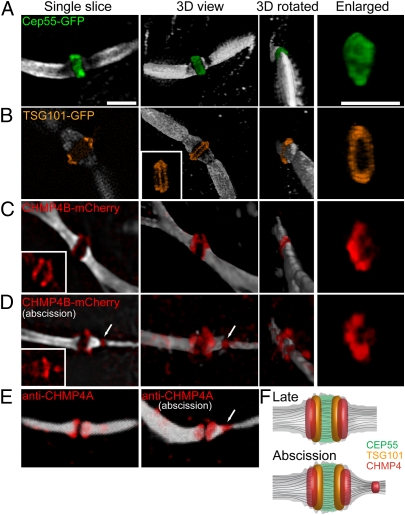
To determine the spatial organization of the ESCRT proteins in the midbody, fluorescently tagged versions of the proteins were imaged by SIM. Antibody staining, with the exception of tubulin, generally was avoided because of known difficulties of labeling the midbody dark zone. Only cells expressing low levels of the fluorescently tagged ESCRT proteins were imaged and analyzed to avoid potential artifacts from overexpression. Indeed, in these cells the overall midbody appearance is normal (Fig. 2 A–D). Moreover, under these conditions the fluorescently tagged ESCRT proteins show their expected interphase cytosolic localization (8, 20, 21), and there is no increase in multinucleated cells, indicating that cytokinesis is not disrupted (Fig. S2). SIM images of fluorescently tagged ESCRT proteins in the intercellular bridge of dividing cells revealed that the midbody structural protein, CEP55, concentrates in a 0.75-μm-wide zone in the midbody (Fig. 2A). This observation is consistent with previous immunolabeling results describing CEP55 midbody localization (22). When the 3D image of CEP55 is rotated, a patchy signal is visible in the interior of the midbody, where microtubules are enriched. This localization pattern indicates that CEP55 extends into the interior of the midbody (as a solid disk), as may be expected since CEP55 associate with microtubules (15, 22). A similar disk-like distribution is seen for the CEP55 binding partner MKLP1 (15, 22) (Fig. S3).
Fig. 2.
Spatial organization of the ESCRT complex in the midbody determined by SIM. MDCK cells expressing CEP55-GFP (A), TSG101-GFP (B), or CHMP4B-mCherry (C and D) were synchronized, fixed, stained with anti–α-tubulin antibodies, and imaged by SIM. (A–D) Each panel shows (from left to right) a single slice, a 3D rendering, a 3D rendering rotated 90°, and a zoomed-in image of the structure. Microtubules are colored in white, CEP55-GFP in green, TSG101-GFP in orange, and CHMP4B-mCherry in red. (A) CEP55 form a diffusely filled structure that is 1.4 ± 0.15 μm in diameter and 0.75 ± 0.07 μm in width. n = 10. A similar structure was observed for MKLP1 (Fig. S3A). (Scale bar: 2 μm.) (B) TSG101 forms a tightly packed double-ring structure surrounding the microtubules at the center of the midbody dark zone (width = 0.82 ± 0.03 μm). The rings are 1.7 ± 0.07 μm in their outer diameter and are 0.23 ± 0.02 μm apart. (Inset) A rotated image demonstrating the existence of two separate rings (n = 5). (C and D) CHMP4B concentrates in two broken rings that are 0.43 ± 0.08 μm apart. The diameter of each broken ring is 1.25 ± 0.18 μm. In some cells CHMP4B also shows an additional pool that is located asymmetrically 1.2 μm away from the center of the dark zone (arrow in D) and is perfectly colocalized to the site of microtubule constriction. (Insets) CHMP4B signal alone (n = 14). The average diameter of the microtubules in all the measurements (excluding D) is 0.98 ± 0.12 μm. The larger diameter measured for the proteins localized to the dark zone correlates with the diameter measured for this area using a membrane marker (1.6 μm; Fig. S3). The dark zone (the zone of no microtubule staining) is 0.7 ± 0.1 μm in width. (E) Confocal 3D-rendered images of antibody staining of endogenous CHMP4A (red) and tubulin (white) on the intercellular bridge of dividing MDCK cells. Images are consistent with the CHMP4B-mCherry localization described by SIM (D). (F) A model for ESCRT organization at the midbody integrating the SIM measurements indicated above.
TSG101 binds directly to CEP55 (11–14) and is one of the most upstream components of the ESCRT machinery (10, 23). In SIM images, TSG101 localizes in two distinct, hollow rings located on either side of the midbody center. The rings are 0.28 μm in thickness and about 0.25 μm apart from one another (Fig. 2B). The rings appear to associate with the cortical surface of the midbody, excluded from the interior of the midbody, in contrast to the disk-like shape of CEP55. The inner diameters of the TSG101 rings are ~1.0 μm, suggesting their presence in the outermost layer of the proteinaceous part of the midbody in close association with the surrounding plasma membrane. The outer diameters of the rings are 1.7 μm on average, suggesting that TSG101 also is localized to the membrane outgrowth seen by EM (Fig. 1A). Indeed, SIM images of the intercellular bridge labeled with a membrane marker show a maximum bridge diameter of 1.6 μm (Fig. S4). The distribution of TSG101, therefore, is consistent with its functioning at the plasma membrane of the intercellular bridge on both sides of the midbody center.
CHMP4 is a subunit of the downstream ESCRT-III complex. SIM imaging of the CHMP4 subunit CHMP4B at the intercellular bridge reveals that it, like TSG101, is organized into two distinct ring-like structures. Previous reports also have shown an accumulation of ESCRT III on both sides of the midbody dark zone (12). Our SIM images show the CHMP4B rings extend more peripherally toward the rims of the dark zone than TSG101 (Fig. 2C). The rings are separated by ~0.45 μm, indicating a 50% overlap with TSG101 in the midbody region. The CHMP4B rings colocalize with the outer membrane of the intercellular bridge (Fig. S4), suggesting they are associated with the plasma membrane. ESCRT-I and -III components (i.e., TSG101 and CHMP4B) thus are organized symmetrically in a series of partially overlapping cortical rings adjacent to the plasma membrane on both sides of the midbody center (Fig. 2F).
Interestingly, in SIM images of midbodies corresponding to late time points in the abscission process (i.e., midbodies having a very narrow constriction with a thinned or broken microtubule bridge), CHMP4B additionally localizes to puncta about 1 μm away from the center of the midbody dark zone, where tubulin antibody labeling is excluded [Fig. 2D (arrows) and F]. This position of CHMP4B puncta (representing a second, later pool) closely coincides with the site of constriction in the intercellular bridge, where membrane abscission occurs (Fig. 2D, arrows). A second puncta pool also is seen with antibody staining for the endogenous ESCRT-III subunit CHMP4A using conventional confocal microscopy (Fig. 2E). Thus, both endogenous and fluorescently tagged CHMP4 proteins localize to the membrane cut site at late time points of cytokinesis. This localization raises the possibility that ESCRT-III proteins are involved directly in the final membrane abscission event.
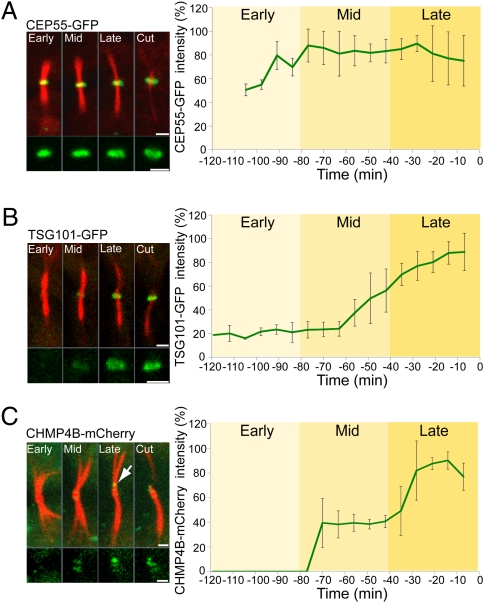
To investigate this scenario further, we compared the SIM data with the temporal kinetics of abscission. Live MDCK cells expressing ultralow levels of fluorescent protein-tagged versions of MKLP1, CEP55, TSG101, CHMP4B, and VPS4B together with fluorescently tagged tubulin were monitored by time-lapse video microscopy during cytokinesis. In these cells, the mean times for cytokinesis were in the normal range (Fig. 3 and Movies S1, S2, S3, S4, and S5), indicating that expression of the proteins did not affect the temporal kinetics of cytokinesis. CEP55 is steadily present at the midbody bulge from the start of cytokinesis (Fig. 3A and Movie S1). A nearly identical kinetic profile is seen for MKLP1 (Fig. S3B and Movie S5). TSG101 levels associated with the midbody bulge, on the other hand, begin to increase only at the midphase, rising continuously until abscission (Fig. 3B and Movie S2). The kinetic profile for levels of the ESCRT-III component CHMP4B at the midbody bulge is different from that of either CEP55 or TSG101: It increases in two steps before abscission. CHMP4B begins incorporating into the midbody at midphase. Then, very close to the timing of the abscission event, there is an acute increase in CHMP4B labeling in the second puncta pool localized at one of the constriction sites (arrow in Fig. 3C and Movie S3). Notably, the peak of CHMP4B association with the constriction site always coincides with an acute decrease in tubulin diameter that is followed by abscission (Fig. 4A). This timing indicates that CHMP4B association at the constriction site correlates with membrane abscission.
Fig. 3.
Live-cell imaging of MDCK cells undergoing cytokinesis reveals sequential recruitment of the ESCRT components to the midbody bulge. Cells expressing low levels of CEP55-GFP (A), TSG101-GFP (B), and CHMP4B-mCherry (C) together with α-tubulin–mCherry (A and B) or α-tubulin–GFP (C) imaged using a spinning-disk confocal microscope at 7-min intervals. Each panel shows maximum intensity projection merged images of the microtubules (red) and the protein specified (green) in different stages during cytokinesis. Maximum intensity projections of the protein of interest alone are shown below. Intensities above background were measured from the sum intensity projection at each time point and are plotted to the right of each panel. Background intensities or lower were set as 0. Time 0 was determined as the time of the first microtubule breakage. The arrow in C indicates the site of acute increase in CHMP4B signal. The mean times for cytokinesis in the cells analyzed were CEP55, 89 ± 9 min (A); TSG101, 105 ± 12 min (B); and CHMP4B, 102 ± 16 min (C). These values are in the normal range for cytokinesis in MDCK cells, which is defined as abscission within 110 ± 30 min (as measured in 50 MDCK cells stably expressing α-tubulin–GFP). The double-ring structure observed for TSG101 by SIM could not be resolved here because it is below the optical resolution of this system. n = 8. (Scale bars: 2 μm.)
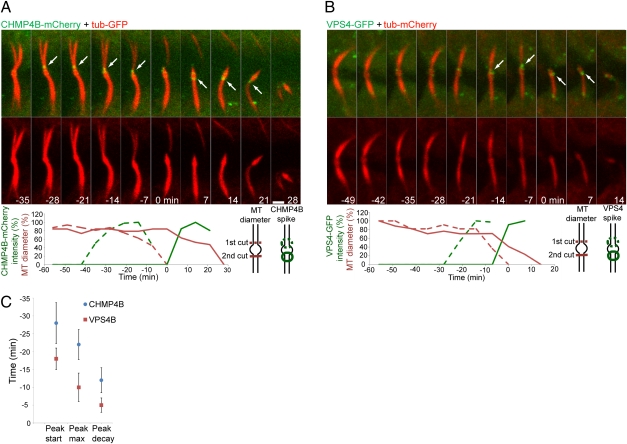
Fig. 4.
Acute increases in CHMP4B and VPS4B levels at the constriction site are tightly correlated in time with acute constriction of the microtubule bridge. MDCK cells expressing CHMP4B-mCherry (green) and α-tubulin–GFP (red) (A) or VPS4B-GFP (green) and α-tubulin–mCherry (red) (B) were imaged during cytokinesis. The images shown are sequential frames of the last steps of abscission. (A and B Upper) The upper rows show an overlay of CHMP4B (A) or VPS4B (B) on the microtubule signal; the lower rows show the tubulin signal alone. (A and B Lower) Plots of the change in microtubule diameter and fluorescence intensity of CHMP4B (A) or VPS4B (B) in the constriction zones (see diagrams at right of graph). (Scale bar: 2 μm.) (C) The average peak time of CHMP4B and VPS4B in the constriction zones relative to the abscission event (time 0) is plotted. n = 5.
Membrane abscission in MDCK cells involves two consecutive cuts on the membrane necks facing either side of the midbody bulge (Fig. 1C and Fig. S1). We therefore compared ESCRT protein recruitment to both cut sites by time-lapse imaging (Fig. 4A). On either side of the midbody bulge, CHMP4B peaks at the constriction site about 20 min before abscission followed by an acute decrease in microtubule diameter that ends in membrane cutting (Fig. 4A and Movie S3). Notably, the microtubule diameter of the second scission site does not change throughout the first scission event and drops only after the appearance of CHMP4B at this second site. This observation implies a direct cause-and-effect relationship between CHMP4B recruitment to the constriction site and membrane abscission.
VPS4B, the ATPase responsible for recycling ESCRT-III and the most downstream component of the ESCRT machinery, shows similar characteristics, but it peaks closer to the time of abscission (~10 min before abscission) (Fig. 4 B and C and Movie S4). Thus, there is a lag in the peak of VPS4B relative to that of CHMP4B, consistent with a function of VPS4 downstream of CHMP4B.
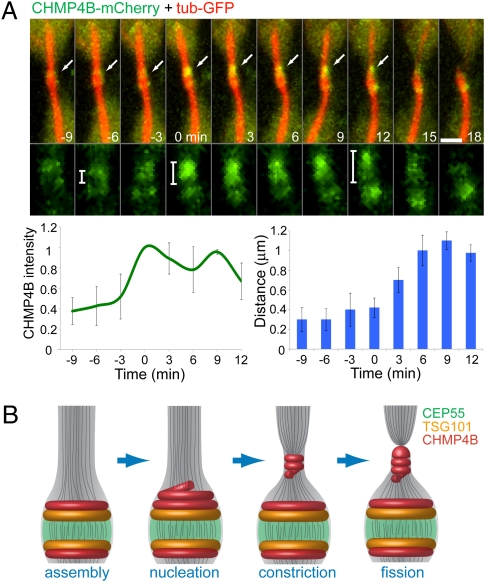
To investigate how the localization pattern observed for the different ESCRT components relates to the event of abscission, we examined more closely the dynamics of the ESCRT-III protein CHMP4B late in cytokinesis. In particular, we focused on the two pools of CHMP4B seen at this time, which, as described above, include an initial pool localizing in rings at positions overlapping with the TSG101 rings and a second pool seen transiently at the abscission sites (Fig. 2D and Fig. 4A). Quantitative imaging of live MDCK cells coexpressing CHMP4B-mCherry and tubulin-GFP close to the time of abscission reveals an acute increase in CHMP4B intensity located asymmetrically on one of the CHMP4B initial rings near the midbody center (Fig. 5A). This new pool of CHMP4B then appears to move outward until it stabilizes at a distance ~1.0 μm away from the center of the midbody, where acute membrane constriction and abscission occur (Fig. 5A, arrows). Although, we cannot not rule out the possibility that the second ESCRT-III pool arises independently of the initial ESCRT-III ring, our finding suggests that these two pools are closely related to one another. Indeed, after the intensity of CHMP4B spikes at the initial ring, no further increase in intensity of the complex occurs as it moves outward, only a loss. ESCRT assembly at the midbody thus could serve as a staging area for nucleation of the ESCRT-III complex, which then relocalizes to the constriction zone where abscission takes place.
Fig. 5.
Suggested model for ESCRT-mediated abscission. (A) MDCK cells expressing CHMP4B-mCherry (green) and α-tubulin–GFP (red) were imaged during late cytokinesis at 3-min intervals. (Upper) The images shown are sequential frames of the merged image (upper row) and the CHMP4B channel alone (lower row). (Scale bars: 2 μm.) (Lower Left) The graph shows the change in CHMP4B intensity on the side that is about to break. (Lower Right) The graph shows the distance between the highest-intensity CHMP4B pixel and the lowest-intensity CHMP4B pixel (located between the two initial rings) measured from a line intensity profile plotted along the midbody bridge on the side that is about break. This measurement was used as an indication of the location of the second CHMP4B pool relative to the center of the midbody. Time 0 was determined as the time CHMP4B intensity reached its maximal value. n = 5. (B) Suggested model for ESCRT-mediated constriction and fission during cytokinetic abscission. Assembly of CEP55 (green), TSG101 (yellow), and CHMP4B (red) at the midbody center forms a platform for initiation of abscission. Closer to abscission, ESCRTIII levels increases at the midbody and then relocalizes to the constriction zones, probably by polymerizing into a spiral. This relocalization induces constriction that is followed by breakage of the intercellular bridge, leading to complete separation of the two daughter cells.
Discussion
The current model for ESCRT function demonstrated in vitro (9, 24) involves sequential membrane recruitment of different ESCRT components to orchestrate fission. As discussed above, a similar sequential recruitment of ESCRTs is seen during cytokinetic abscission. ESCRT-I incorporates into the midbody center after CEP55. The arrival of ESCRT-III at the midbody correlates closely with that of ESCRT-I, but its levels also peak acutely very close to the abscission event. VPS4 follows CHMP4B, with its peak intensity lagging that of CHMP4B and coinciding with membrane cutting. The preservation of the ordered mechanism of ESCRT activity at the cut sites therefore supports a fundamental role for ESCRT machinery in the abscission event itself.
Abscission consists of the nearly simultaneous cleavage of the microtubule bridge and membrane severing. Because these events are coupled by the recruitment of the microtubule-severing enzyme spastin to the late ESCRT-III protein CHMP1B (16, 17), ESCRT-III machinery probably functions both in microtubule severing and membrane scission. That said, our results do not differentiate the extent of direct involvement of ESCRT-III in microtubule severing versus membrane scission, but the latter would be most consistent with the in vitro activity of this complex.
CHMP4B is thought to be one of the two earliest ESCRT-III proteins to assemble and the most important for membrane scission, by analogy to its yeast ortholog (10, 23). CHMP4B also has been shown to form spiral-shaped filaments when overexpressed in cells (25). In addition, in vitro studies have shown that ESCRT-III components spontaneously polymerize into spiral filaments that expose their membrane interaction sites on the surface, allowing them to bind tightly to surrounding membranes (26). A plausible model (Fig. 5B), therefore, is that the first pool of CHMP4B (having a ring-like shape with a large diameter) corresponds to the nuclei of CHMP4 filaments. These filaments subsequently extend away from the center of the midbody to give rise to the second CHMP4B pool, which deforms the membrane neck into a narrow constriction. We do not observe a continuous distribution of CHMP4B from the TSG101 ring region to the narrow constriction, but we might not be able to detect such fluorescence because the cells express very low levels of tagged CHMP4B. An alternative model is that the midbody CHMP4B rings are staging points for the activation of CHMP4 and other ESCRT-III monomers, which then diffuse across the gap to the site of abscission. Whatever the mechanism, the model (Fig. 5B) clarifies the role of the midbody as an organizing center for abscission, in that upstream ESCRT proteins (TSG101) of the midbody serve to recruit and activate ESCRT-III proteins. The ESCRT-III proteins then invade the zone immediately distal to the midbody center, inducing the abscission event.
In conclusion, our central observation is that ESCRT-III and VPS4 concentrate transiently at the narrow constriction sites on either side of the midbody bulge with the appropriate timing to carry out scission. This finding strongly supports the model that ESCRT-III is directly responsible for cytokinetic membrane abscission in mammalian cells using the same type of membrane scission activity observed in vitro for yeast ESCRT-III (9, 24). This mechanism parallels the mechanism for membrane abscission in the Crenarchaea (27, 28), which have ESCRT-III and VPS4 but lack known orthologs of other eukaryotic membrane-trafficking proteins.
Materials and Methods
SIM Imaging.
MDCK cells were seeded at 30% density on number 1.5 coverslips (Zeiss) and were transfected 16 h later with MKLP1-GFP, CEP55-GFP, TSG101-GFP, or CHMP4B-mCherry. As described in SI Materials and Methods, cells then were synchronized with aphidicolin (Sigma) and fixed (4% paraformaldehyde) 10–11 h later. All samples were stained with anti–α-tubulin antibodies (DM1A; Sigma). With the exception of tubulin, we chose to use direct imaging of fluorescently tagged proteins rather than antibody staining to avoid known artifacts of midbody-protein staining that probably are caused by the high density of proteins in this region.
Thin (0.15–0.2 μm) z-stacks of high-resolution images were collected in five rotations for each midbody using an ELYRA PS.1 (Carl Zeiss MicroImaging) microscope. Images then were reconstructed using ZEN software (Carl Zeiss MicroImaging) based on the structured illumination algorithm developed by Heintzmann and Cremer (29). All measurements were performed on reconstructed superresolution images of single z-sections in ZEN. 3D rendering was done in Volocity 4 (Perkin-Elmer).
For CHMP4A antibody staining, cells were synchronized and fixed as described above. Cells then were stained with anti-CHMP4A polyclonal antibodies (20) (kindly provided by Dr. Phyllis Hanson, Washington University, St. Louis, MO) and anti–α-tubulin antibodies and imaged by the Marianas confocal spinning-disk microscope (described below).
Live-Cell Imaging.
MDCK cells were plated in low density on a four-well chamber slide (Nunc) and were cotransfected with a combination of MKLP1-GFP, CEP55-GFP, TSG101-GFP, or VPS4-GFP with tubulin-mCherry or CHMP4B-mCherry with tubulin-GFP. Cells were imaged 16–40 h after transfection. Z-stacks of selected low-expressing cells undergoing cytokinesis were collected at the specified intervals using a confocal spinning disk microscope (Marianas; Intelligent Imaging) and were video recorded on an EM-CCD camera (evolve; Photometrics). Image processing and analysis were done using Slidebook 5 (Intelligent Imaging). Only cells that successfully completed cytokinesis and showed normal kinetics (defined as abscission within 110 ± 30 min as observed in the cytokinesis of 50 MDCK cells stably expressing α-tubulin–GFP) were analyzed. Extra care was taken in imaging CHMP4B-mCherry and VPS4B-GFP because of the known potential dominant negative effect of expressing these proteins. Therefore, only cells that showed a uniform dim cytosolic distribution of these proteins were imaged and analyzed. All intensity measurements were done on sum projection images of the 3D movie series after background subtraction. The microtubule diameter was determined based on the microtubule fluorescence intensity profile along a line that was positioned about 1 μm away from the center of the midbody (from both sides) perpendicular to the intracellular bridge. The position of CHMP4B in time (Fig. 5A) was determined by measuring the distance between the lowest-intensity pixel and the highest-intensity pixel from a line intensity profile positioned along the intercellular bridge. The lowest-intensity pixel was found experimentally to represent the center of the area between the two CHMP4B initial rings and therefore was used as a reference for measuring the position of CHMP4B.
A more detailed description of the methods and descriptions of the EM and AFM methods are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Misha Kozlov for mechanistic insights and fruitful discussion. We thank Carl Zeiss Microimaging, LLC for access to the ELYRA PS.1 microscope and specifically thank Maya Everret for coordinating that access. We also thank Rainer Heintzmann for technical suggestions on SIM acquisition and reconstruction parameters, Mike Davidson for kindly proving the CAAX-tdEOS construct, and Phyllis Hanson for providing the CHMP4A polyclonal antibodies. We thank members of the J.L.-S. laboratory and Jeremy Swan, Angelika Rambold, and Nichole Jonas for help with the illustrations. This research was supported by the Intramural Program of the National Institutes of Health, National Institute of Child Health and Human Development, National Institute of Diabetes and Digestive and Kidney Diseases, and Intramural AIDS Targeted Antiviral Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102714108/-/DCSupplemental.
References
- 1.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 2.Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: From parts list to mechanisms. Annu Rev Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 3.Steigemann P, Gerlich DW. Cytokinetic abscission: Cellular dynamics at the midbody. Trends Cell Biol. 2009;19:606–616. doi: 10.1016/j.tcb.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Sagona AP, Stenmark H. Cytokinesis and cancer. FEBS Lett. 2010;584:2652–2661. doi: 10.1016/j.febslet.2010.03.044. [DOI] [PubMed] [Google Scholar]
- 5.Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gromley A, et al. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 7.Schiel JA, Prekeris R. Making the final cut—mechanisms mediating the abscission step of cytokinesis. ScientificWorldJournal. 2010;10:1424–1434. doi: 10.1100/tsw.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wollert T, et al. The ESCRT machinery at a glance. J Cell Sci. 2009;122:2163–2166. doi: 10.1242/jcs.029884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: It's all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 12.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci USA. 2008;105:10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabbro M, et al. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell. 2005;9:477–488. doi: 10.1016/j.devcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Yang D, et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morita E, et al. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci USA. 2010;107:12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steigemann P, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 21.Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhao WM, Seki A, Fang G. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol Biol Cell. 2006;17:3881–3896. doi: 10.1091/mbc.E06-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 24.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lata S, et al. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindås AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heintzmann R, Cremer CG. Laterally modulated excitation microscopy: Improvement of resolution by using a diffraction grating. Proc SPIE. 1999;3568:185–196. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.