Abstract
Infection-driven inflammation has been implicated in the pathogenesis of ~15–20% of human tumors. Expression of microRNA-155 (miR-155) is elevated during innate immune response and autoimmune disorders as well as in various malignancies. However, the molecular mechanisms providing miR-155 with its oncogenic properties remain unclear. We examined the effects of miR-155 overexpression and proinflammatory environment on the frequency of spontaneous hypoxanthine phosphoribosyltransferase (HPRT) mutations that can be detected based on the resistance to 6-thioguanine. Both miR-155 overexpression and inflammatory environment increased the frequency of HPRT mutations and down-regulated WEE1 (WEE1 homolog-S. pombe), a kinase that blocks cell-cycle progression. The increased frequency of HPRT mutation was only modestly attributable to defects in mismatch repair machinery. This result suggests that miR-155 enhances the mutation rate by simultaneously targeting different genes that suppress mutations and decreasing the efficiency of DNA safeguard mechanisms by targeting of cell-cycle regulators such as WEE1. By simultaneously targeting tumor suppressor genes and inducing a mutator phenotype, miR-155 may allow the selection of gene alterations required for tumor development and progression. Hence, we anticipate that the development of drugs reducing endogenous miR-155 levels might be key in the treatment of inflammation-related cancers.
Keywords: genome stability, LPS, TNF, proliferation, transgenic mice
It now is well established that chronic and persistent inflammation contributes to cancer development (1–3). Infection-driven inflammation is involved in the pathogenesis of ~15–20% of human tumors, and tumor-infiltrating leukocytes, such as monocytes/macrophages, T lymphocytes, and neutrophils, are prime regulators of cancer inflammation (1–3). Furthermore, even tumors that are not epidemiologically linked to pathogens are characterized by the presence of an inflammatory component in their microenvironment.
MicroRNAs (miR) are a class of short, noncoding RNAs that are implicated in many aspects of cell biology. MiR-155 is implicated in hematopoiesis, the innate and acquired immune response, and autoimmune disorders (4–9). A direct link between elevated levels of miR-155 and the formation and development of tumors such as leukemias and breast, lung, or gastric cancers has been established recently (4, 5, 9–12). Thus, transgenic mice overexpressing miR-155 in B cells or hematopoietic stem cells show enhanced proliferative disorder that results in high-grade lymphoma (13, 14). Being oncogenic and implicated in inflammation, miR-155 is the prototype of microRNAs that stand between inflammation and carcinogenesis. However, the molecular mechanisms behind this link remain unclear. Because miR-155 targets transcripts implicated in DNA repair (15), we evaluated the mutator activity of miR-155 and that of the miR-155–linked inflammatory environment as a potential mechanism connecting inflammation and cancer. In this study we report that miR-155 and inflammatory stimuli increase the spontaneous mutation rate.
Results
Overexpression of MiR-155 Results in Enhanced Mutation Rate.
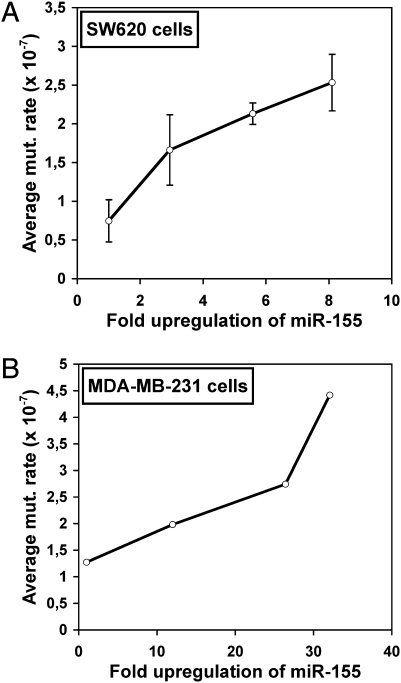
MiR-155 targets core components of the DNA mismatch repair (MMR) machinery, among other mutator pathways (15), suggesting that elevated levels of miR-155 might enhance the rate of spontaneous mutations. To measure the mutation rate, we took advantage of the hypoxanthine phosphoribosyltransferase (HPRT) locus, which is a well-established method for estimating mutation rate. The HPRT enzyme catalyzes the conversion of guanine into guanine monophosphate and hypoxanthine into inosine monophosphate in the purine salvage pathway (16, 17). The loss of HPRT function confers resistance to 6-thioguanine (6-TG), because 6-TG becomes cytotoxic only after phosphoribosylation by HPRT. This resistance can be used to identify cells that have acquired mutations at the HPRT locus (16, 17). Because the acquired mutations are thought to occur randomly, the HPRT gene can be used as a reporter gene, and the frequency of mutation at the HPRT locus can be used as an estimate of global genomic instability. To measure the effects of miR-155 on mutation rate, we first developed stable clones of SW620 colorectal adenocarcinoma cells and MDA-MB-231 breast adenocarcinoma cells expressing mature miR-155 under the control of the Tet-On inducible system. Incubation of SW620 clones 8A, 22C, and 23A with doxycycline increased miR-155 expression by 2.94 ± 0.23-, 5.58 ± 0.43-, and 8.10 ± 0.65-fold, respectively (mean ± SD) (Fig. S1 and Table S1). Similarly, doxycycline treatment increased miR-155 expression by 12.01 ± 2.34-, 26.42 ± 1.11-, and 32.07 ± 3.27-fold in MDA-MB-231 clones 9C, 2B, and 19B, respectively.
The cell growth-adjusted HPRT mutation rate, estimated based on a modified version of fluctuation analysis (18), increased with miR-155 levels in both SW620 and MDA-MB-231 cell clones (Fig. 1 A and B). Constant elevated expression of miR-155 the enhanced mutation rate by up to 3.39-fold in SW620 clones and up to 3.47-fold in MDA-MB-231 clones (Table S1). Moreover, HCT116 colorectal carcinoma cells transiently overexpressing miR-155 by 19.57 ± 0.62-fold under the control of Tet-On inducible system (Fig. S2) showed a 2.81-fold higher mutation rate (Table S1). These results establish a direct link between mutation rate and miR-155 levels.
Fig. 1.
The average mutation rate in SW620 and MBA-MD-231 cells increases with the rate of miR-155 expression. (A and B) Average mutation rates of 6-TG–resistant colonies from SW620 (A) and MDA-MB-231 clones (B) stably expressing miR-155 and either mock-treated or treated with doxycycline, plotted against the fold upregulation of miR-155 values. Fold upregulation 1 value corresponds to mock-treated controls.
As expected, the basal spontaneous cell growth-adjusted mutation rates of SW620 and MDA-MB-231 cells was comparable (0.75 ± 0.27 × 10−7 and 1.28 × 10−7 mutations per cell, respectively) and were ~440-fold lower than the spontaneous mutation rate of HCT116 cells (560 × 10−7 mutations per cell) (Table S1), because HCT116 cells contain a deletion of the hMLH1 (human MutL homolog 1) MMR gene (19). The deletion of the hMLH1 MMR gene decreased the miR-155-induced mutator activity only partially, suggesting that miR-155–induced mutator activity is not very sensitive to the basal level of mutation rate (i.e., to the integrity of the DNA safeguarding machinery) and that miR-155 targets additional transcripts implicated in DNA repair and/or genome stability.
Inflammatory Stimuli Up-Regulate MiR-155 in Breast Cancer Cells.
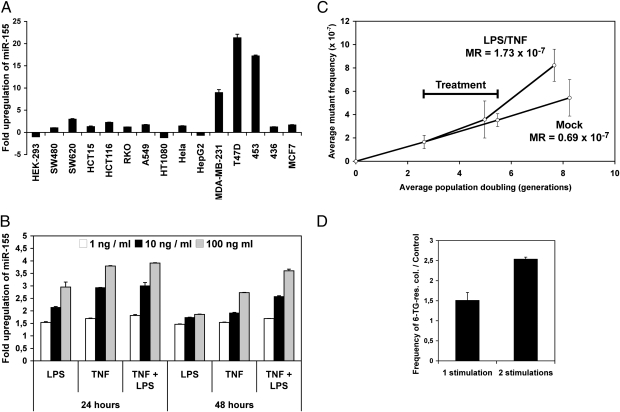
Infection-driven inflammation has been associated with cancer (20). A number of studies have shown that miR-155 is a key component of the inflammatory response (4–7). We thus screened human cell lines for the effects of proinflammatory environment on miR-155 expression. We included colon cancer cell lines (SW480, SW620, HCT15, HCT116, and RKO), because a fraction of colorectal cancers appear linked to the inflammatory environment (20), breast (MDA-MB-231, T47D, 453, 436, and MCF7) and lung cancer (A459) cell lines, because miR-155 is up-regulated in these types of cancers (12), and four other cell lines for comparison. Cells were treated overnight with the supernatant of LPS-stimulated human THP-1 monocytic cells, namely LPS-stimulated macrophage-conditioned medium (LSMCM), which contains many inflammatory cytokines such as TNF, IL-6, IL-8, and IL1-β (1, 3). Based on quantitative RT-PCR (qRT-PCR) analyses, miR-155 expression in colon and lung cancer cell lines was affected only slightly by LSMCM (Fig. 2A). In contrast, miR-155 levels increased by 9-, 17-, and 21-fold in MDA-MB-231, BC-453, and T47D breast cancer cell lines, respectively. We also analyzed the expression of miR-146a, a microRNA that is up-regulated in certain tumors (10, 11) and in LPS-challenged THP-1 cells. It has been proposed that miR-146a controls the termination of the immune response (21). In sharp contrast with miR-155, the highest miR-146a levels were found in HCT15 and HCT116 colon cell lines (not shown), indicating that the up-regulation of miR-155 and miR-146a by inflammatory stimuli occurs independently in the above cancer cell lines and probably is tissue specific. Because both TNF (22) and LPS can induce miR-155 (6, 7), we analyzed the effects of these two molecules in MDA-MB-231 cells. As expected, stimulation with either TNF or LPS or with both increased miR-155 expression (Fig. 2B). We therefore used TNF/LPS to mimic the effects of a proinflammatory environment.
Fig. 2.
Proinflammatory environment increases the mutation rates in MDA-MB-231 cells and the frequency of mutant colonies in T47D cells. (A) The relative up-regulation of miR-155 in the indicated cell lines either mock treated or treated with LSMCM for 48 h were determined using qRT-PCR. The figure gives the ratios LPS-stimulated/unstimulated. Values represent mean ± SD (n = 3). (B) The relative up-regulation of miR-155 in MDA-MB-231 cells treated as indicated was determined using qRT-PCR. (C) The mutation rates (MR) in MDA-MB-231 cells were estimated by calculating the slopes of the curves following mock and TNF/LPS treatment (n = 4 estimations of mutation frequency). (D) Ratios of 6-TG–resistant colonies in T47D cells treated with LSMCM for 48 h versus control mock-treated cells.
Inflammatory Stimuli Enhance the Mutation Rate.
In MDA-MB-231 cells without stimulation, the mutation rate was calculated to be 0.69 × 10−7 mutations per cell per generation, a value similar to that previously found in SW480 cells (0.75 × 10−7) (23). The estimated mutation rate was based on the average mutant frequency and population doubling (23, 24). Of note, both MDA-MB-231 and SW480 cells have intact DNA-repair machinery, unlike HCT116 cells. Mutant frequencies already had became significantly different (P = 0.038) 3 d after treatment, with mutation rate increasing by 2.52-fold to 1.73 × 10−7 mutations per cell per generation (Fig. 2C) in TNF/LPS-treated MDA-MB-231 cells vs. untreated control cells. The treatment lowered the rate of cell proliferation, probably because TNF induces growth arrest in breast cancer cells (25). Accordingly, one or two LSMCM stimulations of T47D cells increased the frequency of 6-TG–resistant colonies by 50% and 150%, respectively (Fig. 2D). Thus, proinflammatory signals resulting in the up-regulation of miR-155 expression induce a significant, although moderate, mutator phenotype, which might be enhanced with chronic inflammation.
Characterization of HPRT Mutants.
We then analyzed HPRT mutations found in cDNAs prepared from RNA extracted from T47D, HCT116, and MDA-MB-231 6-TG–resistant colonies to determine the mutation signature (Table 1 and Table S2). As expected, HPRT mutations from doxycycline-treated HCT116 cells displayed single base deletions or insertions of the type generally found in DNA MMR-deficient cells. The majority displayed a frameshift, transition, and transversion mutation signature consistent with an MMR defect. There also was an increase in insertions and exon deletions with miR-155 overexpression; this increase generally has been ascribed to altered recombination repair (26). In contrast, deletion mutations consistent with recombination repair defects accounted for the majority of HPRT mutations in LSMCM-stimulated T47D cells and doxycycline-treated MDA-MB-231 cells, regardless of conditions (Table 1 and Table S2). These results are consistent with a role for recombination repair, exemplified by breast cancer 1 (BRCA1) and breast cancer 2 (BRCA2) mutations, in these breast tumor cell lines (27). Of note, these types of mutations have been found previously in several T-cell leukemic cell lines (16, 28). However, we noted a modest increase in transitions and transversions consistent with decreased MMR in these cells.
Table 1.
Mutations found in HPRT cDNAs prepared from 6-TG–resistant colonies of T47D, HCT116, and MDA-MB-231 cells
| Cell lines | Treatment | Number of clones analyzed | Clones without mutation in the coding sequence | Clones with frameshift mutation(s) | Clones with deletion(s) | Clones with only a single deletion | Clones with one or more exons lacking | Clones with other types of deletions | Clones with insertion(s) | Clones with transition(s) | Clones with transversion(s) |
| T47D | Mock | 24 | 0 | 12 | 14 | 17 | 24 | 4 | 1 | 4 | 0 |
| Del ex. 3: 11 | Del 1: 7 | ins 1: 1 | Transitions: 5 | ||||||||
| Del ex. 2+3: 9 | |||||||||||
| Del ex. 4+5: 4 | |||||||||||
| T47D | LSMCM | 40 | 0 | 20 | 40 | 25 | 38 | 11 | 3 | 7 | 1 |
| Del l ex. 3: 14 | Del 17: 3 | Ins 1: 3 | Transitions: 7 | Transitions: 3 | |||||||
| Del ex. 2+3: 16 | |||||||||||
| Del ex. 4 + 5: 5 | |||||||||||
| Del ex. 2+3+4: 1 | Del 1: 21 | ||||||||||
| Del ex. 6+7+8: 1 | |||||||||||
| Del ex. 5: 1 | |||||||||||
| HCT116 | − Doxycycline | 14 | 1 | 13 | 13 | 0 | 0 | 13 | 4 | 0 | 2 |
| Del 2: 1 | Ins 1: 5 | Transitions: 7 | Transversions: 3 | ||||||||
| Del 1: 12 | |||||||||||
| HCT116 | − Doxycycline | 14 | 1 | 13 | 13 | 0 | 0 | 13 | 4 | 0 | 2 |
| Del 2: 1 | Ins 1: 5 | Transversions: 2 | |||||||||
| Del 1: 12 | |||||||||||
| HCT116 | + Doxycycline | 25 | 4 | 18 | 11 | 0 | 2 | 9 | 13 | 6 | 6 |
| Del ex. 3: 1 | Del 1: 16 | Ins 1: 17 | Transitions: 6 | Transversions: 8 | |||||||
| Del ex. 2+3: 1 | |||||||||||
| MDA-MB-231 | − Doxycycline | 11 | 3 | 8 | 10 | 5 | 8 | 3 | 0 | 1 | 0 |
| Del ex. 2+3: 1 | Del 1: 5 | Transitions: 1 | |||||||||
| Del ex. 7: 8 | |||||||||||
| MDA-MB-231 | + Doxycycline | 24 | 2 | 17 | 21 | 11 | 17 | 9 | 0 | 4 | 2 |
| Del ex. 2: 1 | Del 1: 17 | Transitions: 6 | Transversions: 2 | ||||||||
| Del ex. 2 + 3: 1 | |||||||||||
| Del ex. 7: 16 |
Ex., exon; LSMCM, LPS-stimulated macrophage-conditioned medium; Mock, unstimulated macrophage-conditioned medium.
Overexpression of miR-155 Enhances Cell Proliferation.
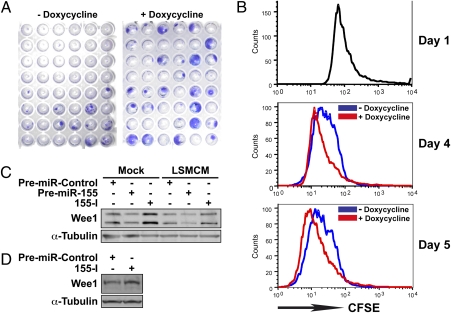
Remarkably, miR-155 up-regulation increased the size of HCT116 (Fig. 3A) and MDA-MB-231 HPRT mutant colonies and allowed them to appear earlier during the selection process. Based on a forward scatter comparison, larger colonies of MDA-MB-231 clone 19B, that presented a 32-fold up-regulation of miR-155 after doxycycline treatment, did not arise from the presence of larger cells (not shown). In contrast, carboxyfluorescein succinimidyl ester (CFSE) staining suggested that these cells underwent at least one extra round of cell division within 4–5 d as compared with untreated cells (Fig. 3B). These results correlate with reports showing that miR-155 promotes proliferation in transgenic mice (13, 14). Thus, the larger size of HPRT mutant colonies overexpressing miR-155 probably arises from enhanced cell proliferation.
Fig. 3.
Elevated miR-155 levels increase the rate of proliferation by targeting WEE1 transcripts. (A) Phenotypes of 6-TG–resistant HCT116 colonies following 48-h mock treatment (− doxycycline) or 48-h doxycycline treatment (+ doxycycline). HCT116 cells were transiently infected with pRetroX-Tight-Pur-miR-155 and Tet-On constructs before doxycycline treatment. (B) Cells from MDA-MB-231 clone 19B were stained with CFSE before induction of miR-155 expression by doxycycline treatment. The proliferation rate was analyzed by flow cytometry 4 and 5 d later. The experiment was repeated two times with similar results. (C) The levels of WEE1 in T47D cells transfected with premiR-Control, premiR-155, or antisense miR-155 inhibitory RNA (155-I) and subsequently either mock treated or treated with LSMCM for 48 h were determined by Western blotting. (D) The levels of Wee1 in primary B cells isolated from the spleen of Eμ-miR-155 transgenic mice after transfection with premiR-Control or antisense miR-155 inhibitory RNA (155-I).
MiR-155 and Inflammatory Environment Down-Regulate WEE1, a Cell-Cycle Inhibitor.
MiR-155 might enhance cell proliferation by targeting cell-cycle regulators. Indeed, in T47D cells, both LSMCM treatment and miR-155 overexpression reduced the levels of WEE1, a kinase that catalyzes the inhibitory tyrosine phosphorylation of Cdc2/cyclin B, blocking cell-cycle progression at the G2/M phase (Fig. 3C) (29). In contrast, an antisense miR-155 inhibitory RNA (155-I) increased WEE1 accumulation. Both LSMCM and miR-155 overexpression also reduced the expression of a luciferase reporter construct containing the WEE1 3′ UTR (Fig. S3). Accordingly, 155-I increased Wee1 levels in primary B cells isolated from Eμ-miR-155 transgenic mice that overexpress miR-155 in B-cell lineage (13), thus confirming that Wee1 is a bona fide miR-155 target (Fig. 3D). Taken together, these results show that inflammatory stimuli down-regulate WEE1 through up-regulation of miR-155. Because WEE1 depletion rapidly induces DNA damage in newly replicated DNA (30), these results suggest that miR-155 overexpression may shorten the period required for selection of cancer-associated mutations. Furthermore, Affymetrix microarrays revealed that transcripts coding for several factors controlling cell cycle, DNA repair, and genome stability were affected by LSMCM in both T47D and MDA-MB-231 cell lines (Table 2). This result suggests that the ability of inflammatory stimuli to induce defective checkpoints and genomic instability, similar to miR-155, might contribute to tumorigenesis.
Table 2.
Transcripts encoding factors related to DNA replication and maintenance whose levels decrease significantly following treatment of T47D and MDA-MB-231 cells
| Symbol | Gene name | Fold change |
| RFC5 | Replication factor C (activator 1) 5, 36.5kDa | 0.72 |
| NEIL3 | Nei endonuclease VIII-like 3 (E. coli) | 0.71 |
| AURKB | Aurora kinase B | 0.71 |
| GTSE1 | G-2 and S-phase expressed 1 | 0.64 |
| RAD54L | RAD54-like (S. cerevisiae) | 0.75 |
| ARL3 | ADP ribosylation factor-like 3 | 0.81 |
| BCCIP | BRCA2 and CDKN1A interacting protein | 0.77 |
| SKP2 | S-phase kinase-associated protein 2 (p45) | 0.65 |
| RDM1 | RAD52 motif 1 | 0.75 |
| PARP2 | Poly (ADP ribose) polymerase 2 | 0.86 |
| RAD54B | RAD54 homolog B (S. cerevisiae) | 0.67 |
| ERCC6L | Excision repair cross-complementing rodent repair deficiency, complementation group 6-like | 0.61 |
| DCTPP1 | dCTP pyrophosphatase 1 | 0.78 |
| DDB2 | Damage-specific DNA binding protein 2, 48kDa | 0.84 |
| CDKN2C | Cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 0.64 |
| AK3L1 | Adenylate kinase 3-like 1 | 0.84 |
| APRT | Adenine phosphoribosyltransferase | 0.84 |
| DDB2 | Damage-specific DNA binding protein 2, 48kD | 0.84 |
| BRCC3 | BRCA1/BRCA2-containing complex, subunit 3 | 0.83 |
| TOP2A | Topoisomerase (DNA) II alpha 170kD | 0.77 |
| CDT1 | Chromatin licensing and DNA replication factor 1 | 0.74 |
| RECQL4 | RecQ protein-like | 0.72 |
| PARP1 | Poly (ADP ribose) polymerase 1 | 0.86 |
After Affymetrix microarray analyses, comparisons were done between two pools of cells treated with either unstimulated macrophage-conditioned medium or with LPS-stimulated macrophage-conditioned medium. The two pools contained four independent replicates from both T47D and MDA-MB-231 cell lines. Transcripts are arranged according to the decreasing values of P (all <1 × 10−7).
Discussion
In this study we analyzed the mutator activity of miR-155 and of the miR-155–related proinflammatory environment. Cells in which inflammatory stimuli resulted in the up-regulation of miR-155 showed a two- to threefold increase in the mutation rate as deduced by HPRT assay. Furthermore, inducible expression of miR-155 resulted in a similar increase in mutation rate, suggesting that the up-regulation of the mutation rate by the inflammatory stimuli is miR-155 dependent. The mutation rate was not increased in cells in which inflammatory stimuli up-regulated only miR-146, another microRNA implicated in the innate immune response (data not shown).
Although miR-155 levels in MDA-MB-231 cells were increased consistently by 12- and 32-fold during doxycycline treatment, they increased transiently by only approximately fourfold after LPS/TNF treatment (Figs. 1B and 2B). Nevertheless, the mutation rate increased by 1.56- to 3.47-fold after doxycycline treatment and by 2.52-fold after TNF/LPS treatment (Figs. 1B and 2C). This result suggests that increased miR-155 levels resulting from chronic inflammation (4–7), autoimmune diseases (8, 9), or the deregulation of endogenous genetic circuitries with the onset of cancer (4) might produce a significant mutator phenotype. These results also suggest that other inflammatory signaling pathways may work in synergy or in parallel with miR-155. Of note, miR-155 has been shown to target tumor suppressor genes such as Fas-associated via death domain (FADD), Jumonji AT-rich interactive domain 2 (JARID2), and Src homology 2-containing inositol phosphatase-1 (SHIP1) (Table S3). In addition, other microRNAs with mutator activity might potentially be up-regulated by LPS signaling.
The increased mutation rate in HCT116 cells that lack the hMLHl DNA repair enzyme suggested that this increase occurs through miR-155 targeting of other transcripts involved in DNA repair, recombination, or cell-cycle checkpoints. Because mutations accumulate during the S phase, when the replication of the DNA takes place right before the G2/M check point, we looked for transcripts that are predicted targets of miR-155 and act as inhibitors of G2/M transition because their reduced expression might be associated with an increased mutation rate. In addition these transcripts should be targets of LPS/TNF signaling. We concentrated on WEE1 kinase, because it fulfilled all these criteria. In T47D cells, overexpression of miR-155 or treatment with LSMCM resulted in the down-regulation of WEE1 expression. By targeting WEE1 and consequently facilitating G2/M transition, miR-155 potentially allows cells that have not yet repaired the DNA to proceed to mitosis, resulting in accumulated mutations. Akt kinase also is known to function as a G2/M initiator and to inactivate WEE1 by phosphorylation, thus promoting the cell-cycle transition. It was reported recently that Akt (v-akt murine thymoma viral oncogene homolog 1) is implicated in LPS signaling by modulating the levels of miR-155, among other microRNAs (31). It is possible that oncogenic Akt and onco-inflammatory miR-155 cross talk at the level of WEE1 during inflammation. We consider that the increased mutation rate associated with inflammatory signals is a combinatorial effect of miR-155 targeting of WEE1 and other DNA repair enzymes that are down-regulated by LPS and are either direct or indirect targets of miR-155. It is believed that cancer results from the accumulation of mutations in somatic cells, and this study suggests that, by increasing the mutation rate, the inflammatory miR-155 might be the key player in inflammatory-induced cancers in general.
The control of cell-cycle progression and DNA repair in eukaryotes are highly conserved. However, in the event of an infection the cells must respond quickly by producing cytokines, chemokines, and other inflammatory components of the immune defense. During this robust response, it is possible that the DNA repair machinery and cell-cycle checkpoints are put on hold. At this stage the up-regulation of miR-155 by inflammatory stimuli to clear the antigen quickly also results in an increased mutation rate. Furthermore, regardless of the primary cause of a mutation, there is a high probability that, in the event of an infection, the mutation will be fixed. In conclusion, we believe that simultaneous miR-155–driven suppression of a number of tumor suppressor genes combined with a mutator phenotype allows the shortening of the series of steps required for tumorigenesis and represents a model for cancer pathogenesis (Fig. S4). Thus, the up-regulation of miR-155 by chronic inflammation appears to indicate at least one of the missing links between cancer and inflammation.
Materials and Methods
Cell Culture, Transfection, and Treatment.
Cells were grown following standard procedures. T47D cells were transfected using lipofectamine (Invitrogen). Unstimulated LSMCM were prepared from the supernatant of human THP-1 monocytic cells mock stimulated or stimulated with Salmonella enteritidis-derived LPS (100 ng/mL, Sigma) for 6 h. THP-1 cells subsequently were centrifuged, and the supernatant was filtrated to eliminate any remaining cells. T47D cells then were cultivated in the presence of unstimulated medium or LSMCM for 48 h. When needed, a second stimulation was conducted in the same way, after the cells had been allowed to recover 4 d in regular medium. TNF was obtained from Invitrogen. The B-cell line was established by purifying B cells from the spleen of an Eμ-miR-155 transgenic mouse using the isolation kit from R & D Systems. B cells subsequently were cultured for 2 wk in 100 ng/mL RPMI/15% FBS/LPS and for 3 more weeks without LPS. They were electroporated using the Amaxa kit (Lonza).
Preparation of Retroviral Expression Constructs and Retroviral Infection.
See SI Materials and Methods for information.
Selection of 6-TG–Resistant Colonies, Estimation of Mutation Rates, and Analysis of HPRT cDNA Mutations.
See SI Materials and Methods for information.
Isolation of RNAs and qRT-PCR.
RNA was extracted with TRIzol (Invitrogen) and subsequently subjected to DNase digestion (Turbo-DNase; Ambion). MiR-155 qRT-PCR was performed using TaqMan MicroRNA Assays (Applied Biosystems). Values were normalized using RNU-44. Real-time PCR was run in triplicate from three different cDNAs.
Flow Cytometry Analysis.
CFSE was purchased from Molecular Probes/Invitrogen. CFSE staining was carried out using manufacturer's protocol. Cells were fixed in 1% paraformaldehyde before analyses. Flow cytometry analyses were performed at the Flow Cytometry facility of Ohio State University. Data were analyzed using the software program FlowJo (Tree Star, Inc.).
Western Blots.
Cells were lysed 48 h after transfection or electroporation. Anti-WEE1 and anti–α-tubulin antibodies were from Cell Signaling Technology.
Affymetrix Microarray Analyses.
RNAs extracted with TRIzol (Invitrogen) were subsequently subjected to DNase digestion (Turbo-DNase; Ambion). Affymetrix microarray analyses were done at the Ohio State University microarray facility.
Luciferase Assays.
Cells plated in 12-well plates (1 × 106 cells per plate) were transfected with 0.4 μg of DNA (pGL3-control vector or WEE1 reporter constructs; Promega), 20 ng of Renilla luciferase control vector (pRL-TK; Promega), and 50 nM of either a premiR control (premiR Precursor Molecule-Negative Control #1; Ambion), premiR-155 (miR-155 precursor; Ambion), or 155-I (an antisense miR-155 inhibitory RNA; Ambion). Assays were performed 48 h after transfection using the Dual Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity.
Supplementary Material
Acknowledgments
This work was funded by National Institutes of Health Grant CA123541.
Footnotes
The authors declare no conflict of interest.
Data deposition: Microarray data have been submitted to the MIAMExpress (Minimum Information About a Microarray Experiment, www.ebi.ac.uk/miamexpress) with accession no. E-TABM-1118.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101795108/-/DCSupplemental.
References
- 1.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 2.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 3.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: Interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tili E, Croce CM, Michaille JJ. miR-155: On the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28:264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 5.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 6.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tili E, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 8.O'Connell RM, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonkoly E, Pivarcsi A. Advances in microRNAs: Implications for immunity and inflammatory diseases. J Cell Mol Med. 2009;13:24–38. doi: 10.1111/j.1582-4934.2008.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tili E, et al. miRNAs and their potential for use against cancer and other diseases. Future Oncol. 2007;3:521–537. doi: 10.2217/14796694.3.5.521. [DOI] [PubMed] [Google Scholar]
- 11.Tili E, Michaille J-J, Costinean S, Croce CM. MicroRNAs, the immune system and rheumatic disease. Nat Clin Pract Rheumatol. 2008;4:534–541. doi: 10.1038/ncprheum0885. [DOI] [PubMed] [Google Scholar]
- 12.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Connell RM, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valeri N, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci USA. 2010;107:6982–6987. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albertini RJ. HPRT mutations in humans: Biomarkers for mechanistic studies. Mutat Res. 2001;489:1–16. doi: 10.1016/s1383-5742(01)00064-3. [DOI] [PubMed] [Google Scholar]
- 17.Krenitsky TA, Papaioannou R, Elion GB. Human hypoxanthine phosphoribosyltransferase. I. Purification, properties, and specificity. J Biol Chem. 1969;244:1263–1270. [PubMed] [Google Scholar]
- 18.Hall BM, Ma CX, Liang P, Singh KK. Fluctuation analysis CalculatOR: A web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25:1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papadopoulos N, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 20.Rhodes JM, Campbell BJ. Inflammation and colorectal cancer: IBD-associated and sporadic cancer compared. Trends Mol Med. 2002;8:10–16. doi: 10.1016/s1471-4914(01)02194-3. [DOI] [PubMed] [Google Scholar]
- 21.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen IM, et al. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol Med. 2009;1:288–295. doi: 10.1002/emmm.200900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glaab WE, Tindall KR. Mutation rate at the HPRT locus in human cancer cell lines with specific mismatch repair-gene defects. Carcinogenesis. 1997;18:1–8. doi: 10.1093/carcin/18.1.1. [DOI] [PubMed] [Google Scholar]
- 24.Rossman TG, Goncharova EI, Nádas A. Modeling and measurement of the spontaneous mutation rate in mammalian cells. Mutat Res. 1995;328:21–30. doi: 10.1016/0027-5107(94)00190-g. [DOI] [PubMed] [Google Scholar]
- 25.Shen WH, et al. Proinflammatory cytokines block growth of breast cancer cells by impairing signals from a growth factor receptor. Cancer Res. 2002;62:4746–4756. [PubMed] [Google Scholar]
- 26.Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460:984–989. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 28.Lin YW, Perkins JJ, Zhang Z, Aplan PD. Distinct mechanisms lead to HPRT gene mutations in leukemic cells. Genes Chromosomes Cancer. 2004;39:311–323. doi: 10.1002/gcc.20005. [DOI] [PubMed] [Google Scholar]
- 29.Katayama K, Fujita N, Tsuruo T. Akt/protein kinase B-dependent phosphorylation and inactivation of WEE1Hu promote cell cycle progression at G2/M transition. Mol Cell Biol. 2005;25:5725–5737. doi: 10.1128/MCB.25.13.5725-5737.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck H, et al. Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. J Cell Biol. 2010;188:629–638. doi: 10.1083/jcb.200905059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Androulidaki A, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.