Abstract
The circadian clock is phase-delayed or -advanced by light when given at early or late subjective night, respectively. Despite the importance of the time-of-day–dependent phase responses to light, the underlying molecular mechanism is poorly understood. Here, we performed a comprehensive analysis of light-inducible genes in the chicken pineal gland, which consists of light-sensitive clock cells representing a prototype of the clock system. Light stimulated expression of 62 genes and 40 ESTs by >2.5-fold, among which genes responsive to the heat shock and endoplasmic reticulum stress as well as their regulatory transcription factors heat shock factor (HSF)1, HSF2, and X-box-binding protein 1 (XBP1) were strongly activated when a light pulse was given at late subjective night. In contrast, the light pulse at early subjective night caused prominent induction of E4bp4, a key regulator in the phase-delaying mechanism of the pineal clock, along with activation of a large group of cholesterol biosynthetic genes that are targets of sterol regulatory element-binding protein (SREBP) transcription factor. We found that the light pulse stimulated proteolytic formation of active SREBP-1 that, in turn, transactivated E4bp4 expression, linking SREBP with the light-input pathway of the pineal clock. As an output of light activation of cholesterol biosynthetic genes, we found light-stimulated pineal production of a neurosteroid, 7α-hydroxypregnenolone, demonstrating a unique endocrine function of the pineal gland. Intracerebroventricular injection of 7α-hydroxypregnenolone activated locomotor activities of chicks. Our study on the genome-wide gene expression analysis revealed time-of-day–dependent light activation of signaling pathways and provided molecular connection between gene expression and behavior through neurosteroid release from the pineal gland.
The oscillatory mechanism of the circadian clock is constituted of clock genes and the encoded proteins to generate physiological and behavioral rhythms with daily periodicity (1–3). The intrinsic period length of the circadian clock deviates from 24 h, thus clocks are entrained to (reset by) the environmental changes, among which light is the most common and the strongest time cue. A light stimulus given at early or late subjective night induces phase delay or phase advance, respectively, whereas the light during the subjective daytime has little effect on the clock (4). This time-of-day–dependent response is conserved among species, but the underlying molecular mechanism remains to be elucidated.
Among vertebrate clock-containing cells, the chicken pinealocyte is unique in that it retains intrinsic phototransduction pathways for entrainment of the clock (5, 6). Hence, the chicken pineal clock cell provides a prominent platform for studies on the light-entrainment mechanism. We performed previously a differential display analysis of light-inducible genes in the chicken pineal gland and identified a limited number of transcripts that are induced by light selectively at either early or late subjective night (7). Notably, light at early subjective night caused remarkable induction of a transcript encoding a bZIP transcription factor E4BP4 that acts as a light-dependent suppressor of Per2, particularly in the phase-delaying process (8).
To identify transcription pathways that are light-activated at a particular time of day, we investigated light responses of the chicken pineal transcriptome by high-density oligonucleotide array (GeneChip) analysis. Our study revealed time-of-day–dependent activation of pathways mediated by sterol regulatory element-binding protein (SREBP), X-box-binding protein 1 (XBP1), and heat shock factor (HSF) transcription factors. SREBP activated the transcription of E4bp4, raising SREBP as a light-responsive immediate regulator of the clock phase. On the basis of the light activation of SREBP and the target genes for cholesterol biosynthesis, we found a substantial activity of the pineal gland to synthesize a neurosteroid, 7α-hydroxypregnenolone, that activates locomotor activities of chicks, connecting light-induced gene expression with behaviors.
Results
Light Pulse Induces a Subset of Pineal Genes in a Time-of-Day–Dependent Manner.
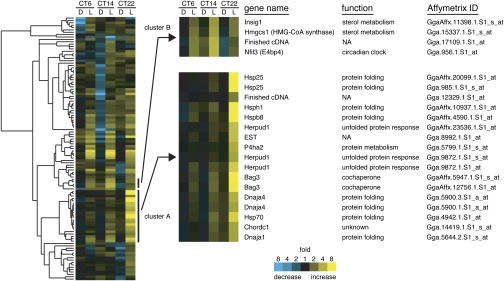
To pursue time-of-day–dependent light responses of the pineal transcriptome, 12-h light/12-h dark cycle-entrained male chicks were transferred to constant darkness and exposed to light for 1 h from circadian time (CT) 6 (representing subjective day), CT14 (early subjective night), or CT22 (late subjective night). The pineal glands were isolated just after the 1-h light pulse or alternative 1-h dark period (as a control) and subjected to gene expression analysis by Affymetrix GeneChip containing 37,700 probe sets. The data from the dark-kept animals revealed circadian expression of the clock genes Per, Cry, Clock, and Bmal (Fig. S1, gray bars) with temporal profiles that are similar to those reported (9–11). Out of ≈21,000 probe sets that detected significant gene expression in the pineal gland, 111 probe sets (corresponding to 62 genes and 40 ESTs) exhibited >2.5-fold increase of the signal intensities when the 1-h light pulse was given at any of the three time points (Fig. S2A). Noticeably, most (89%) of the light-activated genes were strongly induced only at a single time point, indicating strict circadian gating of the light induction.
Light Pulse at Late Subjective Night Induces Stress–Response Genes.
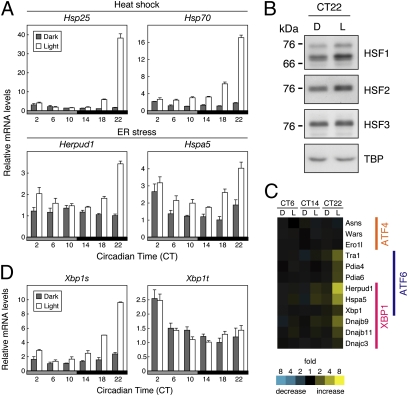
The light-responsive genes detected by the 111 probe sets were first analyzed by a functional classification. Among a variety of functional groups, one related to protein folding formed a predominant group (Fig. S2B). Next, a hierarchical clustering analysis based on the light–response profiles (Fig. 1 and Fig. S3) revealed that most of the protein folding-related genes belonged to a single cluster showing prominent light induction almost specifically at CT22 (Fig. 1 and Fig. S3, cluster A). These genes are known to respond to either heat shock or endoplasmic reticulum (ER) stress (12–14), suggesting that these two pathways are activated specifically at late subjective night. We further explored circadian profiles of the light responses of heat shock–response genes (Hsp25 and Hsp70) and ER stress–response genes (Herpud1 and Hspa5) at 4-h intervals by reverse transcription (RT)-PCR analysis. The light induction of these genes was highly circadian-regulated with the largest response at CT22, whereas their basal expression profiles were less rhythmic in constant darkness (Fig. 2A). Heat shock–response genes are transcriptionally regulated mainly by a family of heat shock factor (HSF) transcription factors, and the activity of HSF is regulated posttranslationally by nuclear localization, oligomerization, and/or phosphorylation (13). We analyzed nuclear levels of HSF1–3 and found that HSF1 and HSF2 protein levels were up-regulated by the 1-h light pulse at CT22, whereas HSF3 level was almost unchanged (Fig. 2B). HSF1 and HSF2 are most likely the key regulators of the light response of the heat shock–response genes (Discussion). As for ER stress, it stimulates gene expression by activating three members of bZip transcription factors: ATF4, ATF6, and XBP1 (14). We found that ATF6- and XBP1-target genes but not ATF4-targets were prominently induced by light at CT22 (Fig. 2C and Fig. S4A). Activity of XBP1 is known to be regulated by frame-switch splicing, which produces a spliced form of Xbp1 mRNA (termed Xbp1s) encoding active XBP1 (14). We found that the formation of pineal Xbp1s showed a striking peak of light response at CT22. In contrast, the total amount of spliced and unspliced forms of Xbp1 (termed Xbp1t) showed no significant light response over the day (Fig. 2D). These results revealed light-dependent activation of HSF1, HSF2, and XBP1 pathways at late subjective night. In the mouse liver, HSF1 is implicated in the feeding-dependent regulation of rhythmic gene expression (15). Here, we found a robust daily rhythm of mouse liver Xbp1s mRNA level in ad libitum feeding condition (Fig. S5A) and the peak in the nighttime was attributed to food intake (Fig. S5B), whereas a recent report showed a low amplitude rhythm of Xbp1s during starvation (16). Because feeding is one of the most important time cues for entrainment of the liver clock (17), it is possible that HSF1 and XBP1 act as common regulators of rhythmic gene expression between the chick pineal gland and the mouse liver when the clock cells respond to resetting signals (Discussion).
Fig. 1.
Light-induced genes in the chicken pineal gland. Dark-kept animals were exposed to a 1-h light pulse (L) from CT6, CT14, or CT22, or kept in the dark (D). The pineal glands were isolated at CT7, CT15, or CT23 for GeneChip analysis (n = 2). (Left) Light-induced genes detected by 111 probe sets (>2.5-fold induction in at least a single time point) were subjected to the hierarchical clustering analysis. Signal intensities obtained from two independent samples were averaged for each probe set at each time point, and the average value of dark samples was set to 1. Genes clustered with stress–response genes (cluster A) and those with E4bp4 (cluster B) are shown at Right.
Fig. 2.
Induction of stress–response genes by a light pulse at late subjective night. (A and D) Effect of light on the mRNA levels. Dark-kept animals were exposed to a 1-h light pulse (open bars) or kept in the dark for 1 h (solid bars) from each time point, and the pineal glands were isolated for RT-PCR analysis. The intensity of each amplified product was normalized to that of Tbp. The lowest value of the dark-kept animals was set to 1. Data are the mean ± SEM (n = 4). (B) Effect of light on nuclear HSF1, HSF2, and HSF3 levels. Dark-kept animals were exposed to a 1-h light pulse (L) from CT22, or kept in the dark (D), and then the pineal glands were isolated at CT23. Nuclear extracts (70 μg of protein for each lane) were subjected to immunoblot analysis. Data are the representatives of two independent experiments. (C) Light–response of ATF4, ATF6, and XBP1–target genes. ATF4, ATF6, and XBP1–target genes commonly identified in previous reports (36–39) were selected, and their expression profiles in the pineal GeneChip analysis are indicated as in Fig. 1. No probe set was assigned to Chop gene, which is a target of both ATF4 and ATF6.
SREBP-Target Genes and E4bp4 Display Similar Light–Response Patterns.
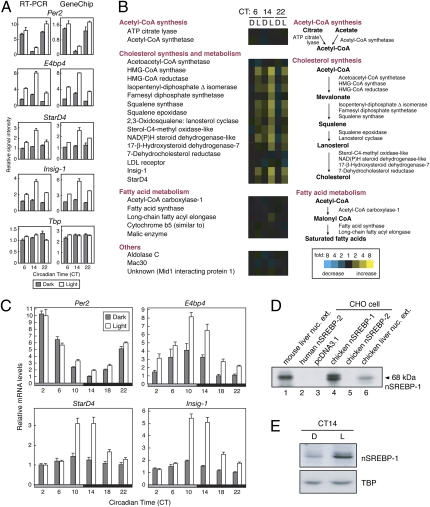
Among the clock genes, E4bp4 was highly light-responsive in the pineal gland (Fig. S1). The hierarchical clustering analysis demonstrated that E4bp4 formed a cluster with Insig-1 and HMG-CoA synthase genes, all of which showed prominent light induction at CT14 (Fig. 1 and Fig. S3, cluster B). By applying our previous differential display analysis of the pineal genes (7), we also found that E4bp4, StarD4, and Insig-1 transcripts were up-regulated by light at early subjective night (Fig. 3A, Left). In the mouse liver, Horton et al. (18) identified StarD4, Insig-1, and HMG-CoA synthase as the target genes of SREBP, a transcription factor activating expression of an array of genes responsible for synthesis and uptake of cholesterol and fatty acids in the liver (19). Giving attention to the SREBP-target genes, we found that genes participating in the cholesterol biosynthesis and metabolism were remarkably induced by the light pulse at CT14 (Fig. 3B and Fig. S4B). In contrast, another subset of SREBP-target genes responsible for synthesis of acetyl-CoA and fatty acids were almost unaffected by the light pulse given at any time point (Fig. 3B and Fig. S4B; see Discussion). Detailed analysis of circadian profiles of the light response of StarD4 and Insig-1 revealed that the transcript levels were markedly up-regulated by the light pulse given at around the day-night transition (CT10 and CT14), a temporal pattern that is very similar to that observed for E4bp4 induction (Fig. 3C). These results suggest a common regulatory mechanism underlying the light induction of E4bp4 and cholesterol-related genes through SREBP-mediated process.
Fig. 3.
Activation of SREBP pathway by a light pulse at early subjective night. (A) Time-of-day–dependent light-induction of E4bp4, StarD4, and Insig-1 genes. Dark-kept animals were exposed to a 1-h light pulse (open bars) or kept in the dark as a control (solid bars) from each time point, and the pineal glands were isolated for RT-PCR and GeneChip analyses. Data are the mean ± SEM (RT-PCR; n = 3) or the mean with variation (GeneChip; n = 2). (B) Light responses of SREBP–target genes in the pineal gland. Expression profiles of SREBP–target genes reported by Horton et al. (18) were extracted from the pineal GeneChip analysis result and indicated as in Fig. 1. The gene expression analysis program indicated that the signals for CYP51, Mevalonate pyrophosphate decarboxylase, Narc-1, and Srebp-1 were “Absent”. No probe set was assigned to Phosphomevalonate kinase, Sterol-C5-desaturase, Sterol C14-reductase-like, and SCALD. (C) Effect of light on the mRNA levels. RT-PCR analysis was performed as in Fig. 2A. (D) Detection of chicken nSREBP-1 by immunoblot analysis. Liver nuclear extracts of mouse (lane 1) or chicken (lane 6) and lysates of CHO-K1 cells transfected with nSREBP-expression vector or empty vector (lanes 2–5) were subjected to immunoblot analysis with anti-SREBP-1 (2A4) antibody. None of available antibodies recognized chicken nSREBP-2 and, hence, we were unable to examine light activation of this protein. (E) Effect of light on nSREBP-1 level in the pineal gland. Dark-kept animals were exposed to a 1-h light pulse (L) from CT14 or kept in the dark (D), and then the pineal glands were isolated at CT15. Nuclear extracts were subjected to immunoblot analysis. Data are the representatives of three independent experiments.
Light Pulse at Early Subjective Night Activates SREBP.
SREBP, a member of bHLH-Zip transcription factors, is encoded by two genes in vertebrates, Srebp-1 and Srebp-2, each of which produces ≈120 kDa inactive precursor bound to ER membrane. The N-terminal bHLH domain of SREBP is proteolytically released from the membrane as a 68-kDa form, which translocates to the nucleus where it acts as an active transcription factor designated nSREBP (19). RT-PCR analysis revealed that Srebp-1 and Srebp-2 are expressed in the chicken pineal gland at levels comparable to those in the liver (Fig. S6A). Among available antibodies, anti-SREBP-1 (2A4) antibody reactive to mouse liver nSREBP-1 (Fig. 3D, lane 1) detected not only chicken nSREBP-1 expressed in CHO-K1 cells (Fig. 3D, lane 4) but also endogenous nSREBP-1 in the nuclear extract of the chicken liver (Fig. 3D, lane 6) with no detectable cross-reactivities to human/chicken nSREBP-2 (Fig. 3D, lanes 2 and 5). By using this antibody, we found that the 1-h light pulse at CT14 caused remarkable accumulation of proteolytically activated ≈68 kDa nSREBP-1 in the pineal cell nuclei (Fig. 3E). These results demonstrate light-induced posttranslational activation of SREBP-1, whereas RT-PCR analysis revealed marginal effects of light on the pineal transcript levels of Srebp-1 and Srebp-2 (Fig. S6B).
SREBP Activates Expression of E4bp4.
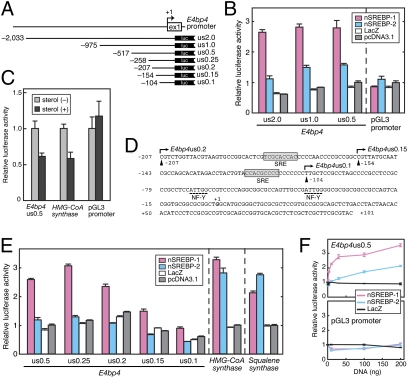
The effect of the active SREBP (nSREBP) on E4bp4 promoter was explored by transcriptional assay in CHO cells. Transcription from a 2.0-kb promoter region of chicken E4bp4 (E4bp4us2.0; Fig. 4A; deposited in GenBank, accession no. EF221611) was activated by nSREBP-1 and to a lesser extent by nSREBP-2 (Fig. 4B). Similar activation profiles were observed even with the shorter promoter regions, us1.0 and us0.5 (Fig. 4 A and B). In support of the idea that SREBP regulates E4bp4 expression, the activity of the E4bp4us0.5 reporter was inhibited by treatment of the cells with a 1:10 mixture of 25-hydroxycholesterol and cholesterol (Fig. 4C), which is known to inhibit SREBP activation (19) as evidenced here by their inhibitory action on HMG-CoA synthase promoter activity (Fig. 4C). We found that the E4bp4us0.5 reporter contained multiple copies of potential SRE (sterol regulatory element) sequence (Fig. 4D, shaded boxes), a binding site of nSREBP (20). The nSREBP-1–mediated activation was largely reduced when the E4bp4 promoter was further shortened from us0.2 to us0.15 and us0.1 (Fig. 4 A and E), in which one and two SRE sequences were eliminated, respectively (Fig. 4D). nSREBP-1 activated the transcription of E4bp4us0.5 reporter more potently than nSREBP-2 (Fig. 4F), contrasting with comparable activation by nSREBP-1 and nSREBP-2 on the promoter of HMG-CoA synthase or Squalene synthase (21, 22) (Fig. 4E). We conclude that SREBP-1 responds to light at early subjective night to operate as a transcriptional activator of E4bp4, a gene important for the photic-input pathway to the circadian clock (Fig. S7).
Fig. 4.
The transcriptional activation of E4bp4 promoter by nSREBP. (A) Schematic structure of chicken E4bp4 promoter-luciferase reporter constructs. The putative transcription start site is indicated as +1. (B, E, and F) The transcriptional assays in CHO-K1 cells using luciferase reporters containing promoter region of E4bp4. The amount of SREBP-expression plasmid was 100 ng (B and E) or 1, 3, 10, 30, 100, and 200 ng (F). The luciferase activity derived from the firefly luciferase reporter was normalized to that from the Renilla luciferase reporter. The mean value obtained from the cells transfected with empty vector and E4bp4us0.5 (B, E, and F, Upper) or pGL3 promoter (F, Lower) was set to 1. Data are the mean ± SEM (n = 3). (C) Regulation of E4bp4 promoter activity by cellular sterol levels. Four hours after transfection, CHO-K1 cells were treated with (+) or without (–) 1 μg/mL 25-hydroxycholesterol and 10 μg/mL cholesterol. After 48-h culture, the cells were subjected to luciferase assay. The mean value obtained from the sterol (–) cells was set to 1. Data are the mean ± SEM (n = 6). (D) Sequence of the promoter region of E4bp4us0.2 reporter vector. Sequences matched 8 of 9 bp with the SRE consensus (YCAYNYCAY, Y = pyrimidine; ref. 20) are indicated by gray boxes. The potential NF-Y binding sites (CCAAT) are underlined with broken lines (see Discussion).
Light Pulse at Early Subjective Night Activates 7α-Hydroxypregnenolone Secretion from the Pineal Gland.
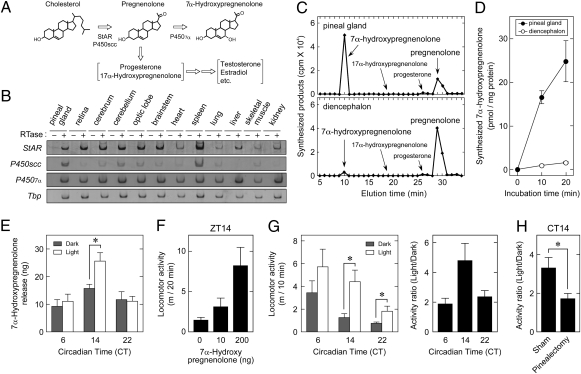
The light-dependent activation of SREBP-target genes for cholesterol biosynthesis (Fig. 3B and Fig. S4B) gave us the idea that light could stimulate pineal production of cholesterol and its derivatives such as neurosteroids, which act as signaling molecules in the brain (23). The rate-limiting step in steroidogenesis is the conversion of cholesterol to pregnenolone, a process that is facilitated by both mitochondrial cholesterol transporter StAR and cholesterol side-chain cleavage enzyme P450scc (24) (Fig. 5A). StAR gene was expressed at similar levels in a wide range of brain regions including the pineal gland, whereas P450scc transcript level was remarkably higher in the pineal gland than the other brain regions tested (Fig. 5B), suggesting active neurosteroidogenesis in the pineal gland. We examined potential neurosteroid biosynthesis by in vitro incubation of the pineal lysate with [3H]-labeled pregnenolone as a substrate. HPLC-based separation of the reaction products demonstrated that pregnenolone was converted primarily to 7α-hydroxypregnenolone with the pineal lysate (Fig. 5C). The synthetic activity of 7α-hydroxypregnenolone was identified in the diencephalon of newts and quail (25, 26), whereas we found that the specific activity of the pineal lysate was ≈15-fold higher than that of the diencephalon of chicks (Fig. 5D). This pineal activity contrasts sharply with that of the adrenal gland showing no detectable activity to synthesize 7α-hydroxypregnenolone (Fig. S8A). We further explored a potential function of the pineal gland to synthesize and secrete 7α-hydroxypregnenolone in organ culture. The pineal glands isolated from the entrained chicks at CT6, CT14, or CT22 were exposed to light (or kept in the dark) under the cultured condition, which maintains the in vivo properties of the pineal gland (5). We found that a significant amount of 7α-hydroxypregnenolone was released from the pineal gland into the culture medium, and the release was stimulated by light at CT14 (Fig. 5E), consistent with the light response of SREBP-target genes for cholesterol biosynthesis (Fig. 3B and Fig. S4B).
Fig. 5.
Generation of 7α-hydroxypregnenolone in the pineal gland. (A) Biosynthetic pathway of 7α-hydroxypregnenolone from cholesterol. (B) Expressions of StAR, P450scc, and P4507α in the pineal gland. Total RNA extracted from the tissues at ZT6 and ZT18 were mixed and subjected to reverse transcription with (+) or without (–) reverse transcriptase (RTase), followed by PCR amplification. (C and D) Production of 7α-hydroxypregnenolone from pregnenolone in the pineal and diencephalon lysates. (C) The homogenate from the pineal gland or the diencephalon were incubated with [3H]pregnenolone for 40 min, and each extract was subjected to HPLC with a reversed-phase column. (D) Radioactivity of 7α-hydroxypregnenolone fraction was plotted against incubation time. Data are the mean ± SEM (n = 6). (E) Effect of light on 7α-hydroxypregnenolone release from the pineal gland. The chicks were exposed to 20-min light pulse (≈300 lx; white fluorescent lamps) from CT6, CT14, or CT22, and then pineal glands were isolated and pooled (eight pineal glands) to culture for 3 h under the light. For dark control group, all procedures were performed in the dark. The amounts of 7α-hydroxypregnenolone released into the medium were measured by gas chromatography/mass spectrometric (GC-MS) analysis. Data are the mean ± SEM (n = 4). (F) Effect of intracerebroventricular injection of 7α-hydroxypregnenolone on chick locomotor activity. 7α-hydroxypregnenolone (0 ng, n = 14; 10 ng, n = 5; 200 ng, n = 9) was injected at ZT14-16, and the locomotor activity of each individual was recorded for 20 min. Data are the mean ± SEM P < 0.005 by one-way ANOVA. (G) Effect of light on locomotor activity. Locomotor activity of individual chick was recorded for 10 min before (under infrared light) and after (under white LED light; ≈300 lx) the 10-min light exposure at CT6–8, CT14–16, or CT22–24 (Left). Data are the mean ± SEM (n = 6–8). Light/dark ratio of the activity of each chick was plotted in Right (P < 0.05 by one-way ANOVA). (H) Effect of pinealectomy on light-dependent stimulation of locomotor activity. Locomotor activity of pinealectomized (n = 6) or sham-operated (n = 7) chick was recorded for 10 min before and after the 10-min light pulse at CT14–16, and light/dark ratio of the activity was plotted as in G. *P < 0.05 by Student's t test.
Light Pulse at Early Subjective Night Strongly Stimulates Locomotor Activity in a Pineal Gland-Dependent Manner.
In newts and quail, 7α-hydroxypregnenolone is reported to activate their locomotor activities (25, 26). In line with these observations, spontaneous locomotor activities of chicks were stimulated in a dose-dependent manner by intracerebroventricular injection of 7α-hydroxypregnenolone (Fig. 5F). The injected dose (10 or 200 ng) is equivalent to or in excess of 7α-hydroxypregnenolone content in the whole brain isolated after 1-h light pulse from CT14 [7.0 ± 1.2 ng (n = 5)]. We then analyzed the effect of light on locomotor activities at CT6, 14, and 22. Light exposure at CT14 strongly stimulated the activities to daytime level (Fig. 5G, Left), and light/dark ratio of the activities was highest at CT14 (Fig. 5G, Right) with a temporal profile very similar to that of the light response of 7α-hydroxypregnenolone release (Fig. 5E). We further investigated the effect of pinealectomy on light stimulation of locomotor activities at CT14 and found that the light response was attenuated by pinealectomy (Fig. 5H) to a level close to the light response of intact animals at CT6 and CT22 (Fig. 5G, Right). Collectively, these results revealed unique functions of the pineal gland, i.e., the biosynthesis and secretion of 7α-hydroxypregnenolone. Light-dependent activation of SREBP pathway in the pineal gland not only transactivates E4bp4 transcription but also appears to contribute to stimulation of the locomotor activity by generating 7α-hydroxypregnenolone in a time-of-day–dependent manner.
Discussion
This study demonstrates that SREBP pathway in the pineal gland was activated within 1 h after the light exposure of chicks, and the light response was pronounced at the early subjective night (Figs. 1 and 3). This gating appears to play an important role in time-of-day–dependent phase delay by light because E4bp4 expression was up-regulated by nSREBP (Fig. 4 and Fig. S7). It should be noted that the light pulse given at early subjective night remarkably activated a subset of SREBP-target genes responsible for biosynthesis and metabolism of cholesterol, whereas it had only marginal effects on those involved in synthesis of acetyl-CoA and fatty acids (Fig. 3B and Fig. S4B). Intriguingly, it has been established that optimal transcriptional activation by nSREBP requires additional contribution of transcription factors such as Sp1 and nuclear factor Y (NF-Y), whose binding sites are positioned close to SRE (27). We searched for Sp1– and NF-Y–binding sites in mammalian SREBP-target genes in the literature (22, 28–30) and found that the NF-Y sites reside near SRE of the light-responsive cholesterol-related genes, such as Farnesyl diphosphate synthase, HMG-CoA synthase, HMG-CoA reductase, Squalene epoxidase, Squalene synthase, and StarD4. In contrast, the NF-Y site was not found in the light-insensitive genes such as Acetyl-CoA synthetase, LDL receptor, Acetyl-CoA carboxylase-1, and Malic enzyme. The presence of two NF-Y sites close to SRE in the chicken E4bp4 promoter (Fig. 4D, broken lines) raises the possibility that NF-Y plays an additional role for the light induction of gene expression in a cooperative manner with SREBP.
In contrast to striking light activation of SREBP pathway at early subjective night, stress–response genes and their regulators were prominently induced by light at late subjective night (Figs. 1 and 2). It is known that nuclear accumulation and activation of HSF1 and HSF3 are stimulated by heat shock, whereas HSF2 is uniquely activated when the ubiquitin-dependent protein degradation machinery is inhibited (13). Our finding that HSF1 and HSF2 but not HSF3 accumulated in the pineal cell nuclei in response to light (Fig. 2B) suggests that the light activation of these factors is not simply due to a heat shock response such as changes in body temperature. Similarly, we found that light induced expression of the target genes of ATF6 and XBP1 but not those of ATF4 (Fig. 2C and Fig. S4A), whereas all of these genes are known to be induced by ER stress (14). It is possible that light activates an unidentified combination of upstream signaling pathways for the selective light response of these stress–response factors, rather than acts as a stress. Interestingly, SREBP (31), HSF1 (15), ATF6 (32), and also XBP1 (Fig. S5) are implicated in the feeding-dependent regulation of rhythmic gene expression in the mouse liver. These factors can be common regulators in various input pathways among clock cells, although the molecular link between the phase advance of the pineal clock and the signaling pathways of ATF6, XBP1, and HSF needs to be established, among which HSF1 is known as a regulator of the mammalian clockwork (15, 33). It is interesting to investigate whether SREBP, ATF6, XBP1, and HSF pathways show time-of-day–dependent responses also in the food-regulated clocks of the liver and in the central clock of the suprachiasmatic nucleus in mammals. Such a comparative genomic analysis will shed light on potential conservation of light-responsive pathways among tissues and/or species.
The brain is not only an important target site of steroid hormones but also capable of converting cholesterol to steroid hormones (neurosteroids) in several types of cells including oligodendrocytes, Purkinje neurons, and hippocampal neurons (23). Here, we found that the pineal gland has high activity to synthesize and secrete 7α-hydroxypregnenolone which enhances locomotor activities of chicks (Fig. 5). To the best of our knowledge, this is the first observation showing neurosteroid synthesis and secretion in the pineal gland in any vertebrate. In contrast, it is established that the canonical pineal hormone melatonin regulates sleep rhythms (34). Therefore, the pineal gland appears to participate in the regulation of sleep/wake state not only by circadian production of melatonin in the dark but also by synthesis and secretion of 7α-hydroxypregnenolone in the light. In this study, we used male chicks for the experiments. Our previous study on the quail diencephalon demonstrated much higher activity of 7α-hydroxypregnenolone synthesis in males than females, corresponding to relatively higher locomotor activities of males (26). Consistent with this observation, we found that the pineal gland of female chicks released far lower amount of 7α-hydroxypregnenolone than males (Fig. S8B), suggesting the conservation of the sex difference in 7α-hydroxypregnenolone system between the chick pineal gland and the quail diencephalon. Further study is necessary to reveal the mechanism underlying the sex difference.
In summary, our genome-wide gene expression analysis revealed multiple signaling pathways that were light-activated in a time-of-day–dependent manner to regulate gene expression. The SREBP-mediated pathway especially should be highlighted as the photic-input mechanism of the circadian clock through regulation of E4bp4, which acts as a light-induced suppressor of Per2 particularly in the phase-delaying process. An intriguing idea is that the combination of these light-activated pathways determines the direction and degree of the time-of-day–dependent phase shift of the circadian clock.
Materials and Methods
Details are provided in SI Materials and Methods.
Animals.
Animal experiments were conducted in accordance with the guidelines of the University of Tokyo. Newly hatched male chicks were maintained under 12-h light/12-h dark cycles for 7 d with the light provided by white fluorescent lamps (≈300 lx at the level of the heads of chicks). They were transferred to constant darkness thereafter and exposed to a 1-h light pulse at various time points. All procedures during the dark period were performed under dim red light.
GeneChip and Quantitative RT-PCR Analyses.
Original analysis data using GeneChip Chicken Genome Array (Affymetrix) were deposited in GEO (accession no. GSE21915). Quantitative RT-PCR analysis was performed as described (35) or by using QuantiTect SYBR Green PCR Kit (Qiagen). The primers and optimal cycle numbers were summarized in Table S1.
Supplementary Material
Acknowledgments
We thank Y. Nakajima, M. Doi, and T. Okano for sharing preliminary results and T. Ubuka, Y. Nobe, and K. Shimizu for technical assistance. M.H. and N.K. were supported by Japan Society for the Promotion of Science Research Fellowships. This work was supported in part by Grants-in-Aid for Scientific Research and by Global Centers of Excellence Program from Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE21915) and in the GenBank database, www.ncbi.nlm.nih.gov/nuccore (accession no. EF221611).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015959108/-/DCSupplemental.
References
- 1.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 2.Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson CH, Elliott JA, Foster R. Entrainment of circadian programs. Chronobiol Int. 2003;20:741–774. doi: 10.1081/cbi-120024211. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi JS, et al. The avian pineal, a vertebrate model system of the circadian oscillator: Cellular regulation of circadian rhythms by light, second messengers, and macromolecular synthesis. Recent Prog Horm Res. 1989;45:279–348. doi: 10.1016/b978-0-12-571145-6.50010-8. discussion 348–352. [DOI] [PubMed] [Google Scholar]
- 6.Okano T, Fukada Y. Chicktacking pineal clock. J Biochem. 2003;134:791–797. doi: 10.1093/jb/mvg221. [DOI] [PubMed] [Google Scholar]
- 7.Doi M, Nakajima Y, Okano T, Fukada Y. Light-dependent changes in the chick pineal temperature and the expression of cHsp90 α gene: A potential contribution of in vivo temperature change to the photic-entrainment of the chick pineal circadian clock. Zoolog Sci. 2002;19:633–641. doi: 10.2108/zsj.19.633. [DOI] [PubMed] [Google Scholar]
- 8.Doi M, Nakajima Y, Okano T, Fukada Y. Light-induced phase-delay of the chicken pineal circadian clock is associated with the induction of cE4bp4, a potential transcriptional repressor of cPer2 gene. Proc Natl Acad Sci USA. 2001;98:8089–8094. doi: 10.1073/pnas.141090998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okano T, et al. Chicken pineal clock genes: Implication of BMAL2 as a bidirectional regulator in circadian clock oscillation. Genes Cells. 2001;6:825–836. doi: 10.1046/j.1365-2443.2001.00462.x. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto K, Okano T, Fukada Y. Chicken pineal Cry genes: Light-dependent up-regulation of cCry1 and cCry2 transcripts. Neurosci Lett. 2001;313:13–16. doi: 10.1016/s0304-3940(01)02227-3. [DOI] [PubMed] [Google Scholar]
- 11.Bailey MJ, et al. Transcriptional profiling of the chick pineal gland, a photoreceptive circadian oscillator and pacemaker. Mol Endocrinol. 2003;17:2084–2095. doi: 10.1210/me.2003-0121. [DOI] [PubMed] [Google Scholar]
- 12.Murray JI, et al. Diverse and specific gene expression responses to stresses in cultured human cells. Mol Biol Cell. 2004;15:2361–2374. doi: 10.1091/mbc.E03-11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimoto M, Nakai A. The heat shock factor family and adaptation to proteotoxic stress. FEBS J. 2010;277:4112–4125. doi: 10.1111/j.1742-4658.2010.07827.x. [DOI] [PubMed] [Google Scholar]
- 14.Mori K. Frame switch splicing and regulated intramembrane proteolysis: Key words to understand the unfolded protein response. Traffic. 2003;4:519–528. doi: 10.1034/j.1600-0854.2003.00112.x. [DOI] [PubMed] [Google Scholar]
- 15.Reinke H, et al. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 2008;22:331–345. doi: 10.1101/gad.453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 hr period rhythmic activation of the IRE1α pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11:47–57. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: Time and food. J Biol Rhythms. 2003;18:250–260. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- 18.Horton JD, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 20.Magaña MM, Osborne TF. Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. J Biol Chem. 1996;271:32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 21.Guan G, Dai P, Shechter I. Differential transcriptional regulation of the human squalene synthase gene by sterol regulatory element-binding proteins (SREBP) 1a and 2 and involvement of 5′ DNA sequence elements in the regulation. J Biol Chem. 1998;273:12526–12535. doi: 10.1074/jbc.273.20.12526. [DOI] [PubMed] [Google Scholar]
- 22.Amemiya-Kudo M, et al. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J Lipid Res. 2002;43:1220–1235. [PubMed] [Google Scholar]
- 23.Mellon SH. Neurosteroid regulation of central nervous system development. Pharmacol Ther. 2007;116:107–124. doi: 10.1016/j.pharmthera.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark BJ, Stocco DM. StAR-A tissue specific acute mediator of steroidogenesis. Trends Endocrinol Metab. 1996;7:227–233. doi: 10.1016/s1043-2760(96)00114-2. [DOI] [PubMed] [Google Scholar]
- 25.Matsunaga M, Ukena K, Baulieu EE, Tsutsui K. 7α-Hydroxypregnenolone acts as a neuronal activator to stimulate locomotor activity of breeding newts by means of the dopaminergic system. Proc Natl Acad Sci USA. 2004;101:17282–17287. doi: 10.1073/pnas.0407176101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsutsui K, et al. 7α-Hydroxypregnenolone mediates melatonin action underlying diurnal locomotor rhythms. J Neurosci. 2008;28:2158–2167. doi: 10.1523/JNEUROSCI.3562-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards PA, Ericsson J. Sterols and isoprenoids: Signaling molecules derived from the cholesterol biosynthetic pathway. Annu Rev Biochem. 1999;68:157–185. doi: 10.1146/annurev.biochem.68.1.157. [DOI] [PubMed] [Google Scholar]
- 28.Lopez JM, Bennett MK, Sanchez HB, Rosenfeld JM, Osborne TF. Sterol regulation of acetyl coenzyme A carboxylase: A mechanism for coordinate control of cellular lipid. Proc Natl Acad Sci USA. 1996;93:1049–1053. doi: 10.1073/pnas.93.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soccio RE, Adams RM, Maxwell KN, Breslow JL. Differential gene regulation of StarD4 and StarD5 cholesterol transfer proteins. Activation of StarD4 by sterol regulatory element-binding protein-2 and StarD5 by endoplasmic reticulum stress. J Biol Chem. 2005;280:19410–19418. doi: 10.1074/jbc.M501778200. [DOI] [PubMed] [Google Scholar]
- 30.Murphy C, Ledmyr H, Ehrenborg E, Gåfvels M. Promoter analysis of the murine squalene epoxidase gene. Identification of a 205 bp homing region regulated by both SREBP'S and NF-Y. Biochim Biophys Acta. 2006;1761:1213–1227. doi: 10.1016/j.bbalip.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 31.Brewer M, Lange D, Baler R, Anzulovich A. SREBP-1 as a transcriptional integrator of circadian and nutritional cues in the liver. J Biol Rhythms. 2005;20:195–205. doi: 10.1177/0748730405275952. [DOI] [PubMed] [Google Scholar]
- 32.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arendt J, Skene DJ. Melatonin as a chronobiotic. Sleep Med Rev. 2005;9:25–39. doi: 10.1016/j.smrv.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Hatori M, Okano T, Nakajima Y, Doi M, Fukada Y. Lcg is a light-inducible and clock-controlled gene expressed in the chicken pineal gland. J Neurochem. 2006;96:1790–1800. doi: 10.1111/j.1471-4159.2006.03712.x. [DOI] [PubMed] [Google Scholar]
- 36.Scheuner D, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 37.Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002;366:585–594. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 39.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.