Abstract
A specific small-molecule inhibitor of p97 would provide an important tool to investigate diverse functions of this essential ATPase associated with diverse cellular activities (AAA) ATPase and to evaluate its potential to be a therapeutic target in human disease. We carried out a high-throughput screen to identify inhibitors of p97 ATPase activity. Dual-reporter cell lines that simultaneously express p97-dependent and p97-independent proteasome substrates were used to stratify inhibitors that emerged from the screen. N2,N4-dibenzylquinazoline-2,4-diamine (DBeQ) was identified as a selective, potent, reversible, and ATP-competitive p97 inhibitor. DBeQ blocks multiple processes that have been shown by RNAi to depend on p97, including degradation of ubiquitin fusion degradation and endoplasmic reticulum-associated degradation pathway reporters, as well as autophagosome maturation. DBeQ also potently inhibits cancer cell growth and is more rapid than a proteasome inhibitor at mobilizing the executioner caspases-3 and -7. Our results provide a rationale for targeting p97 in cancer therapy.
Keywords: apoptosis, autophagy, unfolded protein response
The AAA (ATPase associated with diverse cellular activities) ATPase p97 is conserved across all eukaryotes and is essential for life in budding yeast (1) and mice (2). p97 was first linked to the ubiquitin–proteasome system (UPS) through its role in the turnover of ubiquitin−β-galactosidase fusion proteins via the “ubiquitin fusion degradation” (UFD) pathway (3). Since then, p97 has been shown to play a critical role in the degradation of misfolded membrane and secretory proteins (4) and has also been linked to a broad array of cellular processes, including Golgi membrane reassembly (5), membrane transport (6), regulation of myofibril assembly (7), cell division (8), formation of protein aggregates (9), and autophagosome maturation (10, 11). The broad range of cellular functions for p97 is thought to derive from its ability to unfold proteins or disassemble protein complexes, but the detailed mechanism of how p97 works and is linked to specific cellular processes remains largely unknown.
The structure of p97 comprises three domains: an N-terminal domain that recruits adaptors/substrate specificity factors, followed by two ATPase domains, D1 and D2 (12, 13). p97 monomers assemble to form a homohexamer that is thought to provide a platform for transduction of chemical activity into mechanical force that is applied to substrate proteins. The D1 domain mediates hexamerization (14) and has very low ATPase activity (15). Most of the ATPase activity is contributed by the D2 domain, which is thought to underlie p97’s function as a mechanochemical transducer (16).
The mechanochemical activity of p97 is linked to substrate proteins by an array of 13 UBX (ubiquitin regulatory X) domain adapters that bind the N-terminal domain of p97 (17), as well as the non-UBX domain adaptors Ufd1 and Npl4 (18). The functions and mechanisms of action of these different p97–adaptor complexes remain poorly understood. Recently our laboratory reported a proteomic analysis of UBX cofactors and revealed their interactions with a large number of E3 ligases and a cytosolic substrate, HIF-1α, whose interaction with p97 is mediated by UBXD7 (19). Whereas all UBX proteins interact with p97, only those containing a ubiquitin-associated domain associate with ubiquitin conjugates (19). Thus, some p97–UBX complexes may have functions that do not involve ubiquitin.
A major limitation to current studies on the biological functions of p97/Cdc48 is that there is no method to rapidly shut off its ATPase activity. The available temperature-sensitive (ts) mutants of yeast Cdc48 require long incubations to fully extinguish their activities, and the molecular bases for their effects are not known. Given the range of cellular processes in which Cdc48 participates, it is difficult to determine whether any particular phenotype observed in the existing mutants is due to a direct or indirect effect. Moreover, inhibition of p97 activity in animal cells by siRNA or expression of a dominant-negative version is challenged by its high abundance and is not suited to evaluating proximal phenotypic effects of p97 loss of function. Therefore, a specific p97 inhibitor would be a valuable research tool to investigate p97 function in cells.
Results
High-Throughput Screening to Identify p97 Small-Molecule Inhibitors.
The availability of a powerful suite of high-throughput screening (HTS)-compatible assays to monitor p97 function in cells coupled with the limitations of existing p97 inhibitors (SI Results) encouraged us to pursue development of a potent, selective, and reversible p97 inhibitor that works by a defined mechanism of action. To facilitate HTS, a p97 ATPase assay was devised that relied on using luciferase to measure the amount of ATP that remained after incubation with p97. This assay was miniaturized to a 1,536-well format and used to screen two libraries. Recombinant purified p97 was first screened against a library of 16,000 compounds (at 10 μM each; 16K HTS) from the Maybridge Hitfinder Collection and then against 218,117 compounds (at 8 μM each; 218K HTS) from the National Institutes of Health Molecular Libraries Small Molecule Repository. In the 218K HTS (PubChem AID1481), 925 primary hits were identified, 759 of which were retested in triplicate to confirm activity, yielding 333 active compounds (PubChem AID1517). Fifty-four of these that inhibited activity by ≥50% at 8 μM were subjected to 10-point titration, yielding five active compounds with IC50 ≤50 μM (PubChem AID1534). Subsequent manual retesting of selected compounds suggested that AID1534 yielded high IC50 values for compound potency (i.e., low inhibitory activity). IC50 values from AID1534 and the manual ATPase assay are presented in Table S1.
Identification of DBeQ as a Selective and Potent p97 Inhibitor.
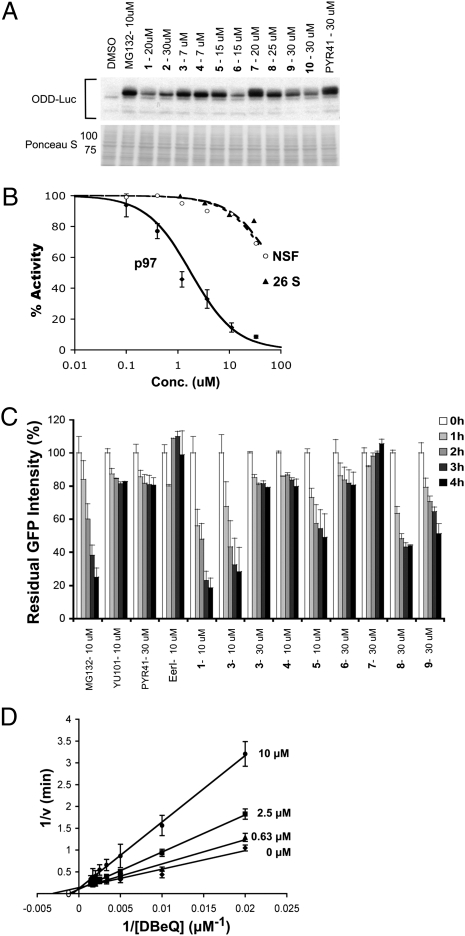
The top compounds to emerge from HTS were next tested for their ability to retard degradation of the UFD pathway and p97-dependent substrate UbG76V-GFP in a cell-based assay (20). On the basis of the results from this assay (Table S1), we narrowed our focus to 10 compounds (Table 1) that inhibited p97 ATPase with IC50 <10 μM in the manual ATPase assay and inhibited degradation of UbG76V-GFP with an IC50 <10 μM. To evaluate the selectivity of the top 10 compounds for targeting p97 in cells as opposed to other ATPases that are generally required for UPS function (e.g., E1 enzyme and the six AAA ATPase subunits of the 26S proteasome), we next assayed them for their ability to inhibit degradation of the p97-independent proteasome substrates ODD-Luc (oxygen-dependent degradation domain of HIF1α fused to luciferase) and Luc-ODC (luciferase fused to the ubiquitin-independent degradation domain of ornithine decarboxylase) (21, 22) (Table 1). ODD-Luc is targeted to the proteasome via the CRL2VHL ubiquitin ligase pathway, whereas Luc-ODC is degraded by the proteasome in a manner that is independent of ubiquitination (23). Some of the compounds could not be evaluated with the luciferase-based substrates because they interfered with measurement of luciferase activity. To classify these compounds, Western blot analysis of ODD-Luc turnover was performed. Compounds 3, 4, 5, 7, and 8 blocked ODD-Luc degradation to a similar extent as MG132 (a reversible proteasome inhibitor) or PYR41 (an E1 inhibitor) (24) (Fig. 1A), suggesting that they inhibited at least one component of the UPS in addition to p97. Of the top 10 compounds, only compound 1 was more than 10-fold less potent in blocking ODD-Luc degradation compared with UbG76V-GFP degradation. As a further test of the selectivity of this compound we evaluated its ability to inhibit AAA ATPase activity of purified N-ethylmaleimide–sensitive factor (NSF) (25) and the ATP-dependent chymotryptic activity of 26S proteasome (26). Compound 1 [which we have renamed as N2,N4-dibenzylquinazoline-2,4-diamine (DBeQ)] was at least 50-fold less potent toward these enzymes (Fig. 1B).
Table 1.
Top 10 hits from two HTS screens for p97 inhibitors
| Assay IC50 (μM)* |
|||||
| Cell-based degradation assay |
ATPase activity |
||||
| Compound no. and ID† | UbG76V-GFP | ODD-Luc | Luc-ODC | WT | C522A |
| (1) CID16472035 CID676352 | 2.6 ± 0.3 | 56 ± 14 | 45 ± 11 | 1.5 ± 0.4 | 1.6 ± 0.3 |
| 2.3 ± 0.5‡ | 1.6 ± 0.2‡ | ||||
| (2) CID886813 | 9.0 ± 1.3 | 43 ± 22 | >30 | 3 ± 0.8 | 2 ± 0.2 |
| 13 ± 2‡ | 1.2 ± 0.5‡ | ||||
| (3) JFD03665 | 1.1 ± 0.5 | 4.4 ± 1.0 | NM | 0.26 ± 0.05 | 8 ± 2 |
| (4) CID6763 | 1.3 ± 0.5 | 3.3 ± 0.9 | NM | 3.5 ± 1.3 | ND |
| (5) JFD02342 | 3.4 ± 1.0 | 5.2 ± 1.8 | 11 ± 4 | 0.2 ± 0.02 | 10 ± 4 |
| (6) CID934321 | 3.6 ± 0.7 | 18 ± 8 | NM | 1.3 ± 0.2 | 12 ± 1 |
| (7) S09756 | 5.0 ± 1.0 | 17 ± 4 | NM | 0.8 ± 0.08 | 20 ± 8 |
| (8) JFD00597 | 6.6 ± 1.9 | NM | NM | 6.1 ± 0.8 | 44 ± 12 |
| (9) SEW05182 | 7.5 ± 2.9 | NM | NM | 13 ± 1 | 30 ± 5 |
| (10) CID1109468 | 10 ± 2 | NM | NM | 2 ± 0.3 | 3 ± 0.2 |
NM, not measured due to interference with luciferase activity.
*Measurements were carried out in triplicate and the result expressed as mean ± SE.
†Compounds with PubChem CID number are hits from the National Institutes of Health Molecular Libraries Small Molecule Repository. All other compounds are from the Maybridge HitFinder collection.
‡IC50 values of the resynthesized hits (SI Methods).
Fig. 1.
DBeQ (1) is a reversible and selective inhibitor of p97. (A) Western blot assays to evaluate specificity of the top 10 hits to emerge from HTS. HeLa cells that stably expressed UbG76V-GFP and ODD-Luc were treated with MG132, washed, and then incubated in the presence of CHX, plus test compound for 2 h before harvest. Samples were immunoblotted to detect ODD-Luc. The Ponceau S-stained filter serves as a loading control. The high level of remaining antigen in lane 2 indicates that MG132 largely blocked ODD-Luc degradation, whereas the relatively low level in lane 3 indicates that 20 μM DBeQ failed to prevent ODD-Luc proteolysis. (B) Inhibition of ATPase activity of p97 (diamonds), NSF (circles), or the ATP-dependent chymotryptic activity of 26S proteasome (triangles) by DBeQ. (C) Reversibility of compound inhibition was determined by first accumulating UbG76V-GFP in the presence of MG132 (4 μM for 1 h), washing out MG132 and exposing cells to CHX plus test compound for 2 h, and then washing out test compound and monitoring decay of GFP signal in CHX for 0–4 h. (D) Lineweaver-Burk plot of the competitive inhibition of p97 ATPase activity by DBeQ.
Inhibition of p97 by DBeQ Is Reversible, ATP-Competitive, and Not Due to Colloid Formation.
The D2 ATPase domain of p97 contains a reactive cysteine (Cys522) in the ATP-binding pocket (27). Because most of the top 10 hits shown in Table 1 contained electrophilic moieties, we further evaluated their mechanism of action by assaying their ability to inhibit the ATPase activity of purified C522A-p97. The reduced activity of compounds 3 through 9 toward C522A-p97 suggested that electrophilic attack of Cys522 was indeed critical for the full potency of these compounds (Table 1). As a test of reversibility in vivo, we evaluated whether UbG76V-GFP degradation resumed upon washout of inhibitor. Whereas UbG76V-GFP was degraded after washing out MG132, it remained stable after preincubation with the covalent inhibitors YU101 (a proteasome inhibitor) (28) or PYR41 (Fig. 1C). Given that all compounds except for DBeQ (compound 2 was not tested) exhibited at least partial irreversibility at concentrations <10 × IC50, we suggest that the mechanism of inhibition by compounds 3–9 was at least partially irreversible (Fig. 1C). Interestingly, the previously described p97 inhibitor EerI (29) was also found to be irreversible.
On the basis of its favorable combination of potency, selectivity, and reversibility, we nominated DBeQ for further characterization. To determine the mechanism by which DBeQ inhibited p97 ATPase, we evaluated rates of ATP hydrolysis at different concentrations of ATP and DBeQ. DBeQ inhibited p97 competitively with respect to ATP, with a Ki of 3.2 ± 0.4 μM (Fig. 1D), suggesting that it binds to the active site of the D2 domain. It has been reported that some HTS hits form colloids that cause nonspecific enzyme inhibition (30). To investigate whether DBeQ acts in this manner, we performed dynamic light scattering analysis (Fig. S1 A–E). At a concentration of DBeQ (11 μM) that causes nearly complete inhibition of ATPase activity, no aggregation was detected (Fig. S1D). Moreover, inhibition of p97 by DBeQ was not affected by 0.01% Triton X-100 (Fig. S1F).
DBeQ Does Not Exhibit Activity Toward Protein Kinases.
The quinazoline scaffold at the core of DBeQ is found in compounds that inhibit protein kinases (31). To uncover potential off-target effects of DBeQ on protein kinases, we evaluated DBeQ using an activity-based proteomics platform (KiNativ; ActivX Biosciences). This assay measures the ability of small molecules to inhibit the covalent labeling of protein kinases by a broadly reactive ATP acyl-phosphate probe directly in native cell lysates. As a positive control, we carried out parallel analyses with a known protein kinase inhibitor based on the pyrazolopyrimidine scaffold (32). The results from these analyses are presented in Dataset S1. Remarkably, DBeQ (15 μM) did not exhibit appreciable inhibition of labeling of any of the ≈170 kinases that were evaluated, whereas pyrazolopyrimidine exhibited >50% inhibition of 13 protein kinases when tested at the same concentration.
Evaluation of DBeQ in ERAD and Autophagy Pathways.
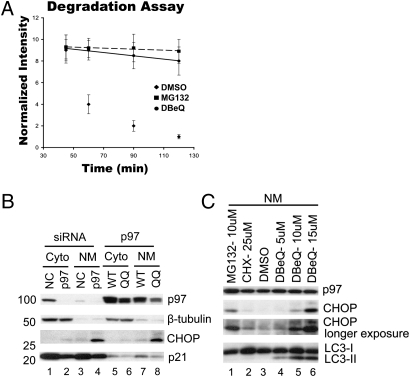
Because p97 has most commonly been studied in the context of endoplasmic reticulum-associated degradation (ERAD) (6), we sought to evaluate the impact of DBeQ on the ERAD reporter TCRα-GFP (α chain of the T-cell receptor fused to GFP). TCRα-GFP overexpressed in non-T cells inserts into the endoplasmic reticulum but behaves as an unfolded protein and is degraded by the proteasome in a p97-dependent manner (33). TCRα-GFP accumulation upon p97 depletion was confirmed by flow cytometry and Western blot (Fig. S2 A and B). DBeQ potently blocked degradation of TCRα-GFP at 10 μM (Fig. 2A and Fig. S2C). Blockade of ERAD ultimately leads to activation of the unfolded protein response (UPR), which contributes to the anticancer effects of bortezomib (34). A well-established marker of the UPR is the transcription factor CHOP (GADD153), which mediates endoplasmic reticulum stress-induced apoptosis (35). CHOP is up-regulated in bortezomib-treated cells within 4 h (34). CHOP accumulated in the nucleus and membrane-enriched fractions of HeLa cells that were either depleted of p97 (Fig. 2B, lane 4) or expressed an ATPase-inactive mutant (Fig. 2B, lane 8). CHOP induces apoptosis in part by down-regulating p21 (36). Consistent with this finding, we also observed a decrease of p21 protein level in cells that were depleted of p97 or overexpressed the ATPase-inactive mutant (Fig. 2B, lanes 2, 4, and 8). DBeQ induced CHOP within 3 h in a concentration-dependent manner (Fig. 2C, lanes 4–6) but did not increase p21 level (Fig. S3, lanes 3 and 6). By contrast, MG132 increased both CHOP (Fig. 2C, lane1) and p21 (Fig. S3, lanes 2 and 5), consistent with the documented instability of p21 (37).
Fig. 2.
DBeQ impairs the ERAD pathway. (A) Hek293 cells stably expressing the ERAD reporter TCRα-GFP were used to determine effect of DBeQ on the ERAD pathway. Cells were treated with drug as described in Fig. 1A, and GFP intensity was determined by flow cytometry at the indicated time points. (B) HeLa cells were transfected with negative control siRNA (NC) or p97 siRNA (10 nM) for 72 h (lanes 1–4) or with cDNAs encoding wild-type (WT) or ATPase mutant (QQ) p97 for 24 h (lanes 5–8), and the levels and distributions of the indicated proteins were determined by immunoblotting cytosolic (Cyto) and nuclear plus membrane (NM) fractions. (C) HeLa cells were incubated with 10 μM of MG132, 25 μM of CHX, or 5, 10, or 15 μM of DBeQ for 3 h, and the levels of CHOP and LC3 in the nuclear plus membrane (NM) fractions were determined by immunoblotting.
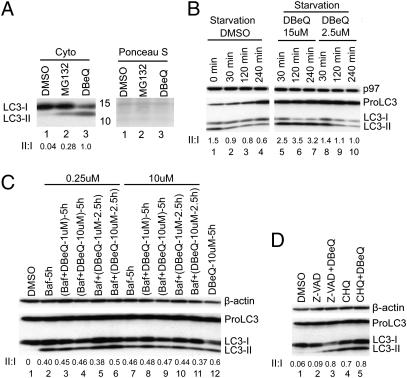
In addition to the well-characterized function of p97 in ERAD, p97 plays an important role in autophagy (10, 11). Inhibition of p97’s function by siRNA or expression of dominant-negative mutants leads to a defect in autophagosome maturation that is manifest as a net conversion of the autophagosome marker LC3-I to the lipidated LC3-II species (10, 11). Consistent with this, we observed 30- to 40-fold accumulation of LC3-II in both cytosol and nucleus/membrane-enriched fractions of p97-depleted HeLa cells (Fig. S4). DBeQ likewise induced a strong accumulation of LC3-II in the nucleus plus membrane-enriched (Fig. 2C, lanes 4–6) and cytosolic (Fig. 3A, lane 3) fractions. LC3-II can accumulate either because DBeQ stimulates basal autophagy (i.e., accelerates conversion of LC3-I to LC3-II) or, like p97 depletion, blocks a late step in autophagosome maturation. To distinguish between these possibilities, we evaluated the effect of DBeQ on cells in which autophagy was induced by nutrient starvation. In nutrient-starved cells treated with DMSO alone or a low dose of DBeQ (2.5 μM), LC3-I was rapidly converted to LC3-II and degraded (Fig. 3B). By contrast, in nutrient-starved cells treated with 15 μM DBeQ, LC3-II was markedly stabilized. Next, we examined the effect of DBeQ on LC3-II level in combination with known inhibitors of autophagy. Coincubation or sequential treatment with bafilomycin A1 (Baf) and DBeQ did not result in a significant increase in LC3-II compared with the sample treated with only Baf (Fig. 3C). A similar observation was made with chloroquine (CHQ; Fig. 3D, lanes 4 and 5). Taken together, these data indicate that DBeQ acted by blocking autophagic degradation of LC3-II instead of inducing autophagy.
Fig. 3.
DBeQ impairs the autophagy pathway. (A) Cells were incubated with 20 μM of MG132 or DBeQ for 3 h, and cytosolic LC3 level was determined by immunoblotting. The Ponceau S-stained filter serves as a loading control. (B) Degradation of LC3-II was monitored in Earle's balanced salt solution (EBSS) nutrient-starvation medium for 0, 30, 120, or 240 min (lanes 1–4). DBeQ (15 μM) completely stabilized LC3-II (lanes 5–7), whereas 2.5 μM DBeQ had little effect (lanes 8–10). (C) Cells were incubated with DMSO (lane 1) or Baf (0.25 μM, 5 h; lane 2) or Baf plus DBeQ (lanes 3 and 4) for 5 h or first treated with Baf for 2.5 h then exposed to DBeQ for an additional 2.5 h (lanes 5 and 6). The same experiments were carried out at 10 μM Baf (lanes 7–11). Lane 12 contains sample from cells treated with DBeQ (10 μM, 5 h) alone. All treatments were evaluated by immunoblotting cell lysates with antibodies to detect the indicated proteins. (D) Cells were treated with Z-VAD (25 μM), Z-VAD plus DBeQ (15 μM), CHQ (20 μM), or CHQ plus DBeQ, followed by immunoblotting cell extracts to detect the indicated proteins.
DBeQ Rapidly Induces Caspases and Inhibits Cell Proliferation.
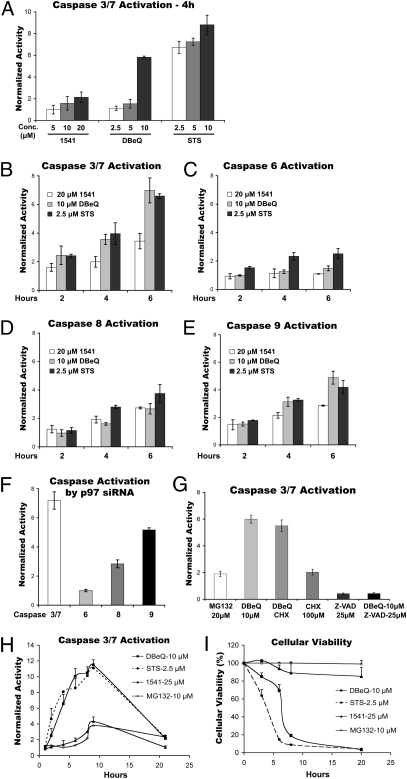
In a prior study, DBeQ was identified as a compound that more potently induces apoptosis in human cancer cells compared with noncancerous cells. However, the underlying mechanism was not determined (38). In our hands, 10 μM DBeQ rapidly promoted activation of the “executioner” caspases-3 and -7 in HeLa cells (Fig. 4A). We benchmarked DBeQ by comparing it with the well-characterized apoptosis inducer staurosporine (STS) (39) and the procaspases-3 and -6 activator 1541 (40). STS induces executioner caspases-3, -6, and -7 via both caspases-8 and -9 in the apoptotic pathway (40, 41). DBeQ activated caspases-3 and -7 by twofold within 2 h (Fig. 4B) but did not activate caspase-6 after 6 h (Fig. 4C). We next evaluated the impact of DBeQ on initiator caspases-8 and -9. DBeQ activated the intrinsic caspase-9 apoptotic pathway more than the extrinsic caspase-8 pathway, whereas STS activated both pathways to a similar extent (Fig. 4 D and E).
Fig. 4.
DBeQ and depletion of p97 induce activation of caspases. (A) HeLa cells were incubated with the indicated concentrations of 1541, DBeQ, or STS for 4 h before determination of caspases-3 plus -7 activities in the cell extract. (B) HeLa cells were incubated with 20 μM of 1541, 10 μM of DBeQ, or 2.5 μM of STS for 2, 4, or 6 h before determination of caspases-3 plus -7 activities in the cell extract. (C–E) Same as B, except caspases-6, -8, and -9 activities were determined, respectively. (F) HeLa cells were transfected with negative control siRNA (NC) or p97 siRNA (10 nM) for 72 h, after which the activities of the indicated caspases were measured in total cell lysate. (G) HeLa cells were incubated with the indicated concentrations of drugs for 3 h before determination of caspases-3 plus -7 activities in total cell lysate. DBeQ plus CHX were included at 10 μM and 100 μM, respectively. (H) HeLa cells were incubated with the indicated concentrations of drugs for 1, 2, 4, 6, 8, 9, or 21 h before determination of caspases-3 plus -7 activities in total cell lysate. (I) Same as H, except cellular viability was determined using CellTiter-Glo after 3, 6, 8, or 20 h.
Depletion of p97 by siRNA mimicked the effects of DBeQ on caspase activation (Fig. 4F), suggesting that apoptosis induction may be an “on-target” effect. Caspase activation by DBeQ was blocked by the general caspase inhibitor [Z-VAD(OMe)FMK; Fig. 4G], whereas accumulation of LC3-II was not affected (Fig. 3D, lane 3), suggesting that the block in autophagosome maturation induced by DBeQ was not an indirect consequence of caspase activation (42). Consistent with the speed of caspase induction by DBeQ, we determined that it was insensitive to cycloheximide (CHX) (Fig. 4G).
We next compared the antiproliferative activity of DBeQ and MG132 on normal and cancerous cell lines after 48 h treatment (Table 2). DBeQ was fivefold more active against multiple myeloma (RPMI8226) cells than normal human fetal lung fibroblasts (MRC5), with HeLa and Hek293 cells showing intermediate sensitivities. MG132 was 10- to 20-fold more potent than DBeQ at blocking growth of all four cell lines tested, in keeping with its 10-fold greater potency in blocking UbG76V-GFP degradation (22). We next evaluated the time-dependence of caspase activation and cellular viability for HeLa and RPMI8226 cells incubated with DBeQ, STS, 1541, or MG132. For both cell lines, there was a good correlation between the kinetics of caspases-3 and -7 activation and cell death induced by DBeQ and STS (Fig. 4 H and I and Fig. S5). Interestingly, in contrast to DBeQ, in a 20-h treatment MG132 exhibited potent activity only against the RPMI8226 cells. Thus, although MG132 is a more potent inhibitor of its intended target, DBeQ is more effective at blocking cell growth upon transient exposure to drug. Strong mobilization of caspase activity by DBeQ seemed to be due primarily to caspase-3, because DBeQ induced only modest activation of caspase-7 in caspase-3–deficient MCF7 cells (43) (Fig. S6A). Finally, consistent with our observation that DBeQ induced caspases-3 and -7 activity even in the presence of CHX (Fig. 4G), DBeQ mobilized caspases-3 and -7 activities with equivalent kinetics in wild-type and p53−/− HCT116 cells (Fig. S6 B and C), implying that p53-mediated gene transcription is not required to induced cell death in response to DBeQ.
Table 2.
Cytotoxicity of MG132 and DBeQ after 48 h treatment
| Concentration (in μM) at which growth is inhibited by 50% (GI50)* |
||||
| Compound | MRC-5† | Hek293 | HeLa | RPMI8226‡ |
| MG132 | 0.4 ± 0.1 | 0.2 ± 0.05 | 1.5 ± 0.3 | 0.14 ± 0.03 |
| DBeQ | 6.6 ± 2.9 | 4 ± 0.6 | 3.1 ± 0.5 | 1.2 ± 0.3 |
*Measurements were carried out in triplicate and the result expressed as mean ± SE.
†Normal human fetal lung fibroblast.
‡Multiple myeloma cell line.
Discussion
Motivated by the value of a potent and selective inhibitor of the AAA ATPase p97 as a basic research tool and as a potential starting point for development of a novel cancer therapeutic, we have carried out two HTS campaigns to identify small molecules that inhibit p97 ATPase activity. Secondary screens with cell lines that report on p97-dependent and -independent degradation (20) allowed us to winnow the field of candidate inhibitors to two related compounds (1 and 2) that acted reversibly and exhibited IC50 values of ≤10 μM both in vitro and in cells. The best compound, 1 (DBeQ), inhibited p97 ATPase and degradation of the p97-dependent substrate UbG76V-GFP, with IC50 values of 1.5 μM and 2.6 μM, respectively, but had negligible effects on degradation of p97-independent substrates and ATPase activity of the closely related NSF.
DBeQ is active against two distinct classes of UPS substrates—UFD and ERAD—that depend on p97 for their degradation. Consistent with its effect on ERAD, DBeQ induces accumulation of the UPR effector CHOP. By contrast, DBeQ has little effect on ODD-luciferase and luciferase-ODC, neither of which depends on p97 for degradation. Together, these results suggest that the major target of DBeQ within the UPS is p97. Although the proteasome contains six AAA ATPase subunits, DBeQ did not inhibit proteasome-dependent degradation of the luciferase reporters, nor did it inhibit ATP-dependent cleavage of Leu-Leu-Val-Tyr-aminomethlycoumarin (LLVY-AMC) by purified proteasome. Thus, DBeQ may be a useful tool to identify UPS substrates whose degradation depends on p97. In addition to its role in the UPS, p97 has been linked to multiple other processes, which may or may not involve ubiquitination (see the Introduction). To evaluate the generality of DBeQ-mediated inhibition of p97 function, we investigated the impact of DBeQ on autophagy. As has been reported for p97 depletion, DBeQ promoted accumulation of the autophagic intermediate LC3-II. This observation suggests that DBeQ may be useful to study functions of p97 both within and outside the UPS.
A remarkable feature of DBeQ is that it rapidly and potently induced activation of executioner caspases and cell death. Whereas a procaspase activator was reported to induce caspase when applied to cells at 25 μM for 4–8 h (40), 10 μM DBeQ mobilized substantial caspase activity within 2–5 h. Although the proteasome inhibitor MG132 was more potent than DBeQ in blocking cell growth in a 48-h incubation, the converse was true upon briefer exposure to these drugs. This may be clinically significant. Proteasome inhibitors have shown strong single-agent activity only in cancers of the hematopoietic lineage (44). By contrast, cell lines derived from solid tumors can survive long (up to 20-h) exposures to proteasome inhibitors, and perhaps as a consequence, bortezomib has shown little clinical activity toward solid tumors (44). The ability of DBeQ to induce rapid caspase activation and cell death even in cell lines derived from solid tumors suggests that p97 inhibition may be more efficacious than proteasome inhibition as a strategy for cancer chemotherapy.
We do not know the mechanism by which DBeQ elicits rapid induction of executioner caspases. One possibility is that simultaneous suppression of two parallel mechanisms of protein degradation accounts for the toxicity of p97 inhibitors. Not only is p97 required for autophagy and ERAD, but in yeast, Cdc48 is required for rapid degradation of proteins that fail to mature properly owing to heat or oxidative stress, pointing to a broad role for p97 in multiple mechanisms of protein quality control (45). The full impact of inhibition of ubiquitin-dependent protein degradation (e.g., by proteasome inhibitors) may normally be mitigated by up-regulation of autophagy (46, 47). Simultaneous inhibition of proteasome and histone deacetylase 6 (HDAC6) [which is required for autophagy (48, 49)] results in synergistic killing of multiple myeloma cells (50). Interestingly, more than one dozen human clinical trials (www.clinicaltrials.gov) combine bortezomib with the broad-spectrum HDAC inhibitor vorinostat, which is active toward HDAC6. Targeting p97 may provide an alternative route to achieving the same objective. Cancer cells may be particularly sensitive to killing by suppression of protein degradation mechanisms, because they may exhibit a heightened dependency on these mechanisms to clear an elevated burden of quality-control substrates (51). For example, some cancers produce high levels of a specific protein that is a prominent quality-control substrate (e.g., Ig light chains in multiple myeloma) (34, 52) or produce high levels of reactive oxygen species (53), which can result in excessive protein damage via oxidation. Alternatively, many cancer cells are aneuploid and produce unbalanced levels of subunits of protein complexes, which can heighten dependency on protein folding and clearance mechanisms (54, 55). Finally, the burden of point and deletion mutations in the cancer cell or an altered metabolic environment may lead to an increased rate of protein misfolding or misassembly (56).
An alternative possibility for why DBeQ induces rapid caspase activation is that p97 may regulate the activity of factors in the extrinsic or intrinsic signaling cascades that culminate in activation of executioner caspases. DBeQ may impinge on maintenance of inhibited caspase–IAP (inhibitor of apoptosis) complexes. Finally it is possible that caspase activation and apoptosis are “off-target” effects. However, p97 knockdown also induces caspases and cell death, and a similar phenomenon has been reported for the irreversible p97 inhibitor EerI (57). Together, these results suggest that caspase signaling to cell death is an “on-target” effect of p97 inhibition. Future work will provide molecular insight into how inhibition of p97 activity by DBeQ results in apoptosis and could strengthen the rationale for a p97-targeted cancer therapeutic.
Methods
Materials and detailed methods are available in SI Methods.
Reporter Degradation Assay.
Two dual reporter stable HeLa cell lines were used and are described elsewhere (22). One expressed the UFD reporter UbG76V-GFP (58) and ODD-Luc (21), and the second expressed UbG76V-GFP and Luc-ODC (21).
Supplementary Material
Acknowledgments
We thank A. Brunger for providing plasmids; G. Georg for a useful suggestion regarding high-throughput screen validation; C. Weihl for helpful discussions; P. Baillargeon and L. DeLuca for compound management; F. Parlati for critical reading of the manuscript; and H. Park, R. Oania, and D. Shimoda for technical assistance. National Institutes of Health (NIH) U54 Grant MH074404 funded Scripps personnel. The University of Kansas was supported by Award U54 HG005031-02 administered by the National Human Genome Research Institute on behalf of the NIH Roadmap Molecular Libraries Program. A.C.J. was supported by NIH Grant F32GM082000. T.-F.C. was supported by a 2008 Fellows Grant Program Award from the Multiple Myeloma Research Foundation, the Howard Hughes Medical Institute (HHMI), and the Weston Havens Foundation. R.J.D. is an HHMI Investigator, and this work was funded in part by HHMI and in part by NIH R03 Grant MH085687.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015312108/-/DCSupplemental.
References
- 1.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 2.Müller JM, Deinhardt K, Rosewell I, Warren G, Shima DT. Targeted deletion of p97 (VCP/CDC48) in mouse results in early embryonic lethality. Biochem Biophys Res Commun. 2007;354:459–465. doi: 10.1016/j.bbrc.2006.12.206. [DOI] [PubMed] [Google Scholar]
- 3.Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- 4.Golbik R, Lupas AN, Koretke KK, Baumeister W, Peters J. The Janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. Biol Chem. 1999;380:1049–1062. doi: 10.1515/BC.1999.131. [DOI] [PubMed] [Google Scholar]
- 5.Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- 6.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 7.Janiesch PC, et al. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat Cell Biol. 2007;9:379–390. doi: 10.1038/ncb1554. [DOI] [PubMed] [Google Scholar]
- 8.Cao K, Nakajima R, Meyer HH, Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 2003;115:355–367. doi: 10.1016/s0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- 9.Boyault C, et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21:2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ju JS, et al. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tresse E, et al. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huyton T, et al. The crystal structure of murine p97/VCP at 3.6A. J Struct Biol. 2003;144:337–348. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 13.DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struct Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Song C, Li CC. Hexamerization of p97-VCP is promoted by ATP binding to the D1 domain and required for ATPase and biological activities. Biochem Biophys Res Commun. 2003;300:253–260. doi: 10.1016/s0006-291x(02)02840-1. [DOI] [PubMed] [Google Scholar]
- 15.Song C, Wang Q, Li CC. ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J Biol Chem. 2003;278:3648–3655. doi: 10.1074/jbc.M208422200. [DOI] [PubMed] [Google Scholar]
- 16.Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: Dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuberth C, Buchberger A. UBX domain proteins: Major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexandru G, et al. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wójcik C, et al. Valosin-containing protein (p97) is a regulator of endoplasmic reticulum stress and of the degradation of N-end rule and ubiquitin-fusion degradation pathway substrates in mammalian cells. Mol Biol Cell. 2006;17:4606–4618. doi: 10.1091/mbc.E06-05-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimbrel EA, Davis TN, Bradner JE, Kung AL. In vivo pharmacodynamic imaging of proteasome inhibition. Mol Imaging. 2009;8:140–147. [PubMed] [Google Scholar]
- 22.Chou TF, Deshaies RJ. Quantitative cell-based proteindegradation assays to identify and classify drugs that target the ubiquitin-proteasome system. J Biol Chem. 2011 doi: 10.1074/jbc.M110.215319. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang M, Pickart CM, Coffino P. Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate. EMBO J. 2003;22:1488–1496. doi: 10.1093/emboj/cdg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 25.Zhao C, Matveeva EA, Ren Q, Whiteheart SW. Dissecting the N-ethylmaleimide-sensitive factor: required elements of the N and D1 domains. J Biol Chem. 2010;285:761–772. doi: 10.1074/jbc.M109.056739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H, et al. Physiological levels of ATP negatively regulate proteasome function. Cell Res. 2010;20:1372–1385. doi: 10.1038/cr.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noguchi M, et al. ATPase activity of p97/valosin-containing protein is regulated by oxidative modification of the evolutionally conserved cysteine 522 residue in Walker A motif. J Biol Chem. 2005;280:41332–41341. doi: 10.1074/jbc.M509700200. [DOI] [PubMed] [Google Scholar]
- 28.Elofsson M, Splittgerber U, Myung J, Mohan R, Crews CM. Towards subunit-specific proteasome inhibitors: Synthesis and evaluation of peptide alpha’,beta’-epoxyketones. Chem Biol. 1999;6:811–822. doi: 10.1016/s1074-5521(99)80128-8. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Li L, Ye Y. Inhibition of p97-dependent protein degradation by Eeyarestatin I. J Biol Chem. 2008;283:7445–7454. doi: 10.1074/jbc.M708347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 31.Blair JA, et al. Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat Chem Biol. 2007;3:229–238. doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]
- 32.Bishop AC, et al. Design of allele-specific inhibitors to probe protein kinase signaling. Curr Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- 33.DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 34.Obeng EA, et al. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–4916. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zinszner H, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mihailidou C, Papazian I, Papavassiliou AG, Kiaris H. CHOP-dependent regulation of p21/waf1 during ER stress. Cell Physiol Biochem. 2010;25:761–766. doi: 10.1159/000315096. [DOI] [PubMed] [Google Scholar]
- 37.Sheaff RJ, et al. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 38.Cubedo E, et al. New symmetrical quinazoline derivatives selectively induce apoptosis in human cancer cells. Cancer Biol Ther. 2006;5:850–859. doi: 10.4161/cbt.5.7.2841. [DOI] [PubMed] [Google Scholar]
- 39.Bertrand R, Solary E, O'Connor P, Kohn KW, Pommier Y. Induction of a common pathway of apoptosis by staurosporine. Exp Cell Res. 1994;211:314–321. doi: 10.1006/excr.1994.1093. [DOI] [PubMed] [Google Scholar]
- 40.Wolan DW, Zorn JA, Gray DC, Wells JA. Small-molecule activators of a proenzyme. Science. 2009;326:853–858. doi: 10.1126/science.1177585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolier M, Decrion-Barthod AZ, Launay S, Prétet JL, Mougin C. Spatiotemporal activation of caspase-dependent and -independent pathways in staurosporine-induced apoptosis of p53wt and p53mt human cervical carcinoma cells. Biol Cell. 2009;101:455–467. doi: 10.1042/BC20080164. [DOI] [PubMed] [Google Scholar]
- 42.Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: An effect rescued by Bcl-xL. Cell Death Differ. 2010;17:268–277. doi: 10.1038/cdd.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jänicke RU, Sprengart ML, Wati MR, Porter AG. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J Biol Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 44.Wright JJ. Combination therapy of bortezomib with novel targeted agents: An emerging treatment strategy. Clin Cancer Res. 2010;16:4094–4104. doi: 10.1158/1078-0432.CCR-09-2882. [DOI] [PubMed] [Google Scholar]
- 45.Medicherla B, Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol. 2008;182:663–673. doi: 10.1083/jcb.200803022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding WX, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 48.Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 50.Hideshima T, et al. Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc Natl Acad Sci USA. 2005;102:8567–8572. doi: 10.1073/pnas.0503221102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solimini NL, Luo J, Elledge SJ. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 2007;130:986–988. doi: 10.1016/j.cell.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 52.Meister S, et al. Extensive immunoglobulin production sensitizes myeloma cells for proteasome inhibition. Cancer Res. 2007;67:1783–1792. doi: 10.1158/0008-5472.CAN-06-2258. [DOI] [PubMed] [Google Scholar]
- 53.Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 55.Torres EM, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Workman P, Burrows F, Neckers L, Rosen N. Drugging the cancer chaperone HSP90: Combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann N Y Acad Sci. 2007;1113:202–216. doi: 10.1196/annals.1391.012. [DOI] [PubMed] [Google Scholar]
- 57.Wang Q, et al. ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proc Natl Acad Sci USA. 2009;106:2200–2205. doi: 10.1073/pnas.0807611106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.