Abstract
Mycobacterium tuberculosis must import iron from its host for survival, and its siderophore-dependent iron acquisition pathways are well established. Here we demonstrate a newly characterized pathway, whereby M. tuberculosis can use free heme and heme from hemoglobin as an iron source. Significantly, we identified the genomic region, Rv0202c–Rv0207c, responsible for the passage of heme iron across the mycobacterial membrane. Key players of this heme uptake system were characterized including a secreted protein and two transmembrane proteins, all three specific to mycobacteria. Furthermore, the crystal structure of the key heme carrier protein Rv0203 was found to have a unique fold. The discovery of a unique mycobacterial heme acquisition pathway opens new avenues of exploration into mycobacterial therapeutics.
Keywords: X-ray crystallography, iron uptake
The human pathogen, Mycobacterium tuberculosis (Mtb), currently infects one-third of the world's population and kills ~1.7 million people annually, a crisis compounded by the emergence of multidrug resistance and the AIDS pandemic, which has resulted in a large increase in persons highly susceptible to tuberculosis. There is an urgent need to understand the pathogen's survival tactics. Iron, essential for pathogen survival, is sequestered from its host by well-characterized siderophore-mediated iron acquisition pathways. Pathogenic mycobacteria including Mtb synthesize two types of siderophores—the cell-bound water-insoluble mycobactins and the secreted water-soluble exochelins, which have a mycobactin-like backbone (1, 2), and are referred to as exomycobactins hereafter. Exomycobactins, the most abundant molecules secreted by Mtb on a molar basis, can remove iron from human transferrin and lactoferrin and transport it to either mycobactins in the Mtb cell wall (3) or the iron transport system, IrtA/B (4). However, there are disadvantages to using siderophore-mediated iron uptake pathways alone. First, mycobactin and exomycobactin production requires at least 11 different enzymes; hence mycobacteria must devote considerable metabolic energy to synthesize them (5). Second, transferrin iron accounts for <1% of the body's total iron whereas heme iron accounts for >80% (6). A recent study suggests that there is an alternative mycobacterial iron uptake pathway. A recombinant bacillus Calmette-Guérin (rBCG) mutant with an interrupted mycobactin/exomycobactin biosynthetic pathway surprisingly multiplied, albeit at a rate considerably less than the parental strain, in SCID mice (7). This result led us to postulate that another type of iron uptake pathway exists. Given that a number of pathogenic bacteria acquire iron from the largest source, heme iron (8), it is highly plausible that mycobacteria may use a heme uptake system.
Over the past two decades, it has become evident that heme is a major source of iron for both Gram-negative and Gram-positive bacteria (9, 10). An extensive bioinformatics search with known membrane-bound and cell wall-associated heme/hemoglobin receptors, heme transporter, or secreted heme scavenging proteins (hemophores) found no homologs in mycobacterial proteomes. However, past studies suggest the presence of an uncharacterized heme acquisition system in mycobacteria. First, the addition of exogenous heme to an Mtb mutant with an interrupted heme biosynthetic pathway, which is essential to many enzymes, restores mutant growth (11). Second, the addition of hemoglobin can increase growth of mycobacteria (12). Third, a gallium substituted heme derivative is toxic to Mycobacterium smegmatis cells, suggesting that gallium-metalloporphyrin is acquired by the mycobacteria and is either used in the cell wall environment, thus preventing electron transfer, or broken down in the cytoplasm to release toxic metal (13).
Here we show that Mtb has a mechanism to use exogenous heme as its iron source. Identification of the genomic region responsible for heme uptake reveals several mycobacterial-specific proteins including a secreted heme-binding protein with a unique fold. Mycobacterial iron acquisition is potentially a therapeutic target (14); the identification of all possible avenues of iron uptake, and thus the discovery of a unique mycobacterial heme uptake system, is central to the development of therapeutic strategies targeting iron acquisition.
Results
Mtb Uses Heme Iron.
To determine whether Mtb has a functional heme uptake system, we constructed an Mtb mutant deficient in iron uptake, MtbΔmbtB, by disrupting mbtB (thereby disrupting mycobactin/exomycobactin biosynthesis) (Fig. S1A). Similar to BCGΔmbtB (7), MtbΔmbtB does not grow in 7H9 media containing ~130 μM Fe3+ (as ferric ammonium citrate), unless supplemented with exogenous mycobactin (Fig. 1 A and B). However, in the absence of mycobactin, MtbΔmbtB displays near maximal growth in the presence of submicromolar concentrations of heme or human hemoglobin (Fig. 1 A and B), confirming that Mtb has a heme-iron uptake system, with the ability to scavenge both free heme and heme bound to hemoglobin as an iron source. This system also functions in wild-type Mtb that produces mycobactins and exomycobactins, as evidenced by its growth in iron-depleted media in the presence of heme or hemoglobin, but not in their absence (Fig. S2).
Fig. 1.
Mtb uses heme as an iron source and secretes a heme-binding protein. (A and B) MtbΔmbtB was grown in 7H9–0.01% tyloxapol media containing mycobactin J (myc) (10 ng/mL), heme (Hm), (0.04, 0.2, or 1 μM), human hemoglobin (Hb), (0.04, 0.2, or 1 μM; concentration of the monomer), or no added supplement (0 μM Hm or Hb). Growth was determined by measuring absorbance at 750 nm. Three independent experiments were performed with similar results. (C) SDS/PAGE of Mtb culture filtrate fractions eluted from a hemin–agarose column. Lane 1, molecular weight marker; lane 2, purified recombinant Rv0203; lanes 3 and 4, eluted fractions.
Identification and Characterization of a Putative Hemophore.
In an attempt to identify proteins involved in mycobacterial heme uptake, we used heme–agarose affinity chromatography to pull down secreted heme-binding proteins that may be potential hemophores. We incubated Mtb culture filtrate containing secreted proteins with heme–agarose beads, washed the beads, and eluted bound proteins under denaturing conditions. SDS/PAGE showed only one eluted protein band, which we analyzed by liquid chromatography tandem mass spectrometry and found to correspond to Rv0203, an uncharacterized mycobacteria-specific protein (Fig. 1C). The gene Rv0203 encodes for a protein with a signal peptide (15). To investigate the effect of Rv0203 on the ability of Mtb to scavenge heme, we deleted Rv0203 from MtbΔmbtB to create a double-deletion mutant, which was constructed by replacing Rv0203 with an apramycin resistance cassette and its own promoter (Fig. S1C). Growth of MtbΔmbtBΔRv0203 in 7H9 medium supplemented with heme was reduced 83% compared with MtbΔmbtB and reduced 61% in 7H9 medium supplemented with hemoglobin (Fig. 2A and Table 1). Also, in the presence of heme or hemoglobin, the growth rate of MtbΔmbtBΔRv0203 was ~50% slower than that of MtbΔmbtB (Table S1). Analysis of mRNA expression of genes immediately flanking deleted Rv0203 demonstrated that the deletion had no negative polar effect, as the genes were expressed and even up-regulated compared with MtbΔmbtB (Table S2). Furthermore, expression of Rv0203 from a plasmid fully restored growth of MtbΔmbtBΔRv0203 in the presence of heme and hemoglobin (Fig. 2B, Fig. S3, and Table S3) and restored secretion of Rv0203 protein into the culture filtrate (Fig. S3C). Together, these results demonstrate that Rv0203 plays an important role in heme uptake, where the Rv0203 mutation alone is responsible for the attenuated phenotype.
Fig. 2.
(A) Secreted protein, Rv0203, and membrane protein, MmpL11, facilitate heme uptake. MtbΔmbtB, MtbΔmbtBΔRv0203, and MtbΔmbtBΔmmpL11 were grown in 7H9–0.01% tyloxapol media containing mycobactin J (myc) (10 ng/mL), 0.4 μM heme (0.4 μM Hm), 0.2 μM human hemoglobin (0.2 μM Hb), or no added supplement (No Sup.). Growth was determined by measuring absorbance at 750 nm. Experiments were performed three to five times with similar results. (B) Complementation analysis of MtbΔmbtBΔRv0203 and MtbΔmbtBΔmmpL11 in studies in which the noncomplemented strains contain the control plasmid pNBV1. Growth and measurement conditions were the same as in A although the medium contained 50 μg/mL hygromycin. Three independent experiments were performed, and growth was compared as described in Table 1. The data are displayed as percentage of growth (mean ± SE) relative to MtbΔmbtB pNBV1.
Table 1.
MtbΔmbtBΔRv0203 and MtbΔmbtBΔmmpL11 are attenuated for growth in the presence of heme and hemoglobin
| % growth compared with MtbΔmbtB, mean ± SE* |
|||
| Strain | Condition | Nonnormalized† | Normalized‡ |
| MtbΔmbtB | Heme | 100 | 100 |
| Hemoglobin | 100 | 100 | |
| MtbΔmbtB ΔRv0203 | Heme | 17 ± 6 (P = 0.002)§ | 19 ± 7 (P = 0.002) |
| Hemoglobin | 39 ± 8 (P = 0.01) | 40 ± 9 (P = 0.04) | |
| MtbΔmbtB ΔmmpL11 | Heme | 13 ± 3 (P = 0.0001) | 13 ± 3 (P = 0.0001) |
| Hemoglobin | 30 ± 8 (P = 0.03) | 33 ± 12 (P = 0.06) | |
*Mean of four or five experiments (heme) or three experiments (hemoglobin).
†Growth of MtbΔmbtBΔRv0203 or MtbΔmbtBΔmmpL11 in the presence of heme or hemoglobin (0.2 or 0.4 μM) compared with growth of MtbΔmbtB under the same conditions measured at maximal difference, with the data fit to a standard sigmoid growth model (SI Methods).
‡Growth of each strain in the presence of heme or hemoglobin after normalization to growth in the presence of mycobactin J.
§Statistical analysis performed using a paired, two-tailed Student's t test.
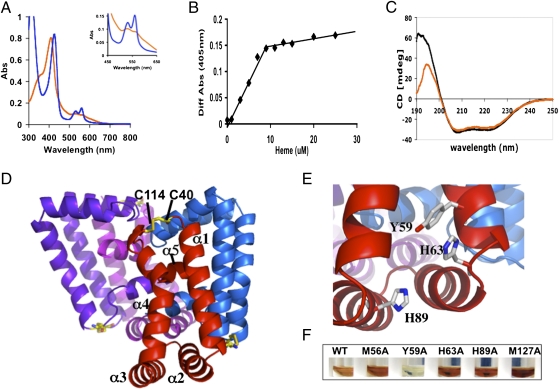
To confirm that Rv0203 binds heme, we purified the recombinant mature protein (lacking the signal peptide) from Escherichia coli and determined its heme binding capabilities. Apo-Rv0203 was incubated with 1.5-M equivalents of heme, and excess heme was removed by gel filtration chromatography. The protein eluted in one brownish-yellow colored peak. The heme–Rv0203 complex was confirmed by absorption spectroscopy, which revealed a maximum Soret band (γ-peak) at 407 nm that upon reduction with sodium hydrosulfite shifted to 421 nm, with the emergence of distinct β- and α-peaks at 526 and 557 nm, respectively, indicative of a heme binding protein (Fig. 3A). Titration of heme into Rv0203 resulted in a 1:1 stoichiometry of heme to Rv0203 (Fig. 3B), and its heme-binding affinity was determined to have a Kd of 5.4 × 10−6 M by isothermal calorimetry. In comparison, hemophores from Bacillus anthracis IsdX1 and Porphyromonas gingivalis HmuY have Kds of ~10−6 M (16, 17), whereas the hemophore from Serratia marcescens, HasA, has a heme affinity Kd of ~10−10 M (18). Thus, the heme binding affinity observed for Rv0203 is consistent with that of other known bacterial hemophores. Also, circular dichroism (CD) analysis shows no appreciable change in secondary structure rearrangement in Rv0203 upon the addition of heme (Fig. 3C). Thus, we have shown that Rv0203 is a heme-binding protein.
Fig. 3.
Mycobacteria-specific Rv0203 binds heme and has a unique structural fold. (A) The UV/vis absorbance spectra of heme-Rv0203 (orange line) and reduced heme-Rv0203 (blue line). The Inset shows the magnified Q-band region between 450 and 650 nm. (B) Titration of heme to apo-Rv0203 (10 μM) is observed by the spectral change at 405 nm between protein plus heme and heme alone. The protein saturates at 10 μM heme. (C) Circular dichroism spectrum of apo-Rv0203 (black line) and heme-Rv0203 (orange line). (D) Tetrameric structure of Rv0203. Each polypeptide chain is colored differently and the disulfide bond is in stick representation, where the carbon and sulfur atoms are colored orange and yellow, respectively. (E) The putative heme-binding site with Tyr59, His63, His89 in stick representation, where carbon, nitrogen, and oxygen atoms are colored white, blue, and red, respectively. (F) Wild type and mutants of heme-bound Rv0203 after gel-filtration chromatography. Rv0203-Y59A is colorless, indicating that Y59 is critical for heme binding.
Structure of the Putative Hemophore.
To further characterize Rv0203, we solved the crystal structure by multiwavelength anomalous diffraction (MAD) phasing to 2.5 Å, yielding an Rfree/Rwork of 19.5/25.6 (Table S4). A structural homology search using the Dali server indicates that Rv0203 has a unique fold and is highly atypical among heme transport proteins (19). The predominately α-helical structure consists of a dimer of dimers, where each monomer consists of five α-helices (Fig. 3D). Within each monomer, the two longest α-helices (α1 and α4) are tethered at the N terminus of α1 and the C terminus of α4 by a disulfide bond between Cys40 and Cys114 and capped by two short helices (α2 and α3) with the α5 helix (α5) lying perpendicularly across α1 and α4 helices. The dimer is formed by antiparallel interactions between the α1 helices, and the four helices of the dimer form an antiparallel helical sheet, with the two shorter helices capping each end of the dimer, forming a catcher's mitt-like structure. Two dimers associate to produce an off-tilt cage-like structure, where the α5 helices form a weak hydrophobic core in the center of the cage (Fig. 3D and Fig. S4A). The monomers in the dimer superimpose on each other to give a root mean square of 2.0 Å. The most obvious differences between these two polypeptide chains are (i) the loop region connecting α2 and α3 and (ii) the loop region connecting α4 to α5, causing the α5 helices not to superimpose upon one another (Fig. S4B).
The Mtb hemophore, Rv0203, is a mycobacteria-specific protein and has a unique fold with an unusual self-association. However, comparison with the structurally divergent S. marcescens hemophore, HasA (20) (Fig. 3D and Fig. S4C), reveals a similar heme-binding motif in Rv0203 (Fig. 3E and Fig. S4E), which is indicative of convergent evolution akin to the catalytic triads found in various, otherwise unrelated, serine proteases (21). The ligands that coordinate heme iron in HasA are His32 and Tyr75 (4.8 Å apart), which is hydrogen-bonded to His83 (Fig. S4E). Within the structure of Rv0203, there is a similar potential heme-binding motif, where Tyr59 and His89 are 7.8 Å apart and Tyr59 is hydrogen bonded to His63 (Fig. 3E). To investigate the effect of potential heme-binding residues on Rv0203, we introduced point mutations Tyr59Ala, His63Ala, and His89Ala (and Met56Ala and Met127Ala as controls) into Rv0203. Each Rv0203 mutant protein was purified, bound with heme, and further purified by gel filtration chromatography. Compared with Rv0203 and other mutants, Rv0203-Tyr59Ala does not bind heme (Fig. 3F). Additionally, absorbance spectra of Rv0203 and mutants revealed Soret bands of 407 nm for all but Rv0203-Tyr59Ala (Fig. S4F). CD analysis confirmed that Rv0203-Tyr59Ala is mainly α-helical and folded as observed for Rv0203 (Fig. S4G). Thus, Tyr59 appears to play an important role in heme binding in Rv0203, and binding of heme predominately through a tyrosine axial ligand is not unprecedented. Biochemical and structural analysis of a periplasmic heme-binding protein, ShuT, indicates that one tyrosine residue is the axial heme-iron ligand (22, 23), and heme iron is five-coordinated by ShuT. In addition, the heme iron in two other heme-binding proteins, IsdA and IsdC, involved in Staphylococcus aureus heme uptake, are both five-coordinated by one tyrosine residue (24, 25). These results infer that Tyr59 is critical for heme binding within the mycobacterial potential hemophore, Rv0203.
Identification of Transmembrane Heme Transporters.
Unlike other organisms, such as Yersinia pestis, S. aureus, B. anthracis, and Pseudomonas aeruginosa, where most of the genes encoding proteins involved in heme uptake are contained within a single operon (16, 26–28), in Mtb, Rv0203 is not located within an operon. However, Rv0203 is surrounded by genes that encode for predicted transmembrane proteins from Rv0201c to Rv0207c (Fig. 4A). Of particular interest are Rv0202c (MmpL11) and Rv0206c (MmpL3), which both belong to a family of 13 mycobacterial transmembrane proteins (MmpLs) and are proposed to be involved in transport of molecules across the mycobacterial membrane (29–31). Thus, we postulate that MmpL11 and MmpL3 are both heme transporters. Previously, it was shown that mmpL3 is likely to be essential (32), and our attempts to delete the potential heme uptake region (Rv0201c–Rv0207c) or mmpL3 alone from MtbΔmbtB were also unsuccessful. To evaluate the involvement of MmpL11 in heme uptake, a double mutant with deleted Rv0202c (mmpL11) was constructed by replacing most of the mmpL11 coding region with an apramycin resistance cassette and its promoter in-frame (Fig. S1D). Similar to MtbΔmbtBΔRv0203, growth of MtbΔmbtBΔmmpL11 in 7H9 media supplemented with heme or hemoglobin was reduced compared with MtbΔmbtB (heme, 87% reduction; hemoglobin, 70% reduction; Fig. 2A and Table 1) and the growth rate was ~50% slower than that of MtbΔmbtB grown in the presence of heme or hemoglobin (Table S1). Analysis of mRNA expression of the genes immediately flanking deleted mmpL11 revealed that they were expressed and up-regulated compared with MtbΔmbtB (Table S2). Also, expression of mmpL11 from a plasmid partially restored growth of the MtbΔmbtBΔmmpL11 strain in the presence of heme and hemoglobin (Fig. 2B, Fig. S3, and Table S3), which may be a consequence of the non-iron–regulated promoter on the plasmid controlling the expression of mmpL11. Taken together, these results suggest that MmpL11 plays a significant role in heme uptake. MmpL3 has 25% sequence identity to MmpL11. Given the sequence homology and their close genomic vicinity, we hypothesize that both MmpL11 and MmpL3 are involved in the transport of heme across the mycobacterial membrane.
Fig. 4.
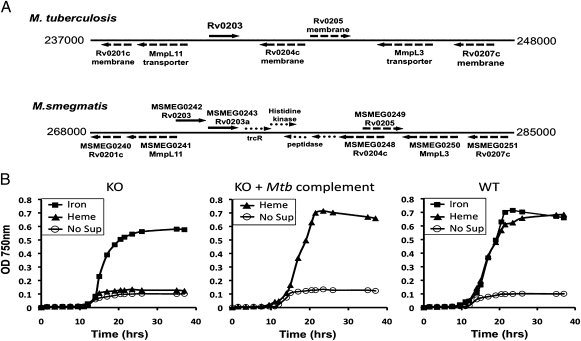
Use of M. smegmatis to identify Mtb genes involved in heme uptake. (A) Mtb and M. smegmatis genomic regions surrounding Rv0203. Rv0203 and M. smegmatis homologs, Msmeg0242 and Msmeg0243, are depicted with solid arrows. The dashed arrows depict genes with high sequence homology between the two genomes. Note that the corresponding genomic region in M. smegmatis contains all of the genes in the genomic region of Mtb surrounding Rv0203. (B) Growth curves of M. smegmatis mc2155 (WT), MsmegΔhemeuptake (genes inclusive of Msmeg0240–Msmeg0251, designated KO) and MsmegΔhemeuptake::Rv0202c-Rv0207c (complemented with Mtb genes inclusive of Rv0202c–Rv0207c, designated KO+Mtb complement) in 7H9–0.01% tyloxapol-NoFe medium supplemented with 1 μM FeCl3, 1 μM heme, or no supplement (No Sup).
Genomic Region Essential for Heme Uptake.
To further investigate the potential heme uptake region, we turned to the model organism, M. smegmatis, which is a soil-dwelling Mycobacterium (33) with alternate iron pathways compared with intracellular pathogenic mycobacteria. M. smegmatis has a similar genomic organization to the corresponding proposed heme uptake region in Mtb (Fig. 4A). To determine whether these genes are essential for heme uptake in M. smegmatis, we constructed a knockout mutant of this region (MsmegΔhemeuptake), whereby M. smegmatis is able to tolerate deletion of MmpL3 and thus the entire proposed heme uptake region. M. smegmatis (WT) and MsmegΔhemeuptake were passaged in iron-deplete 7H9 medium to remove intracellular iron and growth under iron-deplete, 1-μM iron or heme conditions was monitored (Fig. 4B). No growth was observed for either WT or MsmegΔhemeuptake under iron-deplete conditions, although growth was fully restored for WT in the presence of either iron or heme. However, MsmegΔhemeuptake grew substantially in media supplemented with iron but not heme. Importantly, complementation of MsmegΔhemeuptake with a plasmid containing the Mtb genes inclusive of Rv0202c–Rv0207c fully restored growth in the presence of heme to that of WT (Fig. 4B). These results suggest that the Mtb genomic region between Rv0202c and Rv0207c encodes for key proteins involved in mycobacterial heme uptake.
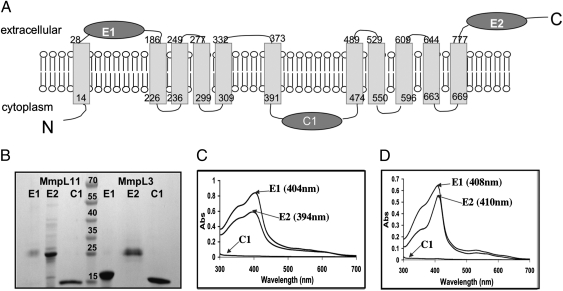
To confirm the hypothesis that MmpL11 and MmpL3 physically bind heme for transportation across the membrane, we purified the soluble domains of each potential heme transporter and determined their heme binding capabilities. We predicted the domain boundaries of the two extracellular (E1 and E2) and cytoplasmic (C1) soluble domains (Fig. 5A and Fig. S4H) and cloned, expressed, and purified them (Fig. 5B). Each domain was incubated with heme, subjected to gel filtration chromatography, and then analyzed by UV/vis spectroscopy. For both MmpL11 and MmpL3, the spectra of extracellular domains had Soret peaks between 399 and 410 nm, indicative of heme-binding protein domains. In contrast, the cytoplasmic domains showed no evidence of heme binding (Fig. 5 C and D). These results imply extracellular to intracellular heme transport and offer concrete support of the previous proposal that several MmpL proteins may be involved in mycobacterial metabolite import (32). The high sequence homology and close genomic proximity of MmpL3 and MmpL11 (and also the hemophore Rv0203), and the similar heme binding properties of their soluble domains, support roles of both MmpL3 and MmpL11 in heme uptake.
Fig. 5.
Examination of heme binding capabilities of the predicted soluble domains of MmpL11 and MmpL3. (A) Predicted topology of MmpL11 (Rv0202c), which is very similar to that of MmpL3 (Rv0206c), Fig. S4H. There are 11 transmembrane helices, two extracellular domains (E1 and E2), and one cytoplasmic domain (C1). (B) SDS/PAGE of purified domains of both MmpL11 and MmpL3. Domains of MmpL11 are to the left of the lane with molecular weight markers and domains of MmpL3 are to the right of the lane with molecular weight markers (kDa), as indicated. (C and D) UV/vis absorbance spectra of domains E1, E2, and C1 from MmpL11 (C) and MmpL3 (D) after each domain was incubated with heme and repurified by gel filtration. For both MmpL11 and MmpL3, both E1 and E2 bind heme whereas C1 does not.
Discussion
Our findings offer strong support that there is a previously uncharacterized mycobacterial heme uptake system. We identified a unique secreted heme-binding protein and several membrane proteins encoded by the region inclusive of Rv0202c–Rv0207c that are implicated in sequestering heme iron from the host and transportation of heme across the cell wall and membrane (Fig. 6). One potential energy source for heme transport is ATP (as seen with other heme uptake systems) (34), as several membrane proteins within the heme uptake vicinity are predicted to bind ATP including MmpL3 and MmpL11 and others to be ATPases (35). Additionally, in a previous study of MmpL proteins, MtbΔmmpL11 established infection normally in a murine low-dose aerosol model of tuberculosis but was significantly less lethal; median survival of C57BL/6 mice infected with wild-type Mtb was 265 d vs. 398 d for mice infected with MtbΔmmpL11 (32). Importantly, this result suggests that heme uptake may contribute to the pathogenesis of Mtb.
Fig. 6.
Proposed model of mycobacterial heme uptake. Depicted is the structure of the hemophore Rv0203 using free heme (Hm, orange circles) or heme from host hemoglobin (Hb, brown tetramer). Rv0203 delivers heme to membrane proteins MmpL11 (pink) or MmpL3 (light blue), where heme is shuttled through the membrane. In the cytoplasm the heme-degrading protein, MhuD (PDB:3HX9), breaks down heme to release iron. Black dashed lines indicate the direction of heme uptake from host to cytoplasm, although the exact sequence of events is still to be determined along with other protein players within this pathway.
Mtb may encounter heme or hemoglobin at both intracellular and extracellular sites of infection. Macrophages degrade senescent red blood cells at a rate of 2 million/s (36). Thus, intracellular Mtb in macrophage phagosomes may interact with heme that diffuses to its phagosome from the phagolysosome, where the erythrocyte is degraded, within the same host cell; alternatively, as some metabolically active Mtb resides in phagolysosomes (37), the bacteria may interact directly with heme or hemoglobin released from erythrocytes. Extracellular Mtb may encounter heme or hemoglobin in the bloodstream during dissemination from primary to secondary sites of infection, as well as in tuberculous lung cavities into which bleeding has occurred, a phenomenon that may result in hemoptysis, a symptom of tuberculosis; in these settings, utilization of heme may be facilitated by hemolysins expressed by Mtb (38).
The discovery of a mycobacterial heme uptake system that has the ability to use heme from human hemoglobin challenges a long existing paradigm that Mtb obtains iron solely via exomycobactins and mycobactins scavenging iron from iron-containing proteins, especially transferrin and lactoferrin. To further support the hypothesis that mycobacteria may use heme as an iron source, a cytoplasmic mycobacterial heme-degrading protein, MhuD, has recently been characterized, suggesting that heme iron traversing the membrane may be degraded to provide iron under iron-deplete conditions (Fig. 6) (39). Moreover, the likely essentiality of mmpL3 (Rv0206c), which encodes a probable heme transport protein, suggests that heme uptake may play a pivotal role in mycobacterial pathogenicity in vivo. In conclusion, we confirmed that a heme uptake pathway exists for Mtb and identified the key players in this pathway. This breakthrough fills in the knowledge gap of iron-sequestering pathways in Mtb and opens new avenues in mycobacterial biology for the development of therapeutics.
Methods
All bacterial strains, plasmids, and primers used in this work are listed in Tables S5–S7.
An MtbΔmbtB mutant (Erdman strain) defective in mycobactin biosynthesis was constructed via an allelic exchange method, which uses a temperature-sensitive sacB plasmid, as previously described for a BCGΔmbtB strain (7). The double mutants, MtbΔmbtBΔRv0203 and MtbΔmbtBΔmmpL11, were constructed via specialized transduction (40), and details are outlined in SI Methods. Mtb wild type and mutants were cultured in 7H9–0.01% tyloxapol containing no supplement, various concentrations of heme or human hemoglobin, or mycobactin J and grown for 20–28 d, until the cultures reached stationary phase. Growth was monitored by removing aliquots for absorbance measurements at 750 nm. The cloning, protein purification (Table S8), and crystallization protocols have been described for other Mtb proteins (41), and crystals of Rv0203 were grown by a hanging-drop, vapor-diffusion method against a reservoir containing 1.7 M LiSO4, 0.1 M Hepes, pH 7.8, at room temperature. Methodology for structure determination has been previously described (42). Heme binding characterization is outlined in SI Methods (43). An M. smegmatis heme uptake knockout mutant was created as described previously (44) and the complement plasmid with Mtb genes was constructed as described in SI Methods (7). M. smegmatis wild type and mutants were passaged in 7H9–0.01% tyloxapol-NoFe and then cultured in media containing no supplement, 1 μM heme, or 1 μM FeCl3. Cell density was measured at 750 nm at time points over a 72-h period.
Supplementary Material
Acknowledgments
The authors thank Dr. John T. Belisle, Colorado State University, National Institutes of Health, National Institute of Allergy and Infectious Diseases Contract N01 AI-75320, for the generous supply of Mtb H37Rv genomic DNA. We also thank all of the staff at Stanford Synchrotron Radiation Lightsource for their invaluable help in data collection. We thank Laleh Rezaei-Homami and Saša Masleša-Galić for technical assistance and Jeffrey Gornbein for assistance with statistical analysis. Finally, we thank Drs. Tom Poulos, Sheryl Tsai, Duilio Cascio, and Morgan Beeby for invaluable discussions and the TB structural genomics consortium for their support. This work was supported by a grant from the National and California American Lung Association RG-78755-N (to C.W.G.) and by National Institutes of Health Grants AI081161 (to C.W.G.), AI068135 (contract to C.W.G.), A1078691 (to L.M.M.), AI068413 (to M.A.H.), AI078691 (to L.M.M.), and HL077000 (to M.A.H.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org/pdb (PDB ID code 3MAY).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009516108/-/DCSupplemental.
References
- 1.Gobin J, et al. Iron acquisition by Mycobacterium tuberculosis: Isolation and characterization of a family of iron-binding exochelins. Proc Natl Acad Sci USA. 1995;92:5189–5193. doi: 10.1073/pnas.92.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong DK, Gobin J, Horwitz MA, Gibson BW. Characterization of exochelins of Mycobacterium avium: Evidence for saturated and unsaturated and for acid and ester forms. J Bacteriol. 1996;178:6394–6398. doi: 10.1128/jb.178.21.6394-6398.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gobin J, Horwitz MA. Exochelins of Mycobacterium tuberculosis remove iron from human iron-binding proteins and donate iron to mycobactins in the M. tuberculosis cell wall. J Exp Med. 1996;183:1527–1532. doi: 10.1084/jem.183.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez GM, Smith I. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J Bacteriol. 2006;188:424–430. doi: 10.1128/JB.188.2.424-430.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez GM. Control of iron metabolism in Mycobacterium tuberculosis. Trends Microbiol. 2006;14:320–327. doi: 10.1016/j.tim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Crichton R. Inorganic Biochemistry of Iron Metabolism: From Molecular Mechansims to Clinical Consequences. West Sussex, UK: Wiley; 2001. [Google Scholar]
- 7.Tullius MV, Harth G, Maslesa-Galic S, Dillon BJ, Horwitz MA. A replication-limited recombinant Mycobacterium bovis BCG vaccine against tuberculosis designed for human immunodeficiency virus-positive persons is safer and more efficacious than BCG. Infect Immun. 2008;76:5200–5214. doi: 10.1128/IAI.00434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilks A, Burkhard KA. Heme and virulence: How bacterial pathogens regulate, transport and utilize heme. Nat Prod Rep. 2007;24:511–522. doi: 10.1039/b604193k. [DOI] [PubMed] [Google Scholar]
- 9.Cescau S, et al. Heme acquisition by hemophores. Biometals. 2007;20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- 10.Wandersman C, Stojiljkovic I. Bacterial heme sources: The role of heme, hemoprotein receptors and hemophores. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 11.Parish T, Schaeffer M, Roberts G, Duncan K. HemZ is essential for heme biosynthesis in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2005;85:197–204. doi: 10.1016/j.tube.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Raghu B, Sarma CR, Venkatesan P. Effect of hemoglobin on the growth of mycobacteria and the production of siderophores. Indian J Pathol Microbiol. 1993;36:376–382. [PubMed] [Google Scholar]
- 13.Stojiljkovic I, Kumar V, Srinivasan N. Non-iron metalloporphyrins: Potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol Microbiol. 1999;31:429–442. doi: 10.1046/j.1365-2958.1999.01175.x. [DOI] [PubMed] [Google Scholar]
- 14.Monfeli RR, Beeson C. Targeting iron acquisition by Mycobacterium tuberculosis. Infect Disord Drug Targets. 2007;7:213–220. doi: 10.2174/187152607782110031. [DOI] [PubMed] [Google Scholar]
- 15.Ben Amor Y, et al. Immunological characterization of novel secreted antigens of Mycobacterium tuberculosis. Scand J Immunol. 2005;61:139–146. doi: 10.1111/j.0300-9475.2005.01557.x. [DOI] [PubMed] [Google Scholar]
- 16.Maresso AW, Garufi G, Schneewind O. Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLoS Pathog. 2008;4:e1000132. doi: 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wójtowicz H, et al. Unique structure and stability of HmuY, a novel heme-binding protein of Porphyromonas gingivalis. PLoS Pathog. 2009;5:e1000419. doi: 10.1371/journal.ppat.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deniau C, et al. Thermodynamics of heme binding to the HasA(SM) hemophore: Effect of mutations at three key residues for heme uptake. Biochemistry. 2003;42:10627–10633. doi: 10.1021/bi030015k. [DOI] [PubMed] [Google Scholar]
- 19.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnoux P, et al. The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat Struct Biol. 1999;6:516–520. doi: 10.1038/9281. [DOI] [PubMed] [Google Scholar]
- 21.Tang J, James MN, Hsu IN, Jenkins JA, Blundell TL. Structural evidence for gene duplication in the evolution of the acid proteases. Nature. 1978;271:618–621. doi: 10.1038/271618a0. [DOI] [PubMed] [Google Scholar]
- 22.Eakanunkul S, et al. Characterization of the periplasmic heme-binding protein shut from the heme uptake system of Shigella dysenteriae. Biochemistry. 2005;44:13179–13191. doi: 10.1021/bi050422r. [DOI] [PubMed] [Google Scholar]
- 23.Ho WW, et al. Holo- and apo-bound structures of bacterial periplasmic heme-binding proteins. J Biol Chem. 2007;282:35796–35802. doi: 10.1074/jbc.M706761200. [DOI] [PubMed] [Google Scholar]
- 24.Grigg JC, Vermeiren CL, Heinrichs DE, Murphy ME. Haem recognition by a Staphylococcus aureus NEAT domain. Mol Microbiol. 2007;63:139–149. doi: 10.1111/j.1365-2958.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 25.Sharp KH, Schneider S, Cockayne A, Paoli M. Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus. J Biol Chem. 2007;282:10625–10631. doi: 10.1074/jbc.M700234200. [DOI] [PubMed] [Google Scholar]
- 26.Létoffé S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 27.Mazmanian SK, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 28.Rossi MS, et al. Identification and characterization of the hemophore-dependent heme acquisition system of Yersinia pestis. Infect Immun. 2001;69:6707–6717. doi: 10.1128/IAI.69.11.6707-6717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Converse SE, et al. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc Natl Acad Sci USA. 2003;100:6121–6126. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domenech P, et al. The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J Biol Chem. 2004;279:21257–21265. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- 31.Jain M, Cox JS. Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in M. tuberculosis. PLoS Pathog. 2005;1:e2. doi: 10.1371/journal.ppat.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domenech P, Reed MB, Barry CE., 3rd Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect Immun. 2005;73:3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratledge C, Ewing M. The occurrence of carboxymycobactin, the siderophore of pathogenic mycobacteria, as a second extracellular siderophore in Mycobacterium smegmatis. Microbiology. 1996;142:2207–2212. doi: 10.1099/13500872-142-8-2207. [DOI] [PubMed] [Google Scholar]
- 34.Köster W. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res Microbiol. 2001;152:291–301. doi: 10.1016/s0923-2508(01)01200-1. [DOI] [PubMed] [Google Scholar]
- 35.Pal D, Eisenberg D. Inference of protein function from protein structure. Structure. 2005;13:121–130. doi: 10.1016/j.str.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 36.Soe-Lin S, et al. Nramp1 promotes efficient macrophage recycling of iron following erythrophagocytosis in vivo. Proc Natl Acad Sci USA. 2009;106:5960–5965. doi: 10.1073/pnas.0900808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee BY, Clemens DL, Horwitz MA. The metabolic activity of Mycobacterium tuberculosis, assessed by use of a novel inducible GFP expression system, correlates with its capacity to inhibit phagosomal maturation and acidification in human macrophages. Mol Microbiol. 2008;68:1047–1060. doi: 10.1111/j.1365-2958.2008.06214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wren BW, et al. Characterization of a haemolysin from Mycobacterium tuberculosis with homology to a virulence factor of Serpulina hyodysenteriae. Microbiology. 1998;144:1205–1211. doi: 10.1099/00221287-144-5-1205. [DOI] [PubMed] [Google Scholar]
- 39.Chim N, Iniguez A, Nguyen TQ, Goulding CW. Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. J Mol Biol. 2010;395:595–608. doi: 10.1016/j.jmb.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bardarov S, et al. Specialized transduction: An efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 41.Goulding CW, Perry LJ. Protein production in Escherichia coli for structural studies by X-ray crystallography. J Struct Biol. 2003;142:133–143. doi: 10.1016/s1047-8477(03)00044-3. [DOI] [PubMed] [Google Scholar]
- 42.Goulding CW, et al. Crystal structure of a major secreted protein of Mycobacterium tuberculosis-MPT63 at 1.5-A resolution. Protein Sci. 2002;11:2887–2893. doi: 10.1110/ps.0219002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izadi N, et al. Purification and characterization of an extracellular heme-binding protein, HasA, involved in heme iron acquisition. Biochemistry. 1997;36:7050–7057. doi: 10.1021/bi962577s. [DOI] [PubMed] [Google Scholar]
- 44.Converse SE, Cox JS. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J Bacteriol. 2005;187:1238–1245. doi: 10.1128/JB.187.4.1238-1245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.