Abstract
The Wnt/β-catenin pathway plays multiple and diverse roles in development by regulating gene expression via T-cell factor/Lymphoid enhancer-binding factor (Tcf/Lef) DNA binding factors. Misregulation of this pathway is thought to initiate colon adenoma formation. It is controversial whether Tcf4 (Tcf7L2) functions as an oncogene or tumor suppressor gene in colon carcinogenesis. We show here that Tcf4 haploinsufficiency results in colon tumor formation in a mouse tumor model that normally only develops small intestinal tumors. Further, we show that loss of Tcf4 early in development and in adult colon results in increased cell proliferation. These findings strongly suggest that Tcf4 normally modulates proliferation of the colonic epithelium and that disruption of Tcf4 activity increases proliferation, leading to colon tumorigenesis. Taken together, our in vivo studies favor a tumor suppressor function for Tcf4.
Keywords: colon cancer, mouse model, differentiation, transit amplifying cells, stem cells
The Wnt/β-catenin signaling pathway plays a critical role during embryonic development and is often exploited in cancer to promote cell growth. The majority of familial [familial adenomatous polyposis (FAP)] and sporadic colorectal cancers (CRCs) feature genetic mutations in Adenomatous polyposis coli (APC), which encodes a key component of the Wingless-type (Wnt) signaling pathway (reviewed in ref. 1). In the absence of Wnt ligands, β-catenin is phosphorylated and targeted for degradation by Apc. Colon tumors form upon homozygous loss of APC, which allows for the accumulation of β-catenin in the nucleus where it binds and transactivates Tcf/Lef proteins (2, 3). A number of mouse models have been developed that contain mutations in Apc, commonly associated with human colon cancer. Consistent with human FAP, mice carrying a specific mutation in the Apc gene (ApcMin) develop intestinal tumors, but unlike human syndromes, mice rarely develop tumors in the colon (4, 5).
The Tcf/Lef transcription factors are cell type-specific downstream effectors of the Wnt/β-catenin signaling pathway. Each contains a DNA-binding high mobility group (HMG) box, as well as an N-terminal β-catenin binding domain, and their transcriptional activity is dependent upon bound corepressors or coactivators. The different Tcf/Lef family members have variant protein structure outside of the β-catenin binding domain with variable exon composition. This likely contributes to the functional diversity and nonredundant functions of Tcf/Lef proteins (reviewed in refs. 6, 7).
Much of our understanding about Tcf/Lef function has come from the studies of model organisms. Data from Tcf4 mutant mice show a loss of proliferative cells, suggesting that Tcf4 is important for stem cell renewal in the small intestine and the general assumption that the formation of the Tcf4/β-catenin complex is cancer-promoting (8). However, the biological function of Tcf4 has recently undergone renewed interrogation, because it has been found to be mutated in clear cell renal cell carcinoma (CCRCC), gastric carcinoma, and breast cancer (9–11). Additionally, Tcf loss of function mutations have been found in primary CRCs and these mutations enhance cell growth in cell lines, suggesting that Tcf4 may function as a tumor suppressor (12–14).
Finding that Tcf4 loss of function mutations result in increased cell growth raises the possibility that the level of Tcf4 protein is important for the switch from active proliferation to terminal differentiation. In the colon, Tcf4 protein levels are lowest in the proliferative cells at the base of the crypt and higher in differentiating cells that are migrating toward the lumen (this study and ref. 15). Similar expression domains are characteristic of other factors implicated in promoting differentiation of the intestinal epithelium, including Caudal type homeobox 1 (Cdx1) and Apc (16, 17). By contrast, nuclear β-catenin protein levels are in the inverse pattern, with higher levels in the proliferative compartment at the base of the crypt and decreased levels in differentiating cells, an expression pattern that correlates with cell proliferation (this study and ref. 15).
Taken together, the experimental evidence suggests two alternative models. Tcf4 could be required for progenitor cell proliferation, and Tcf4 loss would result in a loss of the proliferative cell compartment. Alternatively, Tcf4 could be required for modulating proliferation, and Tcf4 loss would force continued proliferation. To test these models, we have chosen to reexamine Tcf4 function in the mouse. We have developed and characterized a unique Tcf4 allele that resembles the loss of function mutations found in human CRC. With this system, we have defined a role for Tcf4 function in controlling intestinal cell proliferation throughout the gut, both during embryogenesis and in adult colon epithelium. Further, we show that as in human CRC, Tcf4 haploinsufficiency in combination with Apc mutation strongly enhances colon tumor formation. Taken together, our data support a tumor suppressor function for Tcf4 in colon neoplasia.
Results
Model of Tcf4 Function.
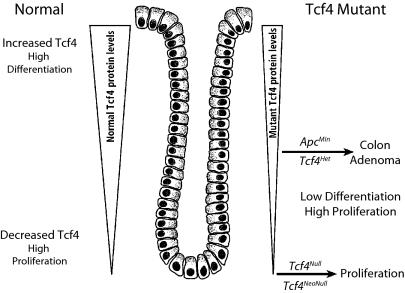
Fig. 1 illustrates a model for the role of Tcf4 in the maintenance of intestinal epithelium homeostasis and colon adenoma tumorigenesis. This model is based on the expression pattern of Tcf4 in the intestinal crypt/villus axis, the role of Tcf4 in the formation of the intestinal epithelium during embryogenesis, and studies of the function of Tcf4 in colon adenoma tumors. The principle feature of the model is that low Tcf4 protein levels favor normal intestinal epithelial and adenoma cell proliferation and thus qualify Tcf4 as a tumor suppressor, whereas high levels of Tcf4 protein promote epithelial cell differentiation. The reason for presenting the model first is that it forms the framework for integrating the results we present on the role of Tcf4 in normal and pathological intestinal etiology.
Fig. 1.
Model of the role of Tcf4 during intestinal development and in tumor formation (in the text).
Tcf4Cre Effectively Marks the Tcf4 Lineage in Early Stages of Development.
To trace Tcf4 cell lineage in the mouse and to generate an effective null allele of this locus, the first exon of Tcf4 was replaced with an EGFP-Cre recombinase fusion (Tcf4Cre; Fig. S1A). Importantly, this exon is also targeted with nonsense and frameshift mutation in human colorectal tumors (12, 13). To allow conditional disruption of Tcf4, we also generated an exon 1 floxed allele (Tcf4Lox; Fig. S1B). The latter allele was used to generate a separate null allele of Tcf4 (Tcf4Δ) by breeding to the HprtCre mouse. Mouse embryos, homozygous for all three mutant alleles (Tcf4Cre/Cre, Tcf4Cre/Lox, and Tcf4Δ/Δ) show the same mutant phenotype, which is distinct from the published loss of function allele (Tcf4Hyg/Hyg; in the text).
Fig. S1C shows Tcf4 lineage in a E14.5 embryo obtained by breeding Tcf4Cre mice with RosamTmG reporter mice, expressing membrane-targeted Tomato fluorescent protein (mTom) before Cre-dependent recombination and membrane-targeted green fluorescent protein (mGFP) following Cre recombination (18). The figure illustrates the broad cell lineage contribution of Tcf4 during embryonic development, including in the intestinal epithelial lining. In newborn Tcf4Cre RosamTmG mice, we observe that intestinal and colonic epithelial cells show Tcf4 lineage (Fig. S1D). In 6-month-old Tcf4Cre RosamTmG mice, Tcf4 lineage is found in many organs (Fig. S2) (19).
Loss of Tcf4 Results in Necrosis Throughout the Small Intestine and Colon.
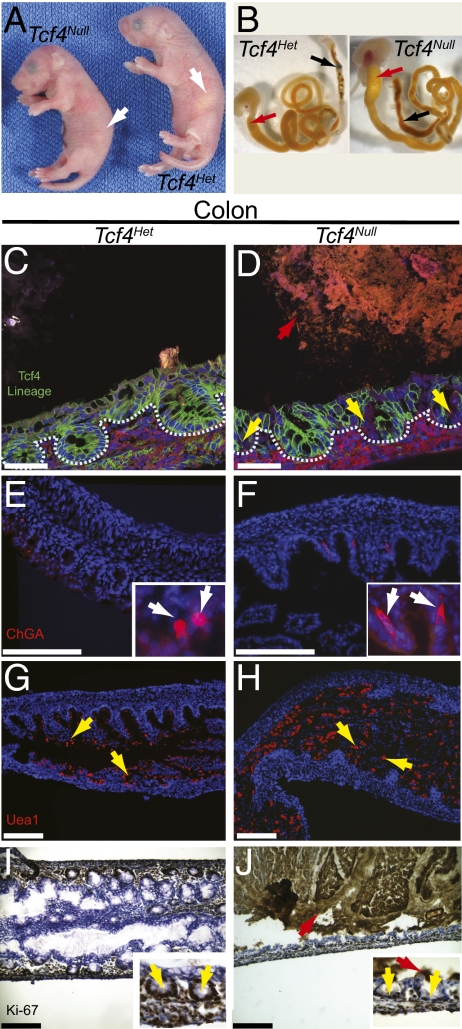
At birth, homozygous Tcf4Cre/Cre, Tcf4Cre/Δ, Tcf4 Δ/Δ, and Tcf4Cre/lox mice are not viable, lack milk in their stomachs, and are on average 21 ± 8% smaller by weight than their Tcf4Het or Tcf4WT littermates (Fig. 2A). Further, there is no Tcf4 protein present in these mutants, as shown by the lack of immunohistochemical Tcf4 antibody staining of small intestine and colon isolated from E16.5 Tcf4Null embryos (Fig. S3).
Fig. 2.
Newborn Tcf4Null mice are not viable and show necrotic death of proliferative progenitors and differentiated epithelial cell types in the colon. (A) Newborn Tcf4Null mice die within 24 h of birth and lack a milk spot (white arrows). (B) Intestines isolated from Tcf4Null mice have a distended duodenum (red arrows) and diffuse meconium (black arrows) compared with the intestines of Tcf4Het newborn mice. The effects of loss of Tcf4 during intestinal development were analyzed in both Tcf4Het and Tcf4Null mice. (C and D) Tcf4 lineage marks the membranes of epithelial cells in cryosections of newborn colon. Red arrow denotes large masses of fluorescent dead cells in the lumen and yellow arrows mark the loss of Tcf4 lineage and crypt structure in mutant crypts (crypts outlined in blue). Loss of Tcf4 impairs enteroendocrine and goblet cell differentiation throughout the gut. Staining of newborn colons with anti-chromogranin A (E and F; enteroendocrine; white arrows) and UEA-1 (G and H; goblet cells; yellow arrows). (I and J) Ki-67 staining marks proliferative cells in the colon of newborn mice. Yellow arrows mark crypts and red arrows mark background peroxidase staining of necrotic cells (scale bar, 10 μm).
The intestine and colon of Tcf4Null mice are fragile and meconium that is normally compact and located distally within the gut is diffuse throughout the colon (Fig. 2B). Histological analysis demonstrated disrupted small intestinal and colonic architecture, decreased epithelial cells, and the absence of crypt structures compared with Tcf4Het or wild-type mice (Figs. 2 C–J, and Fig. S4 A–H). We characterized epithelial cell morphology in the Tcf4Null by following Tcf4 lineage marked by mGFP. In the small intestine, Tcf4 deletion results in epithelial cell morphology changes consistent with necrosis as shown by the loss of the basal and lateral domain of the epithelial cells and the thickening of the apical domain (Fig. S4 A and B; white arrows) (20). Necrosis is apparent throughout the crypt/villus axis. In the colon, large masses of necrotic cells are apparent as well as abnormal crypt morphology and the lack of Tcf4 lineage-positive cells in the crypt (Fig. 2 C and D). This gross necrosis affects all cell types throughout the intestine, enteroendocrine and goblet cells are absent throughout the small intestine (Fig. S4 C–F), and enteroendocrine cells are elongated with large abnormally shaped nuclei (Fig. 2 E and F) and goblet cells are incorrectly localized (Fig. 2 G and H) in the colon.
One prediction of our model is that loss of Tcf4 would result in increased proliferation. However, proliferative cells are completely absent in the small intestine of Tcf4Null newborn mice (Fig. S4 G and H), whereas in the colon, the number of proliferative cells is reduced compared with heterozygous newborn mice (Fig. 2 I and J). Overall, Tcf4 loss affects all cell types throughout the small intestine and colon. It is likely that loss of Tcf4 initiates a cascade of events ultimately resulting in gross intestinal necrosis.
Loss of Tcf4 Results in Increased Proliferation in the Intestine of E13.5 Embryos.
In our model, we predict that loss of Tcf4 should result in increased proliferation. The intestinal defects in Tcf4Null newborns are apparent by E16.5, where epithelium shows signs of gross necrosis and the loss of proliferative cells (Fig. S5 D–K). To further characterize the role of Tcf4 in the intestinal epithelium, we examined E13.5 embryos just at the beginning of the conversion of undifferentiated endoderm into simple columnar epithelium that occurs between E13.5 and E18.5. We found that in the intestines of E13.5 Tcf4Het and Tcf4Null embryos, Tcf4 lineage-positive epithelial progenitors are present along with undifferentiated endodermal cells. We analyzed the ratio of Tcf4 lineage-positive cells (mGFP-positive) to total cells within the epithelial layer (mGFP plus mTom-positive; Fig. 3A). Surprisingly, we found that lineage-negative cells had almost completely been converted to Tcf4 lineage-positive cells in the intestines of Tcf4Null embryos (mean, 90%; P = 2.7 × 10−5), whereas far fewer cells had been converted in Tcf4Het embryos (mean, 58%). To test if this increase in Tcf4 expressing cells reflected increased proliferation in these cells, we stained with Phospho histone H3 (Phh3) antibody and for bromouridine (BrdU) incorporation to analyze cells that are actively in mitosis or that have passed through the S phase of the cell cycle, respectively (Fig. 3 B and C). Consistent with increased proliferation of the Tcf4 lineage in Tcf4Null E13.5 embryos, we found 25–33% increases in proliferative cells (P = 0.002; Phh3, 33%; BrdU, 25%). This increase in proliferation was not maintained and by E14.5 we saw similar levels of Tcf4 lineage in Tcf4Null embryos and proliferation slowed to wild-type levels; however, there were still significantly more cells actively in mitosis (Fig. S5 A–C). These data suggests that in the absence of Tcf4 protein, there is excessive epithelial cell proliferation leading to exhaustion of the system by E16.5.
Fig. 3.
Loss of Tcf4 causes increased proliferation in the intestinal epithelium of Tcf4Null E13.5 embryos. (A) Membrane bound GFP marks Tcf4 lineage and reveals increased proportion of Tcf4 lineage in the epithelium of Tcf4Null E13.5 embryos. Yellow arrows mark red cells that have never expressed Cre and blue dotted line marks the endoderm/epithelium (B). Phh3 staining (blue) reveals increased numbers of mitotic cells in Tcf4Null E13.5 embryos. (C) BrdU staining (red) reveals increased numbers of cells in S phase of the cell cycle in Tcf4Null E13.5 embryos. Unequal variance two-tailed t test. Horizontal line, mean (scale bar, 50 μm).
Conditional Loss of Tcf4 Results in Enlarged Crypts but Is Not Sufficient for Colon Tumor Initiation.
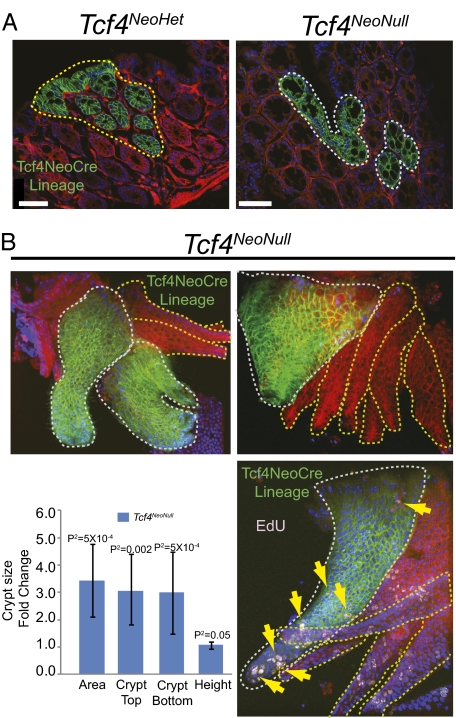
We next wanted to test whether complete loss of Tcf4 results in increased proliferation in the adult colon, a difficult question to address because Tcf4 loss in Tcf4 expressing cells is lethal. To address this question, we used a Tcf4CreNeo/lox line to conditionally delete Tcf4 in a subpopulation of Tcf4 expressing cells (Tcf4NeoNull), because the presence of Neo results in reduced numbers of Cre expressing cells throughout the animal, conveniently providing a hypomorphic allele (21). The Tcf4NeoNull animals are on average 21% smaller than their littermates (n = 5; P2 = 0.0001). In the colon of Tcf4CreNeo/+ (Tcf4NeoHet) adult mice, Cre expression is reduced to <1% of the total number of Tcf4-positive cells in the Tcf4Het in the absence of neo. The Tcf4 NeoCre lineage is dispersed throughout the length of the colon.
Dysplastic aberrant crypt foci (ACF) are generally accepted as the earliest CRC lesions. In Tcf4NeoNull mice, we analyzed colon tissues for the presence of dysplastic crypts or microadenomas by using Tcf4 NeoCre-dependent recombination of RosamTmG as a reporter of Tcf4Lox recombination. Recombined cells were not dysplastic and did not form detectable tumors (Fig. 4A). ACF are subcategorized as hyperplastic or dysplastic based on defining histological features (reviewed in ref. 22). One characteristic of ACF with hyperplasia is enlarged crypts. To directly determine whether Tcf4 loss results in increased proliferation and enlarged crypts, we isolated intact crypts from Tcf4NeoNull animals and compared Tcf4 NeoCre lineage-positive crypts (Fig. 4B; Tcf4 Null crypts, white lines) to adjacent lineage-negative crypts (Fig. 4B; Tcf4 Het Crypts; yellow lines). Consistent with ACF with hyperplasia, we found that Tcf4 Null crypts are enlarged compared with adjacent Tcf4 Het crypts (Fig. 4B).
Fig. 4.
Loss of Tcf4 results in hyperplasia without neoplasia. (A) Loss of Tcf4 in the colon Tcf4CreNeo/lox (Tcf4NeoNull) mice is not sufficient for tumor formation. mGFP marks crypts that are Tcf4Null; mTom marks crypts that are Tcf4Het. (A) Mice were collected between 4 mo and 1 y and did not develop colon tumors. (B) Tcf4NeoNull lineage-positive intact crypts are enlarged and maintain proliferative capacity as demonstrated by EdU labeling of crypts (yellow arrows). Dotted white lines encircle Tcf4Null crypts and dotted yellow lines encircle Tcf4Het crypts. P2-values calculated using paired two-sample t test for means (scale bar, 50 μm).
Our results in E13.5 embryos and adults suggest that loss of Tcf4 results in continued proliferation in the gut. The presence of Tcf4 NeoCre lineage-positive crypts in Tcf4NeoNull animals demonstrates that cells that do not express Tcf4 are still capable of proliferation; otherwise, the Tcf4 NeoCre lineage would not persist. We further tested proliferation in Tcf4NeoNull animals by labeling proliferating cells with the thymidine analog EdU (Invitrogen). We find that EdU is detected in Tcf4 Null crypts isolated from Tcf4NeoNull mice collected 18 h after EdU injection (Fig. 4B, yellow arrows). This demonstrates that cells lacking Tcf4 are still actively proliferating. Enlarged crypts coupled with the lack of tumors in Tcf4NeoNull mice demonstrate that Tcf4 loss is sufficient for hyperplasia, but not for neoplasia at an appreciable rate, suggesting that additional genetic lesions are necessary for tumor formation.
Tcf4 Haploinsufficiency Promotes Colon Tumor Formation in Apc Mutant Mice.
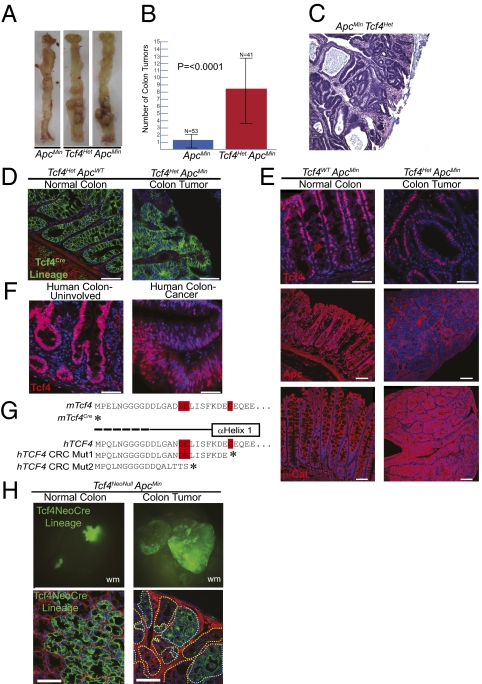
Given the increased proliferation of intestinal epithelial cells in Tcf4 mutants, we examined intestinal tumor formation in Tcf4Het ApcMin mice. Surprisingly, whereas ApcMin mice rarely develop colorectal tumors, Tcf4HetApcMin mice develop an average of 8.3 colon tumors (±5.0) (Fig. 5 A and B) per mouse. There is no statistical difference in the number of small intestinal tumors in Tcf4Het ApcMin mice compared with Tcf4WT ApcMin mice (Fig. S6). Histology revealed that the tumors in Tcf4HetApcMin mice are tubular adenomas, consistent with what is seen in the colons of FAP patients (Fig. 5C). These data support a role for Tcf4 as a tumor suppressor.
Fig. 5.
Tcf4 is a tumor suppressor in normal colonic epithelium. (A and B) Heterozygous loss of Tcf4 causes increased colon tumor formation in Tcf4Het ApcMin mice compared with Tcf4WTApcMin control mice. (C) Hematoxylin and eosin staining of colonic adenoma isolated from Tcf4Het ApcMin mice. (D) Tcf4 lineage analysis in cryosections of colon and adenoma tissue. (E) In colon tumors, Tcf4 antibody staining of tissue shows Tcf4 protein, Apc antibody staining demonstrates loss of heterozygosity of Apc and nuclear β-catenin antibody staining shows β-Catenin activation in colon tumors. (F) Tcf4 antibody staining of paraffin embedded grossly uninvolved human colon and human colon cancer tissue. (G) Heterozygous Tcf4 nonsense Exon 1 truncations have been found in genome-wide sequencing analysis of primary human CRC (12). These mutations are very similar to those created in targeted mTcf4Cre and mTcf4Lox alleles. (H) Tcf4 is not essential for tumorigenesis. Whole mount (wm) and sections of normal colonic epithelium and colon adenomas demonstrate Tcf4 lineage in the absence of Tcf4 in Tcf4NeoNull ApcMin mice. Dotted white lines encircle Tcf4Null tumor cells and dotted yellow lines encircle Tcf4Het tumor cells of a colon adenoma (scale bar, 50 μm).
Tcf4 lineage is found in normal colonic mucosa in the regions where proliferative progenitors and differentiated cells reside (Fig. 5D). However, Tcf4 protein is expressed at very low levels in proliferative cells at the base of the crypt and at much higher levels in differentiated cells (Figs. 1 and 5E (15). In mouse colon tumors, Tcf4 lineage is apparent in cells throughout the epithelial component and is absent from the stromal component (Fig. 5D). Immunohistochemistry of Tcf4 in mouse and human colon tumors indicates similar levels of Tcf4 expression in colon epithelium and tumors (Fig. 5 E and F), suggesting the presence of (at least) one Tcf4 allele in tumors.
Using an antibody to the carboxyl-terminus of Apc, we found that the second allele of Apc is lost in colon tumors, consistent with loss of APC heterozygosity found in FAP and sporadic colon cancer (Fig. 5E). β-Catenin is often found to be nuclear (active) in late human adenomas with significant dysplasia and adenocarcinomas (23). Colon tumors isolated from Tcf4HetApcMin mice exhibit significant dysplasia and are possibly late adenomas. We examined the nuclear accumulation of β-catenin protein in colon tumors and found it throughout the epithelial cell in nontumor tissue and increased in the nucleus in colon tumors isolated from Tcf4HetApcMin mice (Fig. 5E).
Tcf4Het ApcMin mice are an in vivo model for Tcf4 mutations that recently have been identified in genome-wide sequencing of human CRC samples (12, 13). Mutations in TCF7L2 have been found in human CRCs, including heterozygous nonsense or frameshift truncating mutations in exon 1 (Fig. 5G). The 17–24 amino acid amino-terminal fragments lack residues important for interaction with β-catenin and this region is disordered in the hTcf4/β-catenin cocrystal structure (Fig. 5G) (13, 24).
The presence of one intact Tcf4 allele in human CRC raises the question of whether some remaining Tcf4 protein is necessary for tumorigenesis. To test whether complete loss of Tcf4 inhibits colon tumorigenesis, we again used our Tcf4NeoNull allele combined with the ApcMin allele to determine whether colon tumors that form when Tcf4 is heterozygous persist when Tcf4 is conditionally deleted. We found regions in these mice that were Tcf4 Null that appeared normal and did not become adenomas, likely due to the remaining wild-type Apc allele (Fig. 5H). Further, we found polyclonal adenomas with distinct regions that were Tcf4 NeoCre lineage-positive (Tcf4 Null) and regions that were lineage-negative (Tcf4 Het), demonstrating that in the absence of Apc, functional Tcf4 was not essential for colon tumor growth (Fig. 5H).
Tcf4 and Tcf1/Lef1 Play Opposing Roles During Colon Tumorigenesis in Tcf4Het ApcMin Mice.
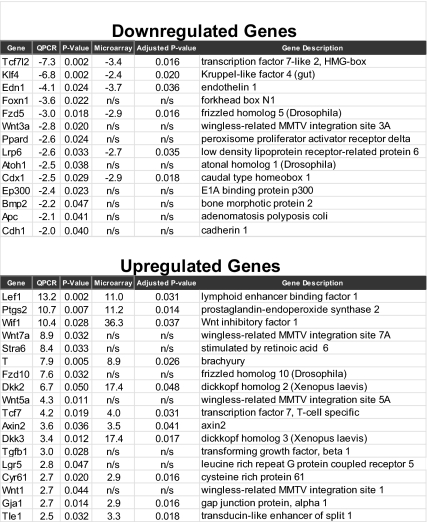
Previous studies have demonstrated that Tcf4 can act both as a transcriptional activator and a repressor (reviewed in ref. 25). It is possible that decreased Tcf4 expression results in the loss of repression and subsequent up-regulation of oncogenes essential for colon tumorigenesis. We used transcriptional profiling analysis to further characterize colon tumors found in Tcf4Het ApcMin mice and found differential expression of many Wnt signaling genes (Fig. 6). Because Tcf4 and Apc are components of the Wnt signaling pathway, we focused on validating by quantitative PCR (QPCR) the differential expression of Wnt signaling components and downstream targets of Wnt signaling in colon tumors compared with normal colon.
Fig. 6.
Lef1 and Tcf1 (Tcf7) are up-regulated in colon tumors. (A) Expression microarray and QPCR analysis of RNA isolated from the colons of Tcf4WTApcMin and Tcf4Het ApcMin littermates reveals misregulation of Wnt signaling components and downstream targets.
Tcf4 is down-regulated in colon tumors isolated from Tcf4HetApcMin mice and we found increased expression of Tcf1 and Lef1 (Fig. 6), whereas there was no significant change in Tcf3 expression. We also found many Tcf/Lef targets with differential expression in colon tumors isolated from Tcf4HetApcMin mice, and of these T-box transcription factor Brachyury (T), Axis inhibition protein 2 (Axin2), Cdx1, and Endothelin 1 (Edn1) have been shown to be directly occupied by Tcf4 using in vivo chromatin immunoprecipitation (26, 27). Therefore, in colon tumors, the increased expression of Lef1 and Tcf1 suggests that loss of Tcf4 activity combined with increased Tcf1 and Lef1 activities are important for the transcriptional changes of Tcf/Lef targets in colon tumors (Fig. 6 and Datasets S1 and S2).
Discussion
Using mouse knockout technology, we uncovered a previously undescribed role for Tcf4 in controlling epithelial cell proliferation in normal colon. Further, we showed that Tcf4 haploinsufficiency in combination with hyperactivation of the Wnt signaling pathway due to loss of Apc results in colon tumors. We suggest that Tcf4 normally functions as a tumor suppressor, mainly through modulating proliferation.
The intestinal phenotype we observed following the loss of Tcf4 is consistent with the loss of the proliferative compartment in Tcf4Hyg/Hyg mice (8). However, our Tcf4Null phenotype was more severe, because there was necrosis throughout the gut epithelium. The apparent disparity between Tcf4Hyg and Tcf4Null mouse phenotypes is likely a consequence of the region of the gene that was targeted. For the Tcf4Hyg allele, a selection marker (Hygromycin) was inserted into the HMG DNA binding domain (8). It has been suggested that a truncated Tcf4 isoform that lacks the DNA binding domain is expressed in targeted Tcf4Hyg mice (28). In support of this possibility, a semifunctional Tcf4 isoform that lacks the HMG domain and regions C-terminal to the domain has been found in the developing pituitary (29). To circumvent the expression of such an alternate isoform, our strategy was to target exon 1 of the Tcf4 locus, to remove the splice site, and add a strong poly(A) signal. If transcription occurs from an alternate promoter, the resulting Tcf4 isoform would lack the β-catenin binding domain and would be expected to be a constitutive repressor (ΔNTcf4) (25). The absence of any evidence of constitutive repressor activity and the lack of Tcf4 protein in the small intestine and colon of our Tcf4Null mice suggests that the Tcf4Cre and Tcf4Δ alleles reflect a complete loss of Tcf4 function. Additionally, the Tcf4Null and Tcf4NeoNull alleles provided added parameters that allowed us to follow the Tcf4 mutant lineage during development and in adult colon. This enabled us to refine the role of Tcf4 during development, intestinal maintenance, and tumorigenesis.
Normally, between E13.5 and E18.5, the mouse gut endoderm is highly proliferative, populating the epithelial mucosa while the intestinal tract grows both in circumference and length. In wild-type E13.5 embryos, the Tcf4 lineage-negative endoderm begins to develop into Tcf4 lineage-positive epithelial cells. However, in Tcf4Null E13.5 embryos, this induction that normally takes place over multiple days is rapidly completed, as shown by the replacement of Tcf4 lineage-negative endodermal cells by Tcf4 lineage-positive epithelial cells. These cells rapidly proliferate, as shown by BrdU and phosphorylated histone H3 staining compared with controls. Therefore, cells turn on Tcf4 Cre, become Tcf4 null, and continue to proliferate. At birth, there is a catastrophic loss of proliferative cells and differentiated cell types throughout the gut of Tcf4Null mice. The loss of epithelium is similar to the intestinal epithelial cell depletion that follows conditional Apc loss in Apcfl/fl mice (17). However, whether loss of Tcf4 directly affects differentiation or whether continued proliferation occurs at the expense of differentiation remains to be determined. We found that adult colonic crypts lacking Tcf4 were enlarged compared with adjacent heterozygous crypts and that there were proliferative and differentiated cell types present, suggesting that at least some proliferative cells are capable of differentiating. However, we propose that the increase in crypt size is likely due to increases in progenitor cells and partially differentiated transit amplifying cells, because terminally differentiated cells do not normally remain in the crypt; instead, they migrate and are ultimately shed into the lumen.
The ApcMin mouse has been useful for studying tumorigenesis in the small intestine, and several modifiers have been found that attenuate or enhance the Apc mutant phenotype in the mouse colon (reviewed in ref. 30). We found that the loss of one copy of Tcf4 dramatically increased the number of colon tumors in ApcMin mice. This result was unexpected, because previous studies implicated Tcf4 in complex with β-catenin in regulating the transcription of growth promoting genes during intestinal development and CRC. However, our data are consistent with genome-wide colon cancer sequencing efforts that found Tcf4 mutated in these tumors (12, 13). Separately, Tcf4 RNAi-targeted CRC cell lines demonstrated an increase in cell growth potential, also suggesting a tumor suppressor role for Tcf4 in CRC (14).
There is an apparent paradox in colon tumorigenesis in our model system. In CRC, β-catenin is well established as an oncogene and its activities are thought to require Tcf4, because Tcf4 is the principle family member expressed in normal colon and colon cancer cell lines. Therefore, how does β-catenin still function in the absence of Tcf4? Indeed, in colon tumors isolated from Tcf4HetApcMin mice, β-catenin is nuclear, suggestive of its role in transactivation of genes involved in proliferation. We propose that Lef1 and Tcf1 have distinct functions from Tcf4, where Lef1 and Tcf1 are obligate partners in the oncogenic activities of β-catenin in colon tumorigenesis. This hypothesis is supported by the increased expression of Lef1 and Tcf1 in Tcf4HetApcMin colon tumors. Further, Lef1 has been shown to be overexpressed in primary human colorectal tumors (31, 32), and Tcf4 and Lef1 are differentially regulated during colon tumor progression (33). We hypothesize that Tcf/Lef family members are fine-tuned for maintaining the equilibrium between normal epithelial cell proliferation and terminal differentiation. Therefore, Tcf/Lef members may function in distinct and opposing roles rather than overlapping roles and are essential for maintaining normal epithelial cell turnover.
Although the Wnt/β-catenin pathway and Tcf/Lef family members have been intensely studied, our data refine the roles of Tcf4 during intestinal development and colon cancer. In addition to CRC and CCRCC, Tcf7L2 polymorphisms have been associated with diabetes, breast cancer, and gastric carcinoma (9, 10, 34). Understanding the role of Tcf4 in normal epithelial cell proliferation and tumor formation provides insights into the etiology of CRC and invaluable information essential for evaluating the role of Tcf4 in other human syndromes. Colon tumors that develop in ApcMinTcf4Cre mice are highly relevant to human disease and provide a unique colon cancer model for studying human CRC. Further, analysis of the molecular mechanisms that underlie colon tumor formation in these mice may reveal targets for therapeutic intervention.
Experimental Procedures
Targeted Mouse Line Production.
Tcf4Cre and Tcf4Δ alleles were generated using standard cloning techniques as described in ref. (35). Tcf4Cre and Tcf4Lox constructs were targeted in R1-45 and G456 mouse embryonic stem cell lines, respectively. Tcf4Lox-targeted mice were crossed with a ubiquitous Cre driver, HprtCre, to generate Tcf4Δ mice. All studies and procedures involving animal subjects were conducted in strict accordance with an animal protocol approved by the University of Utah Institutional Animal Care and Use committee.
Human and Mouse Tissue Immunofluorescence and Immunohistochemistry.
Immunofluorescence was performed on formalin-fixed human and mouse tissues that were paraffin-embedded or frozen using conventional methods. For isolation of intact crypts, colons were incubated with shaking Ca+2 Mg+2-free PBS containing 30 mM EDTA, followed by formalin fixation. Antigen retrieval was done as described (23). Primary antibodies were anti-β-catenin (R&D Systems), anti-Apc (Santa Cruz Biotechnology), anti-Tcf4 (Cell Signaling), anti-BrdU (Caltag Laboratories), Click-iT EdU Cell Proliferation Assay Kit (Invitrogen), anti-Ki-67 (BD Pharmingen), UEA-1 biotin, (Sigma Aldrich), and anti-Chromogranin A (Abcam). Fluorescent images were collected using a Leica TCS SP5 laser-scanning confocal microscope.
Cell Counting and Analysis of Crypt Size.
Using Image J, the percent Tcf4 lineage, Phh3-positive cells, and percent BrdU-positive area were analyzed following randomization of three embryos each of Tcf4Cre/+ or Tcf4Cre/Δ E13.5 and E14.5 embryos. Epithelial and mesenchymal cell area was normalized between fields of view. Phh3 (mitosis)-positive cells and BrdU (S-phase)-positive area were scored separately in two cell types and then combined for total intestinal mitotic cell count per total area or S-phase-positive area per total area. Two sample t tests assuming unequal variances were performed and normalized cell number (or area per total area), genotype specific mean, and two-tailed P-values were reported. Intact crypts were analyzed using confocal microscopy and collection of Z-stack images that were analyzed using Imaris software (IMARIS). Crypt size was determined using the 3-d image generated by Imaris imported into Image J.
Pathway-Specific QPCR Analysis.
For gene expression in tumors, RNA was isolated from the distal 2.5 cm of colon collected from four each of Tcf4Het, ApcMin, and Tcf4WTApcMin littermates. RNA was isolated using an RNAqueous RNA isolation kit (Ambion). cDNA was made from intact RNA using QScript (Quanta Biosciences). QPCR was conducted using either 96-well or 384-well StellARrays (Bar Harbor Biotechnology), including the Mouse Wnt Signaling and Targets of Wnt Signaling arrays, according to the manufacturer's instructions. Data were analyzed by Bar Harbor Biotechnology using Global Pattern Recognition software (Lonza) as previously published (36).
Gene Expression Genomic Array and Analysis.
RNA was labeled and used to probe the Agilent 44K feature mouse genome array (Table 1). Four normal and tumor samples were analyzed. Intensity data from Agilent one-color gene expression arrays was log-transformed (to log base 2) and quantile normalized (37). Data from microarray features with the same probe sequence were averaged together to yield a single normalized intensity value for each unique probe on each array. The normalized data set was loaded into GeneSifter (www.genesifter.net) for analysis. Mutant and wild-type samples were compared using a two-tailed t test. P-values were corrected (adjusted P-value) for multiple testing using the Benjamini-Hochberg procedure (38).
Supplementary Material
Acknowledgments
We thank W. Samowitz for histological tumor analysis, K. Boucher for statistical analysis, M. Hansen for technical assistance, and S. Wu for reagents. We acknowledge technical assistance in cell culture by C. Lenz and S. Barnett and by B. Dalley and B. Milash in the University of Utah Microarray core and Bioinformatics core facilities. Tcf4Cre mice were made in collaboration with G. Kardon, and the GFPCre construct was provided by B. Sauer (National Institute of Diabetes and Digestive and Kidney Diseases). We thank A. Boulet, D. Jones, A. Pozner, S. Wu, and M. Hockin for helpful discussions and critical reading of the manuscript. The project described was supported by National Cancer Institute Grants CA073992 and CA128891. The work was also supported by access to technical cores supported by Cancer Center Support Grant CA042014.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE27522).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102300108/-/DCSupplemental.
References
- 1.Goss KH, Groden J. Biology of the adenomatous polyposis coli tumor suppressor. J Clin Oncol. 2000;18:1967–1979. doi: 10.1200/JCO.2000.18.9.1967. [DOI] [PubMed] [Google Scholar]
- 2.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 3.Morin PJ. beta-Catenin signaling and cancer. Bioessays. 1999;21:1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 4.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 5.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 6.Hurlstone A, Clevers H. T-cell factors: turn-ons and turn-offs. EMBO J. 2002;21:2303–2311. doi: 10.1093/emboj/21.10.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shitashige M, Hirohashi S, Yamada T. Wnt signaling inside the nucleus. Cancer Sci. 2008;99:631–637. doi: 10.1111/j.1349-7006.2007.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korinek V, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 9.Burwinkel B, et al. Transcription factor 7-like 2 (TCF7L2) variant is associated with familial breast cancer risk: a case-control study. BMC Cancer. 2006;6:268. doi: 10.1186/1471-2407-6-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim MS, Kim SS, Ahn CH, Yoo NJ, Lee SH. Frameshift mutations of Wnt pathway genes AXIN2 and TCF7L2 in gastric carcinomas with high microsatellite instability. Hum Pathol. 2009;40:58–64. doi: 10.1016/j.humpath.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Kojima T, et al. FOXO1 and TCF7L2 genes involved in metastasis and poor prognosis in clear cell renal cell carcinoma. Genes Chromosomes Cancer. 2011;49:379–389. doi: 10.1002/gcc.20750. [DOI] [PubMed] [Google Scholar]
- 12.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 13.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 14.Tang W, et al. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci USA. 2008;105:9697–9702. doi: 10.1073/pnas.0804709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregorieff A, et al. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Chan CW, et al. Gastrointestinal differentiation marker Cytokeratin 20 is regulated by homeobox gene CDX1. Proc Natl Acad Sci USA. 2009;106:1936–1941. doi: 10.1073/pnas.0812904106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sansom OJ, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 19.Mathew SJ, et al. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development. 2011;138:371–384. doi: 10.1242/dev.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olkowski AA, Wojnarowicz C, Chirino-Trejo M, Laarveld B, Sawicki G. Sub-clinical necrotic enteritis in broiler chickens: Novel etiological consideration based on ultra-structural and molecular changes in the intestinal tissue. Res Vet Sci. 2008;85:543–553. doi: 10.1016/j.rvsc.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Frank DU, et al. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng L, Lai MD. Aberrant crypt foci as microscopic precursors of colorectal cancer. World J Gastroenterol. 2003;9:2642–2649. doi: 10.3748/wjg.v9.i12.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phelps RA, et al. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137:623–634. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham TA, Ferkey DM, Mao F, Kimelman D, Xu W. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat Struct Biol. 2001;8:1048–1052. doi: 10.1038/nsb718. [DOI] [PubMed] [Google Scholar]
- 25.Hoppler S, Kavanagh CL. Wnt signalling: variety at the core. J Cell Sci. 2007;120:385–393. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- 26.Kim TH, Xiong H, Zhang Z, Ren B. beta-Catenin activates the growth factor endothelin-1 in colon cancer cells. Oncogene. 2005;24:597–604. doi: 10.1038/sj.onc.1208237. [DOI] [PubMed] [Google Scholar]
- 27.Wohrle S, Wallmen B, Hecht A. Differential control of Wnt target genes involves epigenetic mechanisms and selective promoter occupancy by T-cell factors. Mol Cell Biol. 2007;27:8164–8177. doi: 10.1128/MCB.00555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brinkmeier ML, et al. TCF and Groucho-related genes influence pituitary growth and development. Mol Endocrinol. 2003;17:2152–2161. doi: 10.1210/me.2003-0225. [DOI] [PubMed] [Google Scholar]
- 29.Kennell JA, O'Leary EE, Gummow BM, Hammer GD, MacDougald OA. T-cell factor 4N (TCF-4N), a novel isoform of mouse TCF-4, synergizes with beta-catenin to coactivate C/EBPalpha and steroidogenic factor 1 transcription factors. Mol Cell Biol. 2003;23:5366–5375. doi: 10.1128/MCB.23.15.5366-5375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwong LN, Dove WF. APC and its modifiers in colon cancer. Adv Exp Med Biol. 2009;656:85–106. doi: 10.1007/978-1-4419-1145-2_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hovanes K, et al. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 32.Li TW, et al. Wnt activation and alternative promoter repression of LEF1 in colon cancer. Mol Cell Biol. 2006;26:5284–5299. doi: 10.1128/MCB.00105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kriegl L, et al. LEF-1 and TCF4 expression correlate inversely with survival in colorectal cancer. J Transl Med. 2010;8:123. doi: 10.1186/1479-5876-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grant SF, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 35.Wu S, Ying G, Wu Q, Capecchi MR. A protocol for constructing gene targeting vectors: generating knockout mice for the cadherin family and beyond. Nat Protoc. 2008;3:1056–1076. doi: 10.1038/nprot.2008.70. [DOI] [PubMed] [Google Scholar]
- 36.Ruggiero T, et al. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009;23:2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 37.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 38.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.