Abstract
High genetic diversity is a hallmark of the gastric pathogen Helicobacter pylori. We used 454 sequencing technology to perform whole-genome comparisons for five sets of H. pylori strains that had been sequentially cultured from four chronically infected Colombians (isolation intervals = 3–16 y) and one human volunteer experimentally infected with H. pylori as part of a vaccine trial. The four sets of genomes from Colombian H. pylori differed by 27–232 isolated SNPs and 16–441 imported clusters of polymorphisms resulting from recombination. Imports (mean length = 394 bp) were distributed nonrandomly over the chromosome and frequently occurred in groups, suggesting that H. pylori first takes up long DNA fragments, which subsequently become partially integrated in multiple shorter pieces. Imports were present at significantly increased frequency in members of the hop family of outer membrane gene paralogues, some of which are involved in bacterial adhesion, suggesting diversifying selection. No evidence of recombination and few other differences were identified in the strain pair from an infected volunteer, indicating that the H. pylori genome is stable in the absence of mixed infection. Among these few differences was an OFF/ON switch in the phase-variable adhesin gene hopZ, suggesting strong in vivo selection for this putative adhesin during early colonization.
Keywords: comparative genomics, mutation
More than one half of the human population is infected with Helicobacter pylori. Infection with this pathogen causes chronic gastritis, which can give rise to sequelae including peptic ulcers of stomach and duodenum, gastric atrophy, adenocarcinoma, and mucosa associated lymphoid tissue (MALT) lymphoma (1). Extensive allelic diversity and genetic variability are hallmarks of this species. They result from the combination of a high mutation rate (2) and frequent exchange of genetic material during mixed infections of one stomach with multiple H. pylori strains (3–7).
We have previously analyzed the genetic relationships between strains of H. pylori that had been sequentially isolated from the same patient at time intervals ranging from 3 mo to 10 y (8) using multilocus sequence analysis of 10 (4) and 78 (8) loci, mostly from housekeeping genes. Bayesian inference was used to estimate recombination and mutation rates and the length of DNA fragments exchanged through recombination (imports). However, this multilocus approach did not allow conclusions about the chromosomal distribution of import events or about the relative frequencies of imports in different categories of genes.
Here, we have used 454 pyrosequencing technology (9) to analyze the genomes of five sets of sequential isolates of H. pylori. In addition to four pairs of isolates from the earlier studies (with isolation intervals of 3 y), we also included recent follow-up isolates for two of the pairs that were obtained 16 y after the first isolates. Furthermore, we studied one pair of strains from a recent clinical trial where human volunteers were infected with H. pylori challenge strain BCS 100 (10) as part of the evaluation of a vaccine and H. pylori clones reisolated 3 mo later.
Whereas our earlier estimates of import size and frequency were confirmed by this analysis, the whole-genome comparisons showed that the short imports characteristic of H. pylori were frequently part of large clusters, suggesting that several individual imports resulted from the uptake of much larger DNA fragments by transformation and/or conjugation. Imports were frequent in genes of the hop family of paralogous outer membrane protein genes that encodes most known adhesins of H. pylori, providing strong evidence of in vivo selection shaping the adhesin repertoire. No import and very few mutations in the isolate cultured after infection of a human volunteer highlight the importance of mixed infections for the genetic diversification of H. pylori through recombination.
Results
Genome Comparisons Between Sequential H. pylori Isolates Obtained During Established Chronic Infection.
Four pairs of H. pylori strains isolated sequentially from patients living in Nariño, Colombia (NQ315/1712, NQ352/1701, NQ392/1707, and NQ367/1671) were selected from a collection of strain pairs whose genetic relationships were previously characterized by multilocus sequence analysis of 10 loci (4). These included two pairs with evidence of recombination at 1 of 10 loci (NQ315/1712 and NQ367/1671), one pair with recombination at 5 of 10 loci (NQ352/1701), and one pair without differences in the 10 gene fragments (NQ392/1707).
Genomic sequence analysis by 454 technology was performed for these H. pylori isolates with >20-fold coverage. The resulting contigs were assembled in a set-wise fashion and subsequently arranged as virtual genomes using the complete genome sequence of H. pylori strain J99 (11) as a scaffold (Table 1 and SI Appendix, Fig. S1 and Table S1).
Table 1.
SNPs and CNPs in paired sequential genomes of H. pylori
| NQ315/NQ1712 | NQ352/NQ1701 | NQ392/NQ1707 | NQ392/NQ4060 | NQ367/NQ1671 | NQ367/NQ4191 | |
| Length of combined contigs (bp) | 1,601,938/ 1,572,582* | 1,639,151/ 1,639,623 | 1,650,953/ 1,650,485 | 1,650,953/ 1,649,736 | 1,622,411/ 1,626,950 | 1,622,411/ 1,624,247 + 4,818† |
| Time distance between isolates | 3 y | 3 y | 3 y | 16 y | 3 y | 16 y |
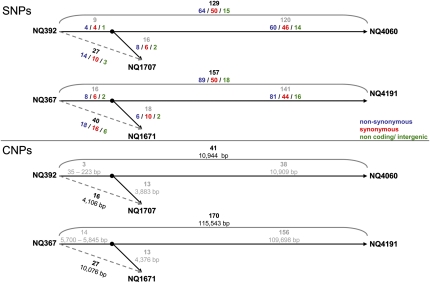
| No. of SNPs | 114 | 232 | 27 | 129 | 40 | 157 |
| (synonymous/non-synonymous/intergenic) | 50/55/10 | 117/97/18 | 10/14/3 | 50/64/15 | 16/18/6 | 50/89/18 |
| No. of CNPs | 121 | 441 | 16 | 41 | 27 | 170 |
| Median length of CNPs (bp) | 295 | 430 | 94 | 151 | 235 | 395 |
| Mean length of CNPs (bp) | 541 | 805 | 257 | 267 | 373 | 680 |
| Length range of CNPs (bp) | 2--3,748 | 2--6,978 | 3--1,887 | 3--3,131 | 2--1,837 | 2--3,942 |
| Total combined length of CNPs (bp) | 65,420 | 355,170 | 4,106 | 10,944 | 10,076 | 115,543 |
*The 29.5 kb difference is consistent with complete deletion of the cag pathogenicity island in NQ1712 (13).
†Isolate NQ4191 harbours a plasmid.
As predicted from the multilocus analysis results, the sets of sequential genomes were highly related with long stretches of complete sequence identity, which are extremely unlikely to occur between unrelated H. pylori isolates (4). We next analyzed differences between the four pairs of genomes with a 3-y interval and determined the number of isolated SNPs (separated from the next sequence difference by >200 bp on both sides) and clusters of nucleotide polymorphisms (CNPs; two or more sequence differences separated by <200 bp and flanked by >200 bp identical sequence on both sides). SNPs are most likely caused by mutations, whereas CNPs are most likely the result of the replacement of fragments of the recipient chromosome by DNA from a different H. pylori strain through homeologous recombination during mixed infection (imports). The pairs NQ392/1707 and NQ367/1671 differed by 27 and 40 SNPs and 16 and 27 CNPs, respectively (SI Appendix, Tables S2, S3, S4, and S5). The other two pairs showed more extensive differences, with 114 SNPs and 121 CNPs for the NQ315/1712 pair and 232 SNPs and 441 CNPs for NQ352/1701 (SI Appendix, Tables S6, S7, S8, and S9). The median lengths of CNPs ranged from 94 (NQ392/1707) to 430 bp (NQ352/1701), with a wide variation between the minimal length of two or three bp in all sets of genomes and a maximum length ranging from 1,837 (NQ367/1671) to 6,978 bp (NQ352/1701) (Table 1 and SI Appendix, Fig. S2). The combined length of CNPs also showed a wide variation between a minimum of ~4 kb for the NQ392/1707 pair and a maximum of ~356 kb for NQ352/1701 (Table 1).
Clockwise Accumulation of Diversity over 16 y of Persistent Colonization.
The interval between the isolations of first and follow-up isolate described above was 3 y. To study evolution further in time, we cultured additional follow-up isolates from endoscopic biopsies taken 16 y after the initial samples, which were available from two of four individuals. The new isolates were first compared with the earlier strains by multilocus sequence analysis and were identical to the second isolates in all seven fragments (NQ1707/4060 and NQ1671/4191). We then sequenced the genomes of these additional isolates. The 16-y isolates differed from the initial isolates by far more SNPs and CNPs than the 3-y isolates, indicating that diversity caused by mutation and recombination has accumulated over time (Fig. 1 and SI Appendix, Tables S2, S3, S10, and S11). Although many SNPs and CNPs differing between initial and 3-y isolates were also found in the 16-y isolates, consistent with a sequential acquisition of changes, this was not the case for all differences. This suggested that the 16-y isolate was not directly derived from the 3-y isolate but that both had descended from a common ancestor, consistent with at least transient coexistence of variant lineages within one stomach.
Fig. 1.
Sequential accumulation of isolated SNPs and clusters of nucleotide polymorphisms in two triplets of sequential isolates, NQ392/1707/4060 and NQ367/1671/4191, cultured after 0, 3, and 16 y of chronic infection. Black numbers indicate the numbers of SNPs or CNPs in pair-wise genome comparisons. Because there was only a partial overlap of differences for the two pair-wise comparisons, a common ancestor of both follow-up isolates was inferred (black dot) based on the overlapping differences, and gray numbers indicate differences between the sequenced isolates and the inferred intermediate clone. Overlapping and nonoverlapping events of the second isolates do not always add up to the total number, because there are cases where the presence or absence of the event in the third isolate could not be determined.
Model-Based Analysis of Recombination over the Chromosome.
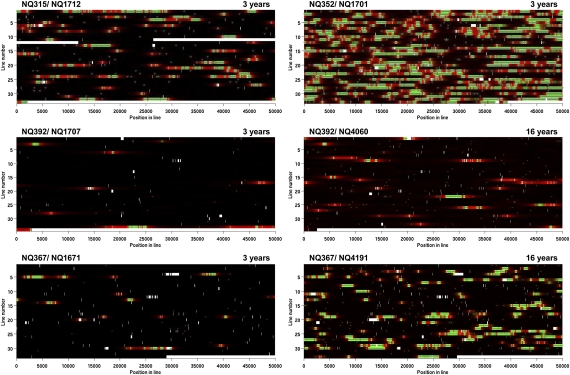
A visual inspection of the aligned virtual genomes immediately suggested that CNPs were not distributed randomly over the chromosome but instead, frequently occurred close to each other. Likewise, the imports detected when using ClonalFrame (12) corresponded roughly to the CNPs and were mostly arranged in groups (SI Appendix, Fig. S3). We, therefore, developed a statistical model of the recombination process, which used a Hidden Markov Model to account for the possibility of imports happening in groups (SI Appendix, Fig. S4). The results of performing Bayesian inference under that model for each of the four sets of isolates are shown in Fig. 2, Table 2 and an extended Table S12 including the 95% credibility intervals.
Fig. 2.
Analysis of the aligned genomes of sequential H. pylori isolate pairs. Each line represents 50 kb of sequence. Nonaligned regions are shown in white. Regions colored in black are clonal, regions in green are imported, and regions in red are not imported but in a cluster of imports. The intensity of each color represents statistical uncertainty. Polymorphism between the paired genomes is shown by gray triangles.
Table 2.
Mean estimates from the Bayesian analyses of the paired H. pylori isolates obtained sequentially during chronic infection
| NQ315/ NQ1712 | NQ352/ NQ1701 | NQ392/ NQ1707 | NQ392/ NQ4060 | NQ367/ NQ1671 | NQ367/ NQ4191 | |
| Length of imports (bp) | 402 | 629 | 283 | 286 | 261 | 519 |
| Length of groups of imports (bp) | 3,362 | 3,686 | 7,922 | 10,088 | 2,599 | 3,359 |
| No. of imports per group | 2.41 | 2.45 | 2.6 | 3.1 | 2.54 | 2.69 |
| Distance between two imports in a group (bp) | 700 | 621 | 1,933 | 2,207 | 542 | 531 |
| Distance between two groups of imports (bp) | 19,438 | 3,157 | 13,4916 | 30,973 | 72,170 | 8,469 |
| Mutation rate* | 2.4 | 6.5 | 0.5 | 0.5 | 0.7 | 0.7 |
| Import rate† | 4.0 | 16.5 | 0.5 | 0.2 | 1.0 | 1.0 |
| Ratio of rates of imports and mutations | 1.7 | 2.5 | 1.2 | 0.4 | 1.4 | 1.5 |
| Ratio of effects of imports and mutations | 26.7 | 62.0 | 12.4 | 4.3 | 13.7 | 29.5 |
*Per site, per year, ×10−5.
†Per initiation site, per year, ×10−5.
Calculated from the four 3-y pairs, the imports had an average length of 394 bp, which is in agreement with our previous estimate based on multilocus data (4). As expected, the imports were significantly grouped, with an average number of 2.5 imports per group (Table 2). The mean distance between two imports within a group was 949 bp, resulting in a mean length of the groups of imports of 4.4 kb (Table 2).
The ratio of the rates of import and mutation varied between the 3-y pairs, ranging from 1.2 to 2.5 (Table 2). Recombination was, however, responsible for the majority of differences in all isolate pairs, with ~12 to ~62 times more substitutions of nucleotides induced by imports than point mutations.
Increased Frequency of Imports in Genes Encoding Outer Membrane Proteins.
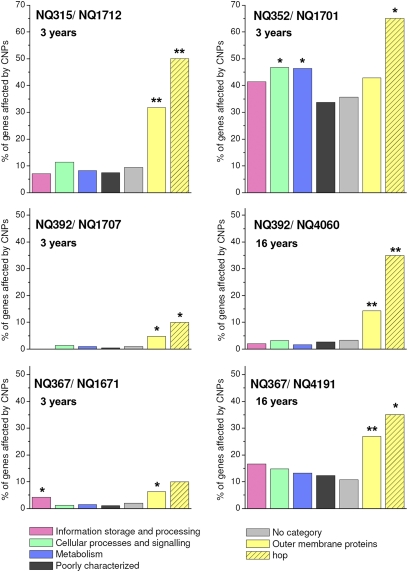
We next asked if imports affected genes coding for different functional classes of proteins with comparable frequencies. In all four sets of isolates, one or multiple functional gene categories showed significantly increased import frequencies (Fig. 3 and SI Appendix, Table S13). The most notable group of genes in this context was the large family of genes encoding outer membrane proteins (13), which, in H. pylori, comprises a total of 64 genes. Outer membrane protein-encoding genes had a significantly increased import frequency in three of four 3-y pairs. This difference became even more pronounced where 16-y follow-up isolates were available (Fig. 3 and SI Appendix, Table S13). Among outer membrane proteins, the hop subfamily (21 proteins) is of particular interest in H. pylori, because it comprises the known adhesion-related genes babA (14), sabA (15), hopZ (16), alpA, and alpB (17). In all sets of sequential isolates, the frequency of imports was highly elevated in the hop family of genes, with up to 65% of hop genes affected by imports after 3 y. Other gene categories showed significantly elevated import frequencies in only one infected individual (for example, the genes involved in cell motility in strains NQ352/1701) (SI Appendix, Table S13).
Fig. 3.
Proportion of genes affected by imports for different functional classes of encoded proteins. Import frequency for a category is calculated as the percentage of genes affected by imports. Color-coding is shown below the graphs. Asterisks indicate significantly increased frequency of imports (Fisher exact test; *P < 0.05; **P < 0.01). The composition of the group of outer membrane proteins (yellow bars) and the hop subfamily (hatched yellow bars) is based on ref. 13. SI Appendix, Fig. S6 has an extended graph including additional outer membrane protein subfamilies.
Genome Comparison Between Challenge Strain BCS 100-H1 and a Reisolate Obtained from an Experimentally Infected Volunteer.
The four sets of strains described above were sampled from adults who had most likely been infected since their childhood. We were, however, also interested in studying the evolution of the H. pylori genome in the first months after infection. We obtained H. pylori strain BCS 100-H1, an isolate lacking a functional cag pathogenicity island that has been used in infection studies in human volunteers, and strain 8A3, a reisolate that was cultured from a biopsy taken from an infected volunteer from the nonvaccinated placebo arm of the trial (18) 3 mo after inoculation. Whole-genome sequence comparison revealed very few differences between the two isolates. Three point mutations were identified, each of which was confirmed by Sanger sequencing (Table 3). All three were nonsynonymous mutations, leading to predicted single amino acid changes in the proteins δ-1-pyrroline-5-carboxylate dehydrogenase (PutA; HP0056), pyridoxal phosphate biosynthetic protein J (PdxJ; HP1582), and HP1181 (a predicted multidrug efflux transporter).
Table 3.
SNPs and repeat length variation after experimental infection of a human volunteer
| HP0056 (putA) | HP1181 | HP1582 (pdxJ) | HP0009 (hopZ) | HP0208 (rfaJ2) | Intergenic region between HP1321 and HP1322 | |
| Alleles | ||||||
| Allele in H1 | A | A | A | OFF (A) | ON (A) | A (5×) |
| Allele in 8A3 | B | B | B | ON (B) | OFF (B) | B (4×) |
| Distribution of alleles in additional single-colony isolates | ||||||
| I from input strain (A/B) | 15/0 | 14/1 | 14/1 | 15/0 | 14/1 | 15/0 |
| II from antrum (A/B) | 1/7 | 1/7 | 1/7 | 0/8 | 1/7 | 2/6 |
| III from corpus (A/B) | 5/4 | 5/4 | 4/5 | 1/8 | 4/5 | 7/2 |
In addition to point mutations, we noted repeat length differences (RLDs) in two different dinucleotide repeat sequences and one repeat consisting of multiple copies of an 8-bp motif. These RLDs were located in the outer membrane protein gene hopZ (HP0009) (16), an ORF encoding a putative glycosyl transferase, rfaJ2 (HP0208) (19), and one intergenic region (Table 3). hopZ contained a cytosine thymine (CT) × 6 repeat in the challenge strain H1 and a (CT) × 7 repeat in the reisolate 8A3, causing a frame shift and an early stop codon in hopZ of H1, whereas an intact hopZ gene is present in strain 8A3. By contrast, rfaJ2 is predicted to be active in H1 [(CT) × 7] and inactive in 8A3 because of an internal frame shift [(CT) × 8]. The functional significance of the intergenic RLD is unknown. Like the SNPs, these length changes were confirmed by Sanger sequencing.
Not a single CNP was identified in the H1/8A3 pair, suggesting that no recombination event had occurred during the experimental infection.
Diversity of Challenge Strain and Reisolates.
The genome sequencing of strains H1 and 8A3 was performed on bacteria that had been propagated from single colonies derived from the original inoculum (H1) or an antral biopsy taken from the volunteer 3 mo after inoculation (8A3). We sequenced the six variable loci from additional single-colony isolates that had been cultured from the input strain (15 isolates termed H2–H16) and two sets of antrum and corpus biopsies taken from volunteer 8 at the endoscopy 6 wk and 3 mo after inoculation (17 additional isolates) (Table 3). Two of three point mutations and one of the CT repeat variants identified in the reisolate 8A3 were also found in 1 (H3) of 16 single-colony isolates of the input strain, whereas one point mutation (in putA), one dinucleotide repeat length variant (in hopZ), and the repeat length variant in the intergenic region between HP1321 and HP1322 were exclusively found in the reisolates (Table 3). All three point mutations identified in 8A3 were detected in seven of eight of the additional clones from the antral biopsies but only in four of nine (putA and HP1181) or five of nine (pdxJ) clones from the corpus biopsies.
Discussion
Previous studies of genetic changes during chronic infection with H. pylori were restricted to selected genes, most of which had housekeeping functions (4, 8). Although this approach has permitted us to estimate rates of recombination, mutation, and size of imported fragments, it did not reveal the distribution of genetic changes on the chromosome, and their frequencies in different functional gene categories. Here, the genomes of 12 H. pylori isolates were sequenced, including four pairs of sequential isolates from Colombia from earlier studies using multilocus sequencing and comparative genome hybridizations (4, 20), two newly obtained follow-up isolates after 16 y of chronic infection, and one pair of strains consisting of the challenge strain (BCS 100) of a recent vaccine trial (18) and a reisolate (8A3) from a human volunteer (nonvaccinated control group) who had been infected with BCS 100 for 3 mo.
Mutation and Recombination Rates.
The average genome-wide mutation rate for the four 3-y pairs of sequential isolates from chronically infected individuals was 2.5 × 10−5 (range = 0.5–6.5 × 10−5) per year and site. This rate is ~18-fold faster than the mutation rate recently calculated for serial H. pylori isolates based on extended multilocus analysis of 78 housekeeping genes (8). The rate of recombination was 5.5 × 10−5 recombination events per initiation site and year, 122-fold higher than the rate of 4.4 × 10−7 calculated from housekeeping genes. We attribute these differences to multiple factors. Faster rates can be expected for a genome-wide analysis, because housekeeping genes are likely to be under strong purifying selection, whereas the genome-wide analysis comprises noncoding DNA as well as genes under diversifying selection. In addition, the rates of mutation and recombination varied strongly between different infected individuals in both the current (Table 2) and the earlier study, which could be because of strain properties (as shown in vitro) (21), the extent of mixed infections that determines the availability of exogenous DNA, or varying selective forces in infected hosts. Finally, we note that two of four pairs were selected because of their high rate of recombination, which introduced a bias to a higher recombination rate (2).
As described previously (20), a complete deletion of the cag pathogenicity island was observed in the follow-up isolate of strain NQ315 (Fig. 2). In the other sets, few changes were observed in the cagPAI region of the chromosome. All four chronically infected individuals displayed evidence of intestinal metaplasia in their initial gastric biopsies. Of these, two individuals (NQ315 and NQ392) progressed to dysplasia during the first 3 y of observation, whereas the pathology remained unchanged in the other two individuals (NQ352 and NQ367). There was, thus, no obvious correlation between the extent of genomic evolution of the bacteria and the gastric pathology of the infected individuals.
Patterns of Mosaicism.
The genome comparisons of four sets of Colombian isolates confirmed our previous estimates of the length of imported fragments. The estimated lengths of imports ranged from 261 to 629 bp, which is in excellent agreement with our previous estimate of 417 bp. Inspection of the genome alignments immediately revealed that the imports were not randomly distributed over the chromosome but often grouped, stretching over chromosomal regions of up to ~20 kb (median group length = 3,524 bp) (Table 2). Grouped imports could be the result of the uptake of a large DNA molecule into the cell and its subsequent partial integration into the chromosome in multiple small pieces. Similar patterns of chromosomal import were reported after in vitro transduction between strains of Escherichia coli (22). The mechanism generating these mosaics is unknown, but McKane and Milkman (22) have argued that restriction modification systems are likely involved; this is because back-transductions of the mosaic alleles into the original donor strain yielded transductants with uninterrupted imports. Alternatively, a large fragment might initially have been integrated as one piece and subsequently, been disrupted by integration of small pieces of DNA originating from other cells of the recipient strain that had not undergone recombination at this location. We did not, however, observe the acquisition of a large import with subsequent fragmentation in the data, and we note that grouped clusters were also observed in strain NQ392, despite a low rate of recombination; therefore, we currently consider the first hypothesis to be more likely. Lin et al. (23) have recently reported the rare (2 of 20 clones) occurrence of multiple imports separated by several hundred base pairs of recipient sequence after in vitro transformation of H. pylori with DNA from a Streptomycin-resistant donor. This observation is consistent with the grouped imports observed in our genome comparisons, but the mechanism by how these are generated remains unknown.
It is currently also unknown by which process the large DNA fragments enter the H. pylori cells. H. pylori is naturally competent for uptake of exogenous DNA from the environment, a process that depends on an unusual DNA uptake system, termed ComB, and on a ComEC-like protein. ComB is related to a type IV secretion system (24) and mediates uptake of DNA through the outer membrane (25) whereas the ComEC-like protein has been proposed to mediate transfer through the inner membrane (25, 26). We have previously reported that the mean length of imports after transformation in vitro varied between 1,294 and 3,853 bp depending on the combination of recipient and donor strain (21). The maximum length of DNA fragments that can pass this system has not been determined for H. pylori. Studies from Neisseria gonorrheae have suggested that chromosomal fragments of 11 and 13 kb could be imported, whereas fragments of 25 kb were never imported (27). Because the length of several of the groups of imports exceeded 13 kb and in one case, 20 kb (even in the NQ 315/1712, NQ392/1707, and NQ367/1671 pairs where imports are rare and therefore, unlikely to be erroneously pooled together), this suggests that the DNA uptake system of H. pylori could import very long fragments of DNA. Alternatively, conjugation could be responsible for transfer of large DNA fragments from one H. pylori cell to another. Conjugation has been reported to mediate the transfer of plasmids and chromosomal DNA between H. pylori cells (28, 29), but as yet, a role of conjugation in the evolution of H. pylori in vivo has not been documented.
Selection of Recombinants During Chronic Infection.
The family of paralogous genes encoding H. pylori outer membrane proteins (hop family) exhibited an increased import frequency in all sets of isolates. The most likely explanation for this finding is positive/diversifying selection, favoring the survival of recombinant clones with changes of the amino acid sequences (30). The Hop subfamily of outer membrane proteins (13, 31) contains multiple proteins with a role in adhesion of H. pylori to the human gastric epithelium. The blood group binding adhesins BabA (14) and SabA (15) are the best characterized of these, but a direct or indirect role in binding has also been shown for other members of the family, including HopZ (16), AlpA, and AlpB (17). Two genes only, jhp1103 and jhp1164, were affected by recombination in all four sets of genomes. Both of these are members of the hop family, suggesting particularly strong diversifying selection operating on these genes whose functions are not yet known.
Genetic Changes During Experimental Infection in a Human Volunteer.
All previous studies of sequential isolates, including those using 454 technology (32, 33), have been performed with bacteria isolated from chronically infected individuals. With the challenge strain BCS 100 and the reisolate 8A3, we could analyze a pair of isolates for which the time elapsed between the infection and reisolation was exactly known, allowing us to address the rate of genetic changes occurring in the early phase of H. pylori infection. In striking contrast to the pairs from Colombia, no single recombination event was detected, most likely because of the absence of coinfection. H. pylori prevalence has fallen in most Western countries, including Germany (where the study was conducted), reducing the risk of acquiring multiple strains. A recent study of two pairs of sequential H. pylori isolates from Sweden also did not detect any evidence of recombination during chronic infection (34). Two of three point mutations and one of three length changes in repeat sequences were already present as a minor population of the input strain mix, whereas one point mutation in HP0056 (putA) and the length change in hopZ were not detected in any of the 15 additional single colony-purified clones from the input strain, suggesting that these changes occurred de novo after infection.
The analysis of additional reisolates from the antrum and corpus of the same volunteer at the five polymorphic loci suggests that the antral environment has favored growth of 8A3-like bacteria, which contain the three point mutations in putA, pdxJ, and HP1181 as well as an inactive rfaJ2 and an active hopZ (Table 3). By contrast, strains reisolated from the corpus biopsies only showed such selection on one of the loci, hopZ, indicating that cells expressing the HopZ protein were strongly selected during early colonization of both antrum and corpus. The receptor to which HopZ binds is yet unknown, but a role of hopZ for colonization in vivo is in concordance with a recent report that deletion of hopZ reduced the fitness of H. pylori in a murine infection model (33).
Conclusion
Our study of H. pylori genomic evolution during human infection shows genome-wide recombination in H. pylori colonizing humans in Colombia, a high-prevalence area with a high rate of mixed infections. Whereas both the empirical and the model-based genome-wide analyses of the length of individual import events yielded values in good agreement with earlier estimates, two important additional findings have emerged: individual imports were often clustered and frequently affected genes coding for outer membrane proteins of the Hop family. By contrast, there was no evidence of recombination/mixed infection during 3 mo of infection of a volunteer in Germany, providing evidence that H. pylori can establish chronic infection after infection with a single strain and that its genome can be stable in the absence of mixed infection. In future studies, further insight into the population dynamics of H. pylori during infection can likely be gained by application of next generation sequencing to multiple isolates from different parts of the stomach and to isolates from individuals between whom H. pylori has been recently transmitted, such as families (36).
Materials and Methods
Bacteria.
The H. pylori strain sets NQ315/1712, NQ352/1701, NQ392/1707/4060, and NQ367/1671/4191 were isolated from sequential biopsies taken from four individuals in Nariño, Colombia in the course of a clinical trial as described previously (4, 35). The time interval between the isolation of the first isolate and the first follow-up isolates was 3 y. Additional isolates cultured from biopsies taken 16 y after the initial cultures were available for two individuals. H. pylori strain BCS 100 (BAA-945; ATCC) (10) had been used for an infection challenge of human volunteers that was part of the evaluation of vaccine candidates. The design and results of this trial have been previously reported (18); 16 single-colony isolates designated H1–H16 were prepared from the inoculum used to infect the volunteers to permit investigations into the homogeneity of the inoculum. This seemed particularly important, because BCS 100 was not propagated from a single colony but from multiple colonies grown from one infected individual (10) to avoid a potential loss of infectivity because of single-colony purification. The H. pylori reisolates investigated in this study were cultured from volunteer 8, who received placebo. After 6 wk and 3 mo of infection, antrum and corpus biopsies were taken from the stomach of this volunteer, H. pylori was cultivated, and nine single colonies were propagated and analyzed in this study. The reisolate 8A3 was propagated from the antrum biopsy 3 mo after infection.
SI Appendix, SI Materials and Methods has details on DNA preparation, 454 sequencing, PCR and Sanger sequencing, generation of virtual genomes, pair-wise analysis of sequential isolates, statistical model, Bayesian inference, and assignment of imports to functional gene categories.
Supplementary Material
Acknowledgments
We thank Birgit Brenneke, Friederike Kops, Jessika Schulze, and Stella Lamprecht for excellent technical assistance. Support was provided by the German Research Foundation as PhD stipends in the International Research Training Group 1273 (to L.K. and S.K.) as well as Grants SFB 900/A1 (to S.S.) and Ae16/4-4 (to T.A. and T.F.M.). Further support was provided by European Union 6th Research Framework Programme, Project Infections and Cancer (INCA) Grant LSHC-CT-2005-018704 (to T.F.M. and S.S.), and German Ministry for Education and Research ERA-NET PathoGenoMics Project HELDIVNET (to S.S.). The field work in Colombia was supported by National Cancer Institute, National Institutes of Health Grant P01CA028842 (to P.C.), and D.F. was supported by Science Foundation of Ireland Grant 05/FE1/B882 to Mark Achtman.
Footnotes
Conflict of interest statement: M.D. is an employee of Roche Diagnostics GmbH.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database as whole genome shotgun projects with 12 project IDs CADC00000000 to CADN00000000 in alphabetical order. Virtual genomes have been deposited online at http://www.mh-hannover.de/20611.html.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018444108/-/DCSupplemental.
References
- 1.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 2.Björkholm B, et al. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc Natl Acad Sci USA. 2001;98:14607–14612. doi: 10.1073/pnas.241517298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suerbaum S, et al. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falush D, et al. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: Estimates of clock rates, recombination size, and minimal age. Proc Natl Acad Sci USA. 2001;98:15056–15061. doi: 10.1073/pnas.251396098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kersulyte D, Chalkauskas H, Berg DE. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol Microbiol. 1999;31:31–43. doi: 10.1046/j.1365-2958.1999.01140.x. [DOI] [PubMed] [Google Scholar]
- 6.Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5:441–452. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- 7.Kang J, Blaser MJ. Bacterial populations as perfect gases: Genomic integrity and diversification tensions in Helicobacter pylori. Nat Rev Microbiol. 2006;4:826–836. doi: 10.1038/nrmicro1528. [DOI] [PubMed] [Google Scholar]
- 8.Morelli G, et al. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 2010;6:e1001036. doi: 10.1371/journal.pgen.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham DY, et al. Challenge model for Helicobacter pylori infection in human volunteers. Gut. 2004;53:1235–1243. doi: 10.1136/gut.2003.037499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alm RA, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 12.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alm RA, et al. Comparative genomics of Helicobacter pylori: Analysis of the outer membrane protein families. Infect Immun. 2000;68:4155–4168. doi: 10.1128/iai.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ilver D, et al. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 15.Mahdavi J, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325–3333. doi: 10.1093/nar/27.16.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odenbreit S, Till M, Hofreuter D, Faller G, Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol. 1999;31:1537–1548. doi: 10.1046/j.1365-2958.1999.01300.x. [DOI] [PubMed] [Google Scholar]
- 18.Aebischer T, et al. Correlation of T cell response and bacterial clearance in human volunteers challenged with Helicobacter pylori revealed by randomised controlled vaccination with Ty21a-based Salmonella vaccines. Gut. 2008;57:1065–1072. doi: 10.1136/gut.2007.145839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langdon R, Craig JE, Goldrick M, Houldsworth R, High NJ. Analysis of the role of HP0208, a phase-variable open reading frame, and its homologues HP1416 and HP0159 in the biosynthesis of Helicobacter pylori lipopolysaccharide. J Med Microbiol. 2005;54:697–706. doi: 10.1099/jmm.0.45842-0. [DOI] [PubMed] [Google Scholar]
- 20.Kraft C, et al. Genomic changes during chronic Helicobacter pylori infection. J Bacteriol. 2006;188:249–254. doi: 10.1128/JB.188.1.249-254.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulick S, et al. Mosaic DNA imports with interspersions of recipient sequence after natural transformation of Helicobacter pylori. PLoS One. 2008;3:e3797. doi: 10.1371/journal.pone.0003797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKane M, Milkman R. Transduction, restriction and recombination patterns in Escherichia coli. Genetics. 1995;139:35–43. doi: 10.1093/genetics/139.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin EA, et al. Natural transformation of Helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 2009;5:e1000337. doi: 10.1371/journal.ppat.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41:379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- 25.Stingl K, Muller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci USA. 2009;107:1184–1189. doi: 10.1073/pnas.0909955107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh YC, Lin TL, Chang KC, Wang JT. Characterization of a ComE3 homologue essential for DNA transformation in Helicobacter pylori. Infect Immun. 2003;71:5427–5431. doi: 10.1128/IAI.71.9.5427-5431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton HL, Dillard JP. Natural transformation of Neisseria gonorrhoeae: From DNA donation to homologous recombination. Mol Microbiol. 2006;59:376–385. doi: 10.1111/j.1365-2958.2005.04964.x. [DOI] [PubMed] [Google Scholar]
- 28.Backert S, Kwok T, König W. Conjugative plasmid DNA transfer in Helicobacter pylori mediated by chromosomally encoded relaxase and TraG-like proteins. Microbiology. 2005;151:3493–3503. doi: 10.1099/mic.0.28250-0. [DOI] [PubMed] [Google Scholar]
- 29.Fischer W, et al. Strain-specific genes of Helicobacter pylori: Genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 2010;38:6089–6101. doi: 10.1093/nar/gkq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith NH, Maynard Smith J, Spratt BG. Sequence evolution of the porB gene of Neisseria gonorrhoeae and Neisseria meningitidis: Evidence of positive Darwinian selection. Mol Biol Evol. 1995;12:363–370. doi: 10.1093/oxfordjournals.molbev.a040212. [DOI] [PubMed] [Google Scholar]
- 31.Tomb J-F, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 32.Giannakis M, Chen SL, Karam SM, Engstrand L, Gordon JI. Helicobacter pylori evolution during progression from chronic atrophic gastritis to gastric cancer and its impact on gastric stem cells. Proc Natl Acad Sci USA. 2008;105:4358–4363. doi: 10.1073/pnas.0800668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giannakis M, et al. Response of gastric epithelial progenitors to Helicobacter pylori Isolates obtained from Swedish patients with chronic atrophic gastritis. J Biol Chem. 2009;284:30383–30394. doi: 10.1074/jbc.M109.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundin A, et al. Slow genetic divergence of Helicobacter pylori strains during long-term colonization. Infect Immun. 2005;73:4818–4822. doi: 10.1128/IAI.73.8.4818-4822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Correa P, et al. Chemoprevention of gastric dysplasia: Randomized trial of antioxidant supplements and anti-Helicobacter pylori therapy. J Natl Cancer Inst. 2000;92:1881–1888. doi: 10.1093/jnci/92.23.1881. [DOI] [PubMed] [Google Scholar]
- 36.Schwarz S, et al. Horizontal versus familial transmission of Helicobacter pylori. PLoS Pathog. 2008;4:e1000180. doi: 10.1371/journal.ppat.1000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.