Abstract
hsa-mir-483 is located within intron 2 of the IGF2 gene. We have previously shown oncogenic features of miR-483-3p through cooperation with IGF2 or by independently targeting the proapoptotic gene BBC3/PUMA. Here we demonstrate that expression of miR-483 can be induced independently of IGF2 by the oncoprotein β-catenin through an interaction with the basic helix–loop–helix protein upstream stimulatory transcription factor 1. We also show that β-catenin itself is a target of miR-483-3p, triggering a negative regulatory loop that becomes ineffective in cells harboring an activating mutation of β-catenin. These results provide insights into the complex regulation of the IGF2/miR-483 locus, revealing players in the β-catenin pathway.
The multifunctional protein β-catenin is involved in cell–cell adhesion when it is localized to the cellular membrane (1), and in transcriptional regulation by translocation into the nucleus through the Wnt pathway (2). Wnt signaling is an important molecular pathway required for cellular differentiation, tissue homeostasis, and tissue morphogenesis. Wnt/β-catenin signaling is one of the most commonly activated pathways in cancer, and several Wnt signaling-related gene mutations have been described: adenomatous polyposis coli (APC) and protein phosphatase 2 regulatory subunit A (PPP2R1B) mutations in colorectal cancer (3), AXIN1 mutation in hepatocarcinoma (4), and WTX gene mutations in Wilms’ tumor (5), and β-catenin gene (CTNNB1) itself was shown to be mutated (6–8) in the amino-terminal region used for degradation by the GSK3β–APC–AXIN–WTX complex (5, 9). These mutations prevent β-catenin degradation and result in its accumulation in the nucleus, where it acts as a specific transcriptional coactivator of the DNA-binding T-cell factor/lymphoid enhancer factor protein family. Among the targets of this family are important genes involved in tumorigenesis such as MYC, CCND1, CJUN, and FRA1 (10, 11).
MicroRNAs (miRNAs) are small noncoding RNAs that modulate gene expression by base pairing to target messenger RNAs (mRNAs) and by inhibiting their translation and/or promoting their degradation (12). MicroRNAs play a critical role in the normal maintenance of fundamental cellular processes, and their deregulation in human neoplasm has been proven to affect a large number of molecular pathways related to cancer (13–15).
Because the miR-483 locus is dysregulated in tumors involving the β-catenin pathway (16–18), we investigated their possible connection.
Results
miR-483-3p Expression Correlates with the Mutational Status of Wnt/β-Catenin Genes in Hepatocarcinoma.
The Wnt/β-catenin pathway is one of the most important pathways dysregulated in hepatocarcinoma (HCC), colorectal cancer (CRC), and Wilms’ tumor (19–21). Because we previously found that miR-483-3p, which is located in intron 2 of the IGF2 gene, is up-regulated in these cancers as well, we investigated the possible involvement of Wnt/β-catenin in miR-483-3p dysregulation.
We previously found a positive coefficient of correlation (R) between IGF2 and miR-483-3p expression in HCC (R = 0.69, P < 0.0001), CRC (R = 0.86, P < 0.0001), and Wilms’ tumor (R = 0.9, P < 0.0001) (16), suggesting that miR-483-3p transcription occurs from the IGF2 promoter. Conversely, some tumor samples from HCC that have a low coefficient of correlation exhibited a divergent expression of IGF2 and miR-483-3p, suggesting alternative mechanisms regulating these two genes. We analyzed the mutational status of APC, CTNNB1, and AXIN1 in 24 HCC samples in which miR-483-3p and IGF2 expression had already been assessed (16). With an miR-483-3p expression cutoff level of 10-fold over the average expression of the controls, we detected an association between miR-483-3p up-regulation and the mutational status of these genes (P = 0.053; Fisher's exact test) (Table S1), whereas no association between IGF2 expression and the Wnt/β-catenin mutational status was found (expression cutoff = 10, P > 0.5; Fisher's exact test). These data suggest that β-catenin may be involved in the regulation of miR-483-3p separately from IGF2. To prove this point, we calculated the ratio between miR-483-3p and IGF2 expression levels (median value = 1.3) to identify samples in which they were divergent (Table S1). A strong association between the miR-483-3p/IGF2 ratio and the mutational status of the Wnt/β-catenin genes was observed when the ratio is greater than 5 (P = 0.015; Fisher's exact test). Overall, these data strongly suggest that expression of miR-483-3p, but not that of IGF2 is associated with the mutational status of the Wnt/β-catenin pathway.
miR-483 Locus Is Regulated by β-Catenin.
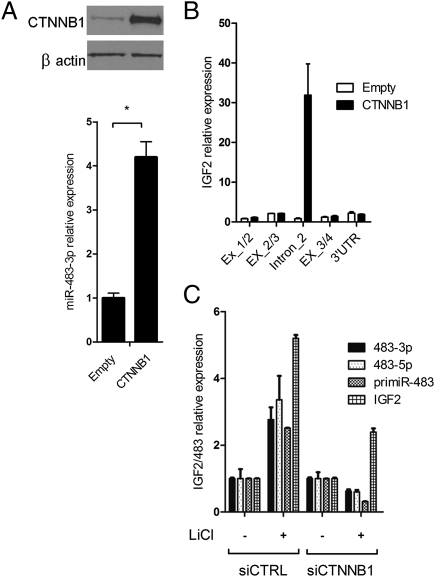
Because β-catenin is the principal transcriptional mediator of the Wnt/β-catenin pathway, we investigated its involvement in the regulation of the IGF2/miR-483 locus. We cloned the coding sequence of the β-catenin gene into the expression vector pIRES-Neo2. Then we assessed the expression level of miR-483-3p in response to β-catenin overexpression in HEK293 cells. A significant increase in miR-483-3p expression was detected in cells transfected with the pIRES-Neo2-β-catenin vector compared with cells transfected with the empty vector (Fig. 1A). To confirm these results, we transiently knocked down β-catenin by using short interfering RNA technology (siRNA) in HCT116 cells that exhibits a higher β-catenin nuclear activity, and miR-483-3p expression, compared with HEK293 cells. Quantitative real-time RT-PCR (qRT-PCR) verified a significant reduction of miR-483-3p expression only in cells transfected with β-catenin siRNA (Fig. S1B).
Fig. 1.
Induction of miR-483-3p expression by β-catenin. (A) miR-483-3p was induced by enforced expression of wild-type CTNNB1. Expression value was related to miRNA expression on empty vector transfected cells (2−ΔΔCt). The expression of CTNNB1 was assessed by Western blot (Upper). *P < 0.02. (B) Expression analyses across the IGF2 locus modulated by the enforced expression of β-catenin. Three PCR products were designed to amplify IGF2 exon junctions (Ex_1/2, Ex_2/3, and Ex_3/4) and one its 3′UTR. One set of primers was located within intron 2 and was used to assess the expression of miR-483 precursor (intron_2). qRT-PCR was carried out with SYBR Green technologies. RNA was previously treated with DNase to avoid genomic contamination. Black bars indicate the expression detected in cells transfected with the CTNNB1-expressing vector; white bars indicate the expression detected in cells transfected with the empty vector. (C) Expression analysis of miR-483-3p, miR-483-5p, pri-miR-483, and IGF2 genes assayed by qRT-PCR using TaqMan probes after LiCl treatment (20 mM for 24 h) with and without siRNA for CTNNB1 gene. IGF2 expression was still induced, although more weakly than the control. This IGF2 induction by LiCl has not been previously described, and is not necessarily due to β-catenin because of the large number of pathways affected by GSK3B inhibition.
Next, we investigated the effect of β-catenin on IGF2 expression. By qRT-PCR, we evaluated the expression of different DNA segments across the IGF2 gene locus using five sets of primers spanning the junctions between the IGF2 cDNA sequence and IGF2 intron 2. The only DNA segment whose transcription was induced by β-catenin was within the second intron of IGF2 that includes miR-483-3p (Fig. 1B). Thus, we concluded that β-catenin activates an miR-483-3p promoter inside the second intron of IGF2.
To further confirm these results, we stabilized β-catenin protein by treating HEK293 cells with lithium chloride (LiCl), an inhibitor of GSK3B which is responsible for β-catenin degradation. Fig. 1C shows that expression of the entire IGF2/miR-483 locus was significantly activated (three- to fivefold) by LiCl treatment. Conversely, treatment with β-catenin siRNA resulted in reduced expression only of miR-483 after LiCl treatment. Taken together, these results suggest that miR-483 locus expression can be driven by β-catenin independently from IGF2.
Zinc Finger CCCTC-Binding Factor CTCF Represses the Genomic Region Upstream of the miR-483 Locus.
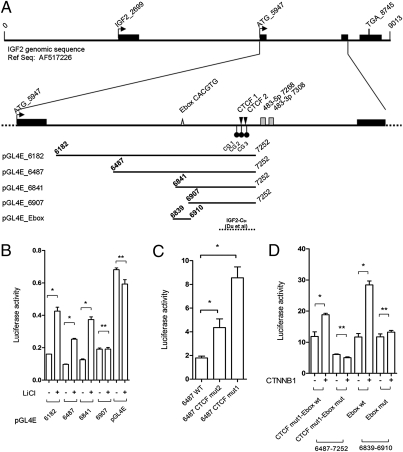
To explore the connection between β-catenin and expression of the miR-483 locus, we cloned four fragments of different lengths, including the putative miR-483 promoter (Fig. 2A) upstream of the luciferase gene, into the pGL4 enhancer vector (pGL4E). Fig. 2B shows LiCl treatment causes significant induction of luciferase activity for all fragments except the small clone pGL4E-6907 (Fig. 2B). Thus, we inferred that the genomic region responsive to LiCl treatment is located between positions 6841 and 6907 of the reference sequence (Gene Bank accession number AF517226).
Fig. 2.
Analysis of the miR-483 minimal promoter region responsive to LiCl/CTNNB1 stimuli. (A) Genomic structure of the IGF2/483 locus from the reference AF517226 genomic sequence. Exons (black bars), start (ATG_5947) and stop codons (TGA_8745), E-box elements (gray triangle), the two predicted CTCF binding sites (black triangle), three CpG dinucleotides around the CTCF binding sites (black circles), and miR-483-3p and miR-483-5p (gray boxes) are shown. The five genomic fragments cloned upstream of the luciferase reporter gene in pGL4E for the analysis of the promoter are indicated at the bottom of the panel. The genomic region with insulator activity studied by Du et al. (22) is indicated (broken line). (B) Luciferase activity of four genomic fragments cloned in pGL4E with and without LiCl treatment; the pGL4E empty vector was used as control. (C) Luciferase activity of the wild-type pGL4E-6487 (WT) and the mutated pGL4E-6487 in the predicted CTCF binding sites (CTCF mut1 and CTCF mut2). (D) Analysis of the wild-type E-box element (Ebox wt) and mutant (Ebox mut) by luciferase assay of the pGL4E-6487 CTCF mut1 and pGL4E_Ebox. Firefly luciferase activity was normalized on Renilla luciferase activity of the cotransfected pGL4R vector. *P < 0.02, **P > 0.02.
Because the luciferase activity in each of these vectors with or without LiCl treatment was always lower than the control (pGL4E empty vector), we suspected the presence of a repressive element within this region. Du et al. have shown that a 151-bp fragment (called IGF2-CBI by the authors) (Fig. 2A), immediately upstream of the miR-483 stem loop, has strong insulator activity and binds to the CTCF repressor (22). CTCF is an important methyl-sensitive regulator of transcription involved in the epigenetic regulation of genomic imprinted loci such as the IGF2/H19 locus in 11p15.5, and is involved in Wilms’ tumor (23, 24), breast cancer (25, 26), and prostate cancer (27). We decided to determine whether CTCF is also involved in the repression of the IGF2/mir-483 genomic regions we cloned. Using bioinformatics tools (http://insulatordb.uthsc.edu), we identified two possible CTCF binding sites (CTCF BS_1 and CTCF BS_2) (Fig. 2A). By transfecting the pGL4 vectors with mutated versions of either CTCF BS_2 or CTCF BS_1 to prevent CTCF binding, we observed a two- or fourfold increase of the luciferase activity compared with the wild-type control (Fig. 2C). Because the CTCF repressor only binds demethylated DNA, we analyzed the methylation status of three CpG dinucleotides close to the CTCF binding sites in a set of 14 cell lines. We found a significant positive correlation between the methylation level of the first CpG (CG_1; Fig. 2A) and miR-483-3p expression (R = 0.682, P = 0.007), whereas the correlation with each of the other two CpGs was less significant (Table S2). These data indicate that CTCF is an important regulator of the miR-483 locus and that this regulation is likely affected by DNA methylation.
Transcription Factor USF1 Serves as a Mediator Between β-Catenin and the miR-483 Locus.
The minimal genomic region responsive to LiCl treatment (between nucleotides 6841 and 6907) contains an E-box motif (CACGTG) that could bind the basic helix–loop–helix (bHLH) protein family. Because one of these bHLH proteins is the MYC transcription factor, a well-known target of the Wnt/β-catenin pathway, it is reasonable to speculate that MYC could be involved in the LiCl regulation of the miR-483 locus. To test this hypothesis, we mutated pGL4E-6487 in the E-box site and also mutated CTCF BS_1 to partially eliminate the repressive activity of this region. After cotransfection of β-catenin and the reporter vectors into HEK293 cells, there was a significant induction of luciferase activity in the wild-type but not in the E-box mutant clone (Fig. 2D). This confirmed the data obtained after LiCl treatment (Fig. 2B) and identified within the E-box motif the sequence responsive to the β-catenin/LiCl stimulation. Similar results were obtained with the vector clone containing only 69 bp around the E-box motif (pGL4E-6841–6910) (Fig. 2D).
Then we determined whether MYC is the β-catenin mediator for miR-483 transcriptional activation. Because MYC is well-expressed in HEK293 cells, we knocked down its expression using a specific siRNA and measured the relative expression of the precursor of miR-483 (pri-miR-483). Unexpectedly, this resulted in an increase in the expression of pri-miR-483 (Fig. S2 A and B). Similar results were obtained by a luciferase assay using the pGL4E-E-box reporter vector in cells cotransfected with siRNA for MYC. These data indicate that MYC is not responsible for the activation of the miR-483 locus after β-catenin activation because of its suggested repressive role on miR-483 transcription.
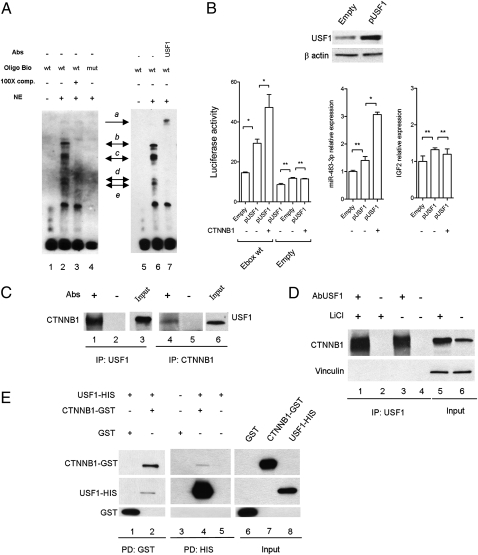
Because the E-box element can also bind to the upstream stimulating transcription factor 1 (USF1), we tested this interaction by electrophoretic mobility shift (EMSA) and supershift assays. USF1 is an evolutionarily well-conserved and ubiquitous transcription factor involved in a wide number of cellular activities such as immune response, cell cycle and proliferation, and lipid and glucose metabolism (28). As shown in Fig. 3A, the EMSA generates a specific band-shift pattern (complexes b–e, lane 2, Fig. 3A) that disappears in the mutant form of the E-box element oligonucleotide (lane 4, Fig. 3A). Moreover, by using the anti-USF1 antibody, a supershift complex was generated (complex a, lane 7, Fig. 3A), suggesting that USF1 recognizes the E-box element upstream of the miR-483 locus.
Fig. 3.
USF1 serves as a mediator between β-catenin and the miR-483 locus. (A) EMSA of nuclear extract (NE) from HEK293 cells using the miR-483 E-box probe (lanes 1–3 and 5–7) or the mutant form (lane 4). The specific complexes are indicated by black arrows (b–e). Lane 7 shows the supershift generated by USF1 antibody (complex a). (B) Luciferase assay of wild-type pGL4E_E box after enforced expression of USF1 and CTNNB1 using the pGL4E empty vector as control (Left). The center and right panels show the expression of miR-483-3p and IGF2 gene, respectively, after enforced expression of USF1 and CTNNB1. The miR-483-3p and IGF2 expression values of the empty vector were the controls. The exogenous expression of USF1 was assessed by Western blot (Upper). *P < 0.02, **P > 0.02. (C) Nuclear extract from HEK293 cells were immunoprecipitated with either USF1 (lanes 1–3) or CTNNB1 antibodies (lanes 4–6). After being washed, samples were run on an SDS/PAGE gel and transferred to nitrocellulose. The blots were probed with USF1 and CTNNB1 antibodies. (D) Nuclear extract from HEK293 cells with (lanes 1 and 2) and without (lanes 3 and 4) treatment with LiCl (20 mM for 24 h) was immunoprecipitated with USF1 antibody and the blot was probed for CTNNB1 protein. The input shows an incremented quantity of CTNNB1 after LiCl treatment (lanes 5 and 6). Vinculin protein expression was used as loading protein control. (E) GST-CTNNB1 and HIS-USF1 fusion proteins were subjected to GST (lanes 1 and 2) and HIS (lanes 3–5) pull-down analysis. Binding reaction products were washed, and proteins were separated by SDS/PAGE. The membrane was probed with anti-CTNNB1, anti-USF1, and GST antibodies.
To confirm this result, we cloned the coding sequence of the USF1 gene into the pCMV-Tag vector and cotransfected it into HEK293 cells along with pIRES-Neo β-catenin and the reporter vector pGL4E-6841–6910 with a wild-type or mutant E box (Fig. 3B Left). USF1 overexpression was able to induce luciferase activity (twofold) that was further increased in the presence of exogenous β-catenin (about threefold) compared with the control. Similar results obtained by qRT-PCR for miR-483-3p showed that miR-483-3p was weakly induced by exogenous USF1 (P = 0.05) but increased about threefold with coexpression of USF1 and CTNNB1 (P < 0.02) (Fig. 3B Center). Note that IGF2 expression was unchanged (Fig. 3B Right). To exclude a transcriptional regulation of USF1 by β-catenin, we also tested that the USF1 protein level after β-catenin enforced expression was not changed (Fig. S3). To further confirm these results, we transiently knocked down USF1 by siRNA in HepG2 cells that show a very high β-catenin activity and miR-483-3p expression. qRT-PCR verified a significant reduction of miR-483-3p expression, compared with the control, after 72 h from siRNA transfection (Fig. S4). These data suggest USF1 is an important mediator of the regulation of the miR-483 locus driven by β-catenin.
β-Catenin and USF1 Directly Interact.
To understand the interplay between β-catenin and USF1, we tested the possibility of interaction between these two proteins. We coimmunoprecipitated from HEK293 nuclear extract lysate with either an anti-USF1 antibody or an anti-β-catenin antibody and then immunoblotted with either anti-β-catenin or anti-USF1, respectively, and found the two proteins coimmunoprecipitated (Fig. 3C). Because LiCl treatment is able to induce miR-483 locus expression, we tested the ability of β-catenin to coimmunoprecipitate with USF1 with and without LiCl treatment. Western blot analysis revealed an increased quantity of β-catenin immunoprecipitated with anti-USF1 (22%) compared with the nontreated control (Fig. 3D), where the induction of β-catenin protein level after LiCl treatment was increased about 60%. Then, we demonstrated the direct interaction by using the purified proteins CTNNB1-GST and USF1-HIS in pull-down assays (Fig. 3E).
miR-483-3p Reveals a Negative Regulatory Loop by Targeting β-Catenin.
Because regulatory feedback loops between microRNAs and their targets have been shown in numerous cases (29–31), we investigated the possibility that β-catenin and/or USF1 are targets of miR-483-3p or -5p. By in silico analysis (http://targetscan.org), we found that CTNNB1 is a predicted target of miR-483-3p (Fig. 4A).
Fig. 4.
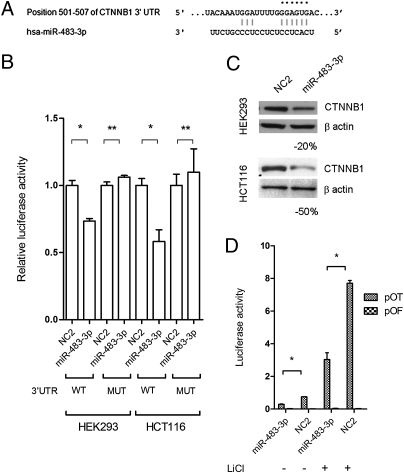
β-Catenin is a target of miR-483-3p. (A) Putative binding site of miR-483-3p in CTNNB1 3′UTRs (TargetScan database). Asterisks indicate nucleotides substituted in 3′UTR miR-483-3p predicted target site to perform luciferase assay. (B) CTNNB1 3′UTRs regulate luciferase activity dependent on miR-483-3p in HEK293 and HCT116 cell lines. MUT, mutant; WT, wild type. (C) Western blot analysis of CTNNB1 after miR-483-3p transfection in HEK293 and HCT116 cell lines. Cells were collected 48 h after miRNA transfection. (D) Luciferase activity of the reporter vectors pOT and pOF in HCT116 cells cotransfected with miR-483-3p and scramble oligo (NC2) with and without LiCl treatment. *P < 0.02, **P > 0.02.
We tested the direct interaction of miR-483-3p with the CTNNB1 3′UTR by luciferase assay as described in SI Materials and Methods. In comparison with the control vector, miR-483-3p caused a decrease in luciferase activity of about 30% and 50% in HEK293 and HCT116 cells, respectively, whereas in the mutated 3′UTR clones, luciferase activities were not perturbed by miR-483-3p (Fig. 4B).
To further confirm β-catenin as a target of miR-483-3p, protein level was assessed by Western blot of HEK293 and HCT116 cells transfected with miR-483-3p. Protein expression was reduced about 20% and 50% compared with the negative control (Fig. 4C). Moreover, in HCT116 cells that harbor a mutated β-catenin, the cotransfection of miR-483-3p and the reporter vector pGL3-OT revealed a functional reduction of nuclear β-catenin of ~60% compared with the controls, with or without LiCl (Fig. 4D). These data prove that miR-483-3p can inhibit β-catenin transcriptional activity.
Mutated Form of β-Catenin Can Evade the miR-483-3p Regulatory Loop.
Our findings suggest that a negative regulatory loop between miR-483-3p and β-catenin exists. However, both genes are overexpressed or have higher activity in tumors, suggesting that β-catenin could escape miR-483-3p regulation. Because it has been described that an SNP within the 3′UTR of the mRNA target could affect the binding of the miRNA (32), we analyzed the mutational status of the β-catenin 3′UTR in eight cell lines (RD, MCF7, RKO, SK-NEP1, G401, A204, U2OS, and HEK293) and 30 Wilms’ tumor samples. We found two SNPs, rs2953 and rs4135387, which we cloned into pGL3-control and tested for their susceptibility to miR-483-3p regulation by a luciferase assay. No significant differences were found (Fig. S5). We also determined that miR-483-3p was not mutated in these samples. This result led us to speculate that mutations in the coding sequence of β-catenin may be affecting this process. Therefore, we determined the extent to which miR-483-3p can regulate the transcriptional activity of either wild-type β-catenin or the activated form of β-catenin.
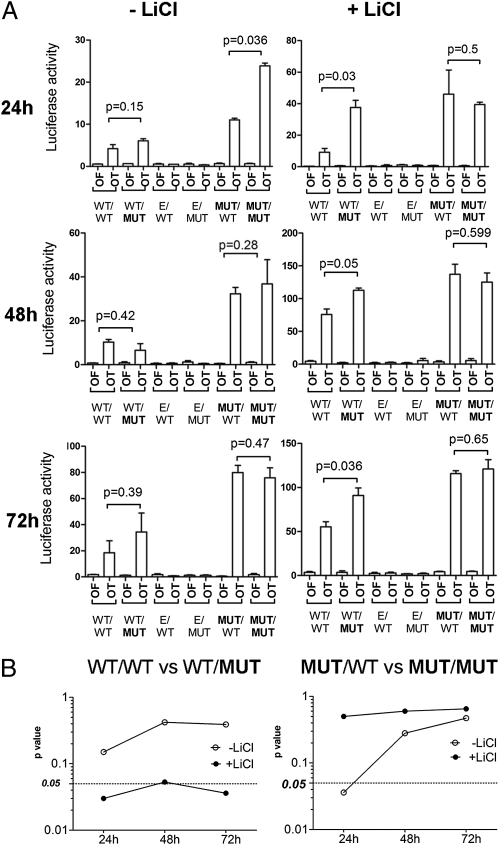
We cloned either the wild-type β-catenin 3′UTR or the 3′UTR with a mutated miR-483-3p target site into two different pIRES-Neo2 β-catenin expression vectors, one with wild-type β-catenin and the other with mutated β-catenin (Ser45 deletion). As control, a vector with only the 3′UTR and no coding sequence was used. The six combinations of vectors were transfected into HEK293 cells and β-catenin nuclear activity was measured by the pGL3-OT luciferase reporter vector after 24, 48, and 72 h. This allowed evaluation of the two different types of negative regulation of β-catenin (GSK3B-mediated degradation and miR-483-3p-mediated translational inhibition). After transfection in HEK293 cells, all forms of β-catenin were able to increase the luciferase activity of the pGL3-OT at 24, 48, and 72 h compared with the controls (Fig. 5A); however, we did not see any significant difference between the WT/WT and WT/MUT forms (CDS/3′UTR). This could be due to the high rate of GSK3B-mediated β-catenin degradation, which is strong in HEK293 cells and could hide the effect of miR-483-3p. To overcome this problem the experiment was conducted in the presence of LiCl, which resulted in a significant increase of activity in the presence of the WT/MUT but not the WT/WT form, suggesting that the wild-type 3′UTR is controlled by miR-483-3p but the mutant form is not (Fig. 5 A and B Left).
Fig. 5.
Mutated form of β-catenin evades the miR-483-3p regulatory loop. (A) Luciferase activity during the time (24, 48, and 72 h) of the reporter vectors pOT and pOF in HEK293 cells cotransfected with the four different CTNNB1 expression vectors (mutational status of the CDS and 3′UTR miR-483-3p target site of CTNNB1: CDS/3′UTR) with (Right) and without LiCl treatment (Left). As experiment controls, wild-type CTNNB1 3′UTR and mutated mir-483-3p target were cloned in the expression vector pIRESNeo2. E/MUT, empty/MUT; E/WT, empty/WT. (B) P values are indicated and graphically represented.
On the other hand, the Ser45 mutant form of β-catenin showed a significant difference in regulation by miR-483-3p at the 3′UTR (MUT/WT vs. MUT/MUT) only at 24 h without LiCl (P = 0.036). Because the mutant form is not degraded and thus accumulates, it is reasonable that at an early time point β-catenin protein levels are still controlled by miR-483-3p but that this regulation is lost over time (Fig. 5B Right). Taken together, these data support that the activating mutation of β-catenin results in loss of regulation by miR-483-3p.
Discussion
We identified a molecular mechanism of autoregulation of β-catenin activity through miR-483-3p, and a unique interaction between the transcription factors USF1 and β-catenin. After genetic analysis of HCC samples, we identified an association between the activation of the β-catenin pathway and the overexpression of miR-483-3p, an association that was stronger in a subset of samples that exhibited a divergent expression between IGF2 and miR-483-3p. This observation was supported by the finding that the miR-483 locus could be up-regulated independently from its host gene IGF2 by enforced overexpression of β-catenin. These findings indicate the existence of at least two mechanisms responsible for the expression of miR-483-3p: coregulation with IGF2 and transcriptional induction by β-catenin. The first mechanism appears to be significantly represented in Wilms’ tumors and possibly other pediatric tumors. In these tumors, the well-known overexpression and loss of imprinting of IGF2 may be responsible for most of the miR-483 up-regulation. Through this mechanism, both cell growth and survival can be simultaneously stimulated by IGF2 and miR-483-3p, respectively. The second mechanism may explain the overexpression of miR-483-3p in adult human cancers, where its up-regulation may occur independently from IGF2 expression.
The mediator of miR-483 stimulation by β-catenin was identified as the basic helix–loop–helix upstream stimulating factor USF1. Here we prove that it can directly interact with β-catenin and recognize the E-box CACGTG element located 400 nucleotides upstream of the miR-483 locus. The interaction between β-catenin and another bHLH protein, MyoD, was previously described (33), and here we describe the direct interaction between β-catenin and USF1. USF1 is a widely expressed transcription factor that plays a crucial role in the regulation of the cell cycle and proliferation (34, 35) and gluco-lipidic metabolism (36, 37). Moreover, it has been shown that USF1 also plays an important role in the maintenance of a chromatin barrier at the insulator elements of the chicken β-globin gene (38).
Our discovery of a negative regulatory loop between miR-483-3p and β-catenin generates an apparent paradox. To explain this discrepancy, we found that the mutated form of β-catenin is able to evade miR-483-3p regulation and the mutated protein can still accumulate in the cell.
In addition to the β-catenin-USF1/miR-483 mechanism, in this study we found that CTCF can also participate in the regulation of miR-483 expression. Indeed, we confirmed data published by Du et al. (22), who found that the region immediately upstream of the miR-483 locus binds the multifunctional protein CTCF. We found evidence that CTCF could play an important role in the regulation of the miR-483 locus as a transcriptional repressor and that repression is indirectly correlated to the methylation status of a CpG dinucleotide in the CTCF binding site. CTCF is a well-known regulator of imprinted regions of the genome such as the IGF2/H19 locus, where it permits the monoallelic expression of these two genes (39, 40) by modulating the conformation of this region, which (41) affects transcriptional activity. It is also important to note that the chromosomal region where CTCF is located (16q21) is often deleted in Wilms’ tumor (42, 43). Our finding highlights the role of the miR-483 locus as a point of junction among the most important players in Wilms’ tumor (IGF2 locus, Wnt/β-catenin pathway, and CTCF).
By unraveling the complex regulation of the miR-483 locus, the present study provides insight into the Wnt/β-catenin pathway and indicates new targets for anticancer therapy.
Materials and Methods
HEK293 and HCT116 cells were transfected using Lipofectamine 2000 (Invitrogen). Mutational and DNA methylation analysis was carried out using primers indicated in Table S3. DNA constructs were cloned using primers listed in Table S3. RNA isolation was performed using TRIzol (Invitrogen) according to the manufacturer's instructions and quantitative real-time reverse transcription–PCR using SYBR Green and TaqMan technologies (Applied Biosystems). Western blot, EMSA, immunoprecipitation, and pull-down analyses were carried out using primary antibodies for CTNNB1 (Cell Signaling; 9562), USF1 (Santa Cruz Biotechnology; sc-229), and MYC (Cell Signaling; 5605). Statistical analysis results are expressed as mean ± SD and significance was accepted at a P value <0.05. More detailed information is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro, Ministero dell'Università e della Ricerca, Ministero della Salute and Fondazione Cariplo (Progetto NOBEL) to M.N. and by National Cancer Institute grants to C.M.C.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101734108/-/DCSupplemental.
References
- 1.Perez-Moreno M, Jamora C, Fuchs E. Sticky business: Orchestrating cellular signals at adherens junctions. Cell. 2003;112:535–548. doi: 10.1016/s0092-8674(03)00108-9. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Wang SS, et al. Alterations of the PPP2R1B gene in human lung and colon cancer. Science. 1998;282:284–287. doi: 10.1126/science.282.5387.284. [DOI] [PubMed] [Google Scholar]
- 4.Satoh S, et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 5.Major MB, et al. Wilms tumor suppressor WTX negatively regulates WNT/β-catenin signaling. Science. 2007;316:1043–1046. doi: 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- 6.Morin PJ, et al. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 7.Devereux TR, et al. CTNNB1 mutations and β-catenin protein accumulation in human hepatocellular carcinomas associated with high exposure to aflatoxin B1. Mol Carcinog. 2001;31:68–73. doi: 10.1002/mc.1041. [DOI] [PubMed] [Google Scholar]
- 8.Kusafuka T, Miao J, Kuroda S, Udatsu Y, Yoneda A. Codon 45 of the β-catenin gene, a specific mutational target site of Wilms’ tumor. Int J Mol Med. 2002;10:395–399. [PubMed] [Google Scholar]
- 9.Rubinfeld B, et al. Binding of GSK3β to the APC-β-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 10.Mann B, et al. Target genes of β-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci USA. 1999;96:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tetsu O, McCormick F. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 14.Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: From research to therapy. J Cell Sci. 2007;120:1833–1840. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- 16.Veronese A, et al. Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer Res. 2010;70:3140–3149. doi: 10.1158/0008-5472.CAN-09-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guled M, et al. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma—A miRNA microarray analysis. Genes Chromosomes Cancer. 2009;48:615–623. doi: 10.1002/gcc.20669. [DOI] [PubMed] [Google Scholar]
- 18.Soon PS, et al. miR-195 and miR-483-5p identified as predictors of poor prognosis in adrenocortical cancer. Clin Cancer Res. 2009;15:7684–7692. doi: 10.1158/1078-0432.CCR-09-1587. [DOI] [PubMed] [Google Scholar]
- 19.Nusse R. Cancer. Converging on β-catenin in Wilms tumor. Science. 2007;316:988–989. doi: 10.1126/science.1143337. [DOI] [PubMed] [Google Scholar]
- 20.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 21.de La Coste A, et al. Somatic mutations of the β-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci USA. 1998;95:8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du M, et al. Insulator and silencer sequences in the imprinted region of human chromosome 11p15.5. Hum Mol Genet. 2003;12:1927–1939. doi: 10.1093/hmg/ddg194. [DOI] [PubMed] [Google Scholar]
- 23.Sparago A, et al. Mechanisms causing imprinting defects in familial Beckwith-Wiedemann syndrome with Wilms’ tumour. Hum Mol Genet. 2007;16:254–264. doi: 10.1093/hmg/ddl448. [DOI] [PubMed] [Google Scholar]
- 24.Prawitt D, et al. Microdeletion of target sites for insulator protein CTCF in a chromosome 11p15 imprinting center in Beckwith-Wiedemann syndrome and Wilms’ tumor. Proc Natl Acad Sci USA. 2005;102:4085–4090. doi: 10.1073/pnas.0500037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan CS, Song JS. CCCTC-binding factor confines the distal action of estrogen receptor. Cancer Res. 2008;68:9041–9049. doi: 10.1158/0008-5472.CAN-08-2632. [DOI] [PubMed] [Google Scholar]
- 26.Butcher DT, Rodenhiser DI. Epigenetic inactivation of BRCA1 is associated with aberrant expression of CTCF and DNA methyltransferase (DNMT3B) in some sporadic breast tumours. Eur J Cancer. 2007;43:210–219. doi: 10.1016/j.ejca.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Paradowska A, et al. Aberrant epigenetic modifications in the CTCF binding domain of the IGF2/H19 gene in prostate cancer compared with benign prostate hyperplasia. Int J Oncol. 2009;35:87–96. doi: 10.3892/ijo_00000316. [DOI] [PubMed] [Google Scholar]
- 28.Corre S, Galibert MD. Upstream stimulating factors: Highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- 29.Petrocca F, et al. E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Yang X, et al. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23:2388–2393. doi: 10.1101/gad.1819009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 32.Nicoloso MS, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim CH, Neiswender H, Baik EJ, Xiong WC, Mei L. β-Catenin interacts with MyoD and regulates its transcription activity. Mol Cell Biol. 2008;28:2941–2951. doi: 10.1128/MCB.01682-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.North S, et al. Regulation of cdc2 gene expression by the upstream stimulatory factors (USFs) Oncogene. 1999;18:1945–1955. doi: 10.1038/sj.onc.1202506. [DOI] [PubMed] [Google Scholar]
- 35.Cogswell JP, Godlevski MM, Bonham M, Bisi J, Babiss L. Upstream stimulatory factor regulates expression of the cell cycle-dependent cyclin B1 gene promoter. Mol Cell Biol. 1995;15:2782–2790. doi: 10.1128/mcb.15.5.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Read ML, Clark AR, Docherty K. The helix-loop-helix transcription factor USF (upstream stimulating factor) binds to a regulatory sequence of the human insulin gene enhancer. Biochem J. 1993;295:233–237. doi: 10.1042/bj2950233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iynedjian PB. Identification of upstream stimulatory factor as transcriptional activator of the liver promoter of the glucokinase gene. Biochem J. 1998;333:705–712. doi: 10.1042/bj3330705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol Cell Biol. 2007;27:7991–8002. doi: 10.1128/MCB.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 40.Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 41.Kurukuti S, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maw MA, et al. A third Wilms’ tumor locus on chromosome 16q. Cancer Res. 1992;52:3094–3098. [PubMed] [Google Scholar]
- 43.Yeh A, et al. Chromosome arm 16q in Wilms tumors: Unbalanced chromosomal translocations, loss of heterozygosity, and assessment of the CTCF gene. Genes Chromosomes Cancer. 2002;35:156–163. doi: 10.1002/gcc.10110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.