Abstract
Maf1 protein is a global negative regulator of RNA polymerase (Pol) III transcription conserved from yeast to man. We report that phosphorylation of Maf1 by casein kinase II (CK2), a highly evolutionarily conserved eukaryotic kinase, is required for efficient Pol III transcription. Both recombinant human and yeast CK2 were able to phosphorylate purified human or yeast Maf1, indicating that Maf1 can be a direct substrate of CK2. Upon transfer of Saccharomyces cerevisiae from repressive to favorable growth conditions, CK2 activity is required for the release of Maf1 from Pol III bound to a tRNA gene and for subsequent activation of tRNA transcription. In a yeast strain lacking Maf1, CK2 inhibition showed no effect on tRNA synthesis, confirming that CK2 activates Pol III via Maf1. Additionally, CK2 was found to associate with tRNA genes, and this association is enhanced in absence of Maf1, especially under repressive conditions. These results corroborate the previously reported TFIIIB–CK2 interaction and indicate an important role of CK2-mediated Maf1 phosphorylation in triggering Pol III activation.
Keywords: RNA polymerase III regulation, transfer RNA, casein kinase II regulation
RNA polymerase (Pol) III is responsible for the transcription of some 300 different genes in yeast (class III genes), mostly tRNA genes (1). In-depth analyses of the yeast Pol III transcription system have revealed a cascade of protein–DNA and protein–protein interactions leading to the recruitment of Pol III to its target tRNA genes: binding of the six-subunit TFIIIC factor to the intragenic promoter, TFIIIC-directed recruitment and assembly of the three subunits of TFIIIB (TBP, Brf1, and Bdp1) and subsequent recruitment of the 17-subunit Pol III enzyme (2). High rate of tRNA transcription is achieved through many rounds of reinitiation by Pol III on stable DNA-bound complexes of the initiation factor TFIIIB (3, 4).
Pol III is under control of the general negative regulator Maf1 (5, 6), which binds to Pol III clamp and rearranges specific subcomplex C82/34/31, which is required for transcription initiation (7). In the repressive complex, Maf1 impairs recruitment of Pol III to a complex of promoter DNA with the initiation factors TFIIB and thus prevents closed-complex formation (4, 7). Maf1 is essential for repressing Pol III transcription in yeast and mediates several signaling pathways (8). In addition to the down-regulation that occurs normally in the stationary phase, Pol III repression accompanying starvation, respiratory growth, as well as oxidative and replication stress, also requires Maf1 (9–11). Maf1 inhibits Pol III transcription via a mechanism that depends on the dephosphorylation and nuclear accumulation of Maf1 followed by its physical association with Pol III at Pol III-transcribed genes genomewide (6, 12). In contrast Maf1 phosphorylation occurs in favorable growth conditions and is linked to cytoplasmic localization of Maf1 (6, 13).
Maf1 was recently found to be phosphorylated by protein kinases PKA (14, 15), Sch9 (16–18), and TORC1 (19), but participation of other kinases in Maf1 regulation is conceivable. First, phosphorylation by PKA prevents Maf1 nuclear import (15) but does not seem to be involved in direct regulation of Maf1 activity by carbon source or nutrient availability (9, 17). Second, tRNA transcription levels were not significantly changed either by Sch9- or TORC1-dependent phosphorylation of Maf1 (16, 17, 19). Finally, mutating the potential phosphorylation sites for Sch9 and PKA still allowed regulation of Pol III, indicating another critical factor (16, 17). The results presented here point to casein kinase II (CK2) as a possible missing kinase.
CK2, a vital, pleiotropic and highly conserved serine/threonine phosphotransferase, is involved in transcription-directed signaling, gene control, and cell cycle regulation and has been implicated in critical cellular processes such as proliferation, apoptosis, differentiation, and transformation (20). CK2 is mostly present as a tetramer composed of two catalytic and two regulatory subunits (in yeast Cka1/2 and Ckb1/2, respectively). The regulatory subunits stimulate the catalytic subunits, stabilize the CK2 heterotetramer, and act as a scaffold for specific kinase partners (21) The enzyme is expressed constitutively at a modest level, is predominantly in the nucleus, where it is a component of several chromatin-associated complexes, and is predicted to phosphorylate a broad spectrum of nuclear as well as cytoplasmic proteins involved in multiple aspects of gene expression (22).
It has been shown previously that CK2 is a positive regulator of Pol III transcription in yeast and humans. CK2 is stably associated with TFIIIB (23, 24) and phosphorylates subunits TBP in yeast, and Brf1 and Bdp1 in humans (24, 25). CK2 activity is required for assembly of Pol III transcription initiation complex on gene promoters in human cells (24, 26). Surprisingly, phosphorylation of Bdp1 by CK2 decreases Pol III activity during mitosis in human cells (27).
We previously demonstrated that nuclear localization of Maf1 is not sufficient for Pol III repression but must be accompanied by Maf1 dephosphorylation to ensure interaction with Pol III and inhibition of tRNA transcription (13). We also concluded that phosphorylation of Maf1 is essential for its export out of the nucleus. We thus postulated the existence of a nuclear kinase which would phosphorylate Maf1, thereby breaking its interaction with the Pol III complex and allowing activation of tRNA transcription. Here we show that CK2 is the postulated kinase involved in this process.
Results
CK2 Phosphorylates Maf1 Protein.
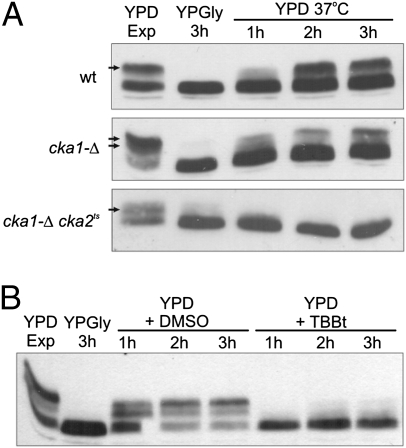
CK2 is the evolutionarily conserved kinase involved in numerous pathways regulating gene expression. To assess whether it might also be involved in Maf1 regulation, we took advantage of yeast strains with decreased catalytic activity of CK2 (28). In wild-type (WT) cells grown in glucose-containing medium, two phosphorylated forms of Maf1 are observed in SDS/PAGE as slower migrating additional bands. This pattern of Maf1 phosphorylation was variably altered in the single cka1-Δ and double cka1-Δ cka2ts mutants. Inhibition of CK2 activity following shift to an elevated temperature resulted in lack of Maf1 phosphorylation of Maf1 in cka1-Δ cka2ts cells unlike in WT or cka1-Δ strains where we observed no change or only a slight decrease in Maf1 phosphorylation, respectively (Fig. S1).
As shown previously, Maf1 is phosphorylated in response to a shift from respiratory conditions to fermentative growth (13). We thus examined a possible role of CK2 in Maf1 phosphorylation in response to carbon source. Cells exponentially growing in a rich glucose medium (YPD) were transferred to a nonfermentable glycerol medium (YPGly) at 30 °C then back to the glucose medium and incubated at 37 °C. Gradual phosphorylation of Maf1 at 37 °C was observed in the control, but phosphorylation was less efficient in cka1-Δ and did not occur in the cka1-Δ cka2ts mutant cells (Fig. 1A). Maf1 phosphorylation was also inhibited when wild-type cells were transferred from YPGly to YPD supplemented with tetrabromobenzotriazol (TBBt) (Fig. 1B). Altogether these results suggest participation of CK2 in Maf1 phosphorylation during the switch between respiration and fermentation. A decrease of CK2 activity concomitantly compromised Maf1 exit from the nucleus (Fig. S2). Noteworthy, TBBt only modestly affected Maf1 phosphorylation in cells growing exponentially in YPD (Fig. S3).
Fig. 1.
CK2 is necessary for regulation of Maf1 activity by carbon source. Wild-type cells and isogenic mutants cka1-Δ, cka2-Δ, and cka1-Δ cka2ts were examined by Western blot using anti-Maf1 antibody. Cells were grown to exponential phase in rich glucose medium (YPD Exp) at 30 °C, transferred to glycerol medium (YPGly), incubated at 30 °C for 3 h, then transferred to (A) prewarmed YPD, and incubated at 37 °C (B) prewarmed rich glucose medium containing 4% vol/vol DMSO or 200 μM TBBt, and incubated at 30 °C. Cells were harvested as indicated. Arrows indicate phosphorylated forms of Maf1.
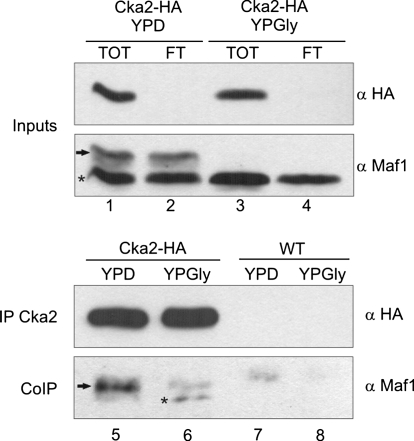
Because active CK2 has been shown to bind directly to the TBP subunit of TFIIIB (23), we used coimmunoprecipitation experiments to test whether Maf1 was able to interact with the CK2 catalytic subunit. Crude extracts were prepared from Cka2–HA-tagged cells and control wild-type cells with untagged Cka2. Cells were grown in glucose medium, transferred to prewarmed glycerol medium, and harvested after 3 h. HA-tagged Cka2 was immunoprecipitated from cell extracts with magnetic beads coated with anti-HA antibodies and the immunoprecipitates were examined for the presence of Maf1 by immunoblotting. As shown in Fig. 2, Cka2 was efficiently immunopurified from crude extracts (compare lane 1 with lane 2 and lane 3 with lane 4) and a hyperphosphorylated form of Maf1 was selectively coimmunoprecipitated with HA-tagged Cka2 from glucose cells (compare lane 5 with control lane 7). Maf1 was also detectable in Cka2-HA immune complexes from glycerol cells (compare lane 6 with control lane 8). This result indicates that Maf1 and Cka2, a catalytic subunit of CK2, interact with each other.
Fig. 2.
Maf1 interacts with CK2 catalytic subunit. YPH499 strain expressing HA-tagged Cka2 and WT control strain were grown in rich glucose medium to exponential phase (YPD), transferred to a nonfermentable glycerol medium (YPGly), and incubated for 3 h. Subsequently, cellular extracts were prepared and subjected to immunoprecipitation with magnetic beads coated with anti-HA antibodies, followed by elution of bound proteins. Total cellular extracts (TOT), flow through (FT), and immunopurified proteins were analyzed by SDS/PAGE and Western blot using anti-HA and anti-Maf1 antibodies. Arrow indicates hyperphosphorylated Maf1 and asterisk indicates hypophosphorylated Maf1.
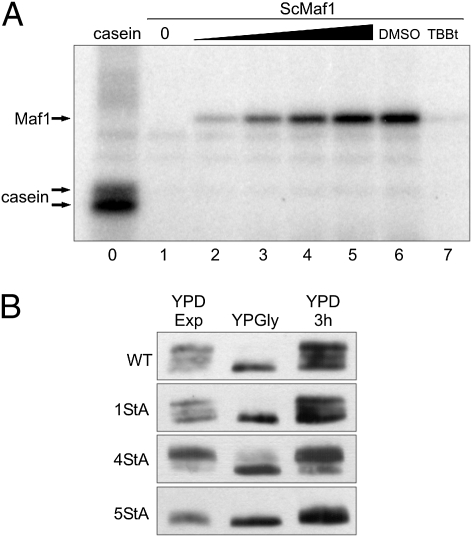
Next we asked whether Maf1 could be directly phosphorylated by CK2 in vitro. Recombinant Saccharomyces cerevisiae Maf1 expressed in Escherichia coli was incubated with [γ32P]-ATP and purified yeast TAP tagged CK2. Analysis of the reaction outcome by SDS/PAGE and autoradiography indicated that Maf1 was phosphorylated (Fig. 3A). Consistent with the idea that CK2 is responsible for the observed phosphorylation of recombinant Maf1, incorporation of [γ32P]-ATP was completely blocked by TBBt, a specific CK2 inhibitor (29). To extend the results obtained using recombinant Maf1, we studied phosphorylation of yeast Maf1 and human Maf1 with highly purified human CK2. Human recombinant Maf1, expressed in insect cells, was mixed with purified human CK2 α-catalytic subunit in the presence of [γ32P]-ATP resulting in successful phosphorylation (Fig. S4). Notably, human CK2 also phosphorylated yeast Maf1. Therefore, the process of Maf1 phosphorylation by CK2 is evolutionarily conserved.
Fig. 3.
Maf1 is phosphorylated by CK2. (A) CK2 phosphorylates recombinant S. cerevisiae Maf1 expressed in E. coli and purified by HIS-tag isolation. Autoradiogram shows [γ32P]-ATP–labeled products of reactions containing 100 ng yeast TAP-tag purified CK2 and 1 μg casein (lane 0); increasing amounts of recombinant yeast Maf1 (0, 0.25, 0.5, 1, and 2 μg; lanes 1–5); 2 μg recombinant yeast Maf1 and 4% (vol/vol) DMSO (lane 6); 2 μg recombinant yeast Maf1, 4% (vol/vol) DMSO and 10 μM TBBt (lane 7). (B) Mutations of CK2 sites prevent Maf1 phosphorylation in vivo. Substitutions in MAF1 cloned in pRS315 vector were made using mutagenic PCR. Yeast mutants 1StA (S388A), 4StA (S159A, S160A, S161A, and S162A), 5StA (S159A, S160A, S161A, S162A, and S388A) and corresponding WT were grown overnight in SC –leu medium, then in glucose-rich medium to exponential phase (YPD Exp), transferred to glycerol medium for 3 h (YPGly), and again transferred to YPD for 3 h. TCA-precipitated proteins were examined by Western blot using anti-Maf1 antibody.
Having established that CK2 phosphorylates Maf1 in vitro, we next asked what the specific phosphorylation sites are. The mass-spectrometry analysis of CK2-phosphorylated recombinant yeast Maf1 revealed phosphorylation of serine S388 and at least one phosphorylation site located in serine tract S159–162 (Table S1, repeated experiments 1–3). Importantly, parallel mass spectrometry analysis of Maf1 immunopurified from cells exponentially growing in glucose identified phosphorylation of both one and two serines within the S159–162 tract (Table S1, experiments 4–6). Unfortunately in this analysis S388-containing peptide was not detected; thus, we were not able to confirm modification at this site. Besides S159–162 sites assigned presumably to CK2, several PKA/Sch9-specific phosphorylation sites were also identified in Maf1 from glucose cells, confirming results published previously (15, 17) (Table S1, repeated experiments 4–6). All possible phosphorylation sites detected in CK2-phosphorylated recombinant Maf1 were inactivated, resulting in 5StA mutant (S159A, S160A, S161A, S162A, and S388A). Without a doubt, Maf1 phosphorylation in vivo was not observed in 5StA mutant (Fig. 3B). Interestingly, concurrent Western analysis of 1StA (S388A) shows no effect on Maf1 phosphorylation, whereas analysis of 4StA (S159A, S160A, S161A, and S162A) mutant revealed differently migrating forms of Maf1 (Fig. 3B). Therefore, contribution of both S159–162 and S388 CK2-phosphorylation sites seems to be similarly important for Maf1 function. Notably, the lack of Maf1 phosphorylation in 5StA mutant seems not to be an effect of protein misfolding because it complemented growth phenotype of maf1-Δ (Fig. S5A). Moreover, inactivation of CK2-phosphorylation sites in Maf1 somewhat decreased Pol III activity (Fig. S5B).
CK2 Controls Maf1 Association with tRNA Genes.
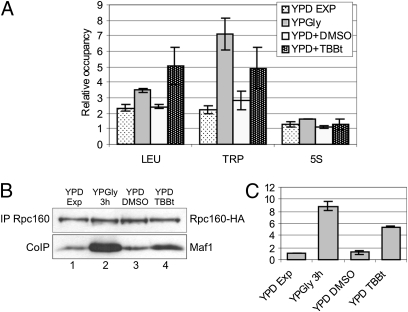
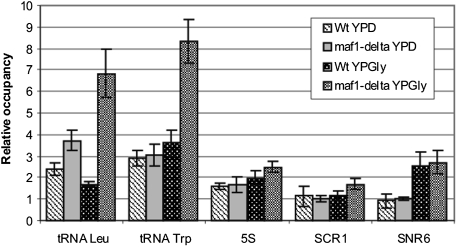
Previous work implicated Maf1 association with tRNA genes in a nutrient-dependent manner (6, 19). Because tRNA transcription is also regulated by carbon metabolism (9), we monitored carbon source-dependent Maf1 occupancy on class III genes and its dependence on CK2. Chromatin immunoprecipitation (ChIP) analyzed by real-time quantitative PCR showed moderate enrichment of Maf1 on tRNA genes when glucose-grown cells were harvested in exponential growth phase (Fig. 4A). In glycerol medium, however, the Maf1 occupancy on tRNA genes increased significantly. Strikingly, we were unable to detect Maf1 on 5S rDNA chromatin in either condition. Shifting the cells back to glucose medium led to the dissociation of Maf1 from tRNA genes and, significantly, supplementation of the medium with the TBBt inhibitor impaired this dissociation dramatically. Altogether, these data suggest that CK2 mediates dissociation of Maf1 from tRNA genes following transfer of yeast from nonfermentable (glycerol) to fermentable (glucose) growth conditions.
Fig. 4.
Dissociation of Maf1 from tRNA genes following transfer of yeast from nonfermentable glycerol growth conditions to glucose is dependent on CK2 activity. Relative occupancy of Maf1 at selected class III genes. Strain expressing Myc-tagged Maf1 was grown in rich glucose medium (YPD) to exponential phase and transferred to a nonfermentable glycerol medium (YPGly). Following 3-h incubation at 30 °C, the culture was split in half; one part was transferred to YPD with 2% (vol/vol) DMSO and the second, to YPD with 200 μM TBBt in DMSO. (A) Cross-linked chromatin was immunoprecipitated with antibodies against Myc epitope, followed by quantitative real-time PCR. Occupancies of Maf1 at tRNA −Leu, tRNA −Trp, and 5S rRNA genes were measured relative to occupancy at 35S rRNA. (B) Cellular extracts were incubated with magnetic beads coated with anti-HA monoclonal antibody followed by elution of bound proteins and Western analysis with anti-HA and anti-Maf1 antibodies. (C) Quantification of the amounts of Maf1 immunopurified with Rpc160. Experiment was performed in triplicate to estimate SD.
To investigate the possible effect of CK2 activity on Maf1 interaction with Pol III we used yeast cells expressing HA-tagged Rpc160, the largest Pol III subunit. Pol III was immunopurified from crude extracts and the immunoprecipitate was analyzed by Western blot (Fig. 4B). Equal amounts of Rpc160 were precipitated from yeast grown in glucose and glycerol media, whereas the levels of coimmunopurified Maf1 varied depending on growth conditions. In glucose medium, little Maf1 is bound to Pol III (Fig. 4B, lane 1), whereas in the glycerol medium the amount is over ninefold higher (Fig. 4B, lane 2 and Fig. 4C). Maf1 dissociated from the Pol III complex when cells were shifted back to the glucose medium supplemented with DMSO (lane 3), but this dissociation was over fivefold less pronounced when the medium was supplemented with TBBt (Fig. 4B, lane 4 and Fig. 4C). We concluded that CK2 activity is required to release Maf1 from the Pol III complex and tRNA genes upon exit from nonfermentable to fermentable growth conditions.
Maf1 Mediates CK2 Signal Transduction to Pol III.
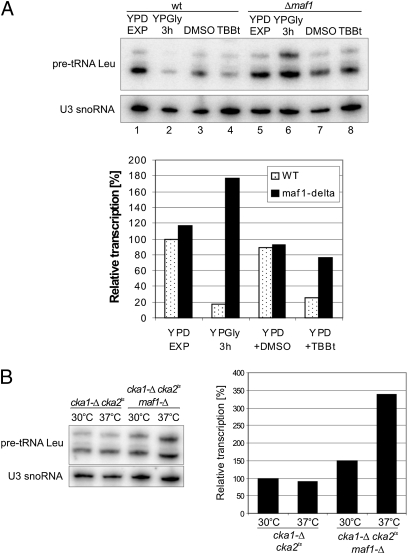
To confirm a role of Maf1 in regulation of Pol III transcription by CK2, total RNA was isolated from WT and maf1-Δ cells grown in glucose medium, transferred to glycerol medium, and transferred back to glucose medium supplemented or not with TBBt. The amounts of newly synthesized pre-tRNALeu were estimated by hybridization, revealing that TBBt precluded pre-tRNA derepresion in the wild-type strain, whereas in the maf1-Δ strain the pre-tRNA level was similar in the glucose medium without TBBt (Fig. 5A, compare lanes 3 and 7 and 4 and 8). To extend these studies, cka1-Δ cka2ts maf1-Δ triple mutant was constructed and examined by hybridization. Deletion of the MAF1 gene led to increased pre-tRNALeu synthesis in the cka1-Δ cka2ts mutant both at 30 °C and 37 °C (Fig. 5B). Altogether, these data suggest that CK2 activates Pol III transcription via Maf1 phosphorylation. In contrast, cka1-Δ cka2ts double mutation leads to the constitutive presence of repressive hypophosphorylated forms of Maf1 (as shown in Fig. S1), that inhibit Pol III transcription.
Fig. 5.
Pol III transcription is activated via Maf1 phosphorylation by CK2. Levels of newly synthesized pre-tRNA were estimated by Northern blot hybridization with tRNALeu intron probe and U3 probe (loading control). Quantification of pre-tRNALeu transcript (Upper band) was normalized to U3 snoRNA transcript. (A) Wild-type and maf1-Δ strains were grown in rich glucose medium (YPD) to exponential phase and transferred to glycerol medium (YPGly). Following 3-h incubation at 30 °C the culture was split in half; one part was transferred to YPD with 4% (vol/vol) DMSO and the second, to YPD with 200 μM TBBt in DMSO. (B) Isogenic mutants cka1-Δ cka2t and cka1-Δ cka2ts maf1-Δ were grown in glucose medium (YPD) at 30 °C to exponential phase and transferred to YPD prewarmed to 37 °C for 1 h.
CK2 Associates with tRNA Genes in a Maf1-Sensitive Manner.
The previously reported CK2 copurification with TFIIIB (23) prompted us to investigate the presence of CK2 in class III genes. We used a control wild-type strain and an isogenic maf1-Δ mutant, both expressing Myc-tagged catalytic subunit of CK2 (Cka1-Myc) to perform ChIP analysis followed by real-time quantitative PCR. Slight but significant association of CK2 with tRNALeu and tRNATrp genes (two- to fourfold above background) was observed for the wild-type strain. The occupancy of CK2 at tRNA genes was, however, significantly increased in the maf1-Δ strain, especially when cells were grown under repressive conditions in glycerol medium (seven- to eightfold above background). To our surprise, little or no association of Cka1-Myc was found with other class III genes tested—RDN5 (5S rRNA gene), SCR1, and SNR6—in the wild type and maf1-Δ strains, regardless of the growth conditions (Fig. 6). These data suggest that CK2 is present at some Pol III-transcribed genes but the level of this occupancy is modulated by Maf1.
Fig. 6.
CK2 binds to tRNA in carbon source and Maf1-sensitive manner. Relative occupancy of Cka1-Myc at selected class III genes. Strains WT and maf1-Δ, both expressing Myc-tagged α-catalytic subunit of CK2 (Cka1-Myc) were grown in rich glucose medium (YPD) to exponential phase, transferred to a nonfermentable glycerol medium (YPGly), and incubated at 30 °C for 40 min. Cross-linked chromatin was immunoprecipitated with antibodies against Myc epitope, followed by real-time quantitative PCR. All occupancies are expressed relative to TEL15 region. The experiment was performed in triplicate to estimate SD.
Discussion
In this work, we present evidence that Maf1 is phosphorylated by CK2 kinase. We show that Maf1 phosphorylation by CK2 contributes directly to activation of Pol III transcription by stimulating Maf1 dissociation from the Pol III complex and from tRNA genes.
We found that yeast Maf1 is phosphorylated by CK2 on S388 and on at least two residues within serine tract S159–S162. Significantly, these sites are located in two highly flexible regions of Maf1. S388 is located in the C-terminal tail, whereas S159–S162 lie in a mobile linker separating two conserved, highly structured A and BC fragments. Notably, S177–180 sites known to be phosphorylated in Maf1 by PKA/Sch9 (15, 17) lie in the second serine tract located between the A and BC boxes (Fig. S6). Moreover, human Maf1 is phosphorylated by mTOR, and, significantly, the phosphorylation sites are within the linker region (30, 31). The phosphorylation state of the linker, therefore, appears to have a conserved role in control interaction between the A and BC regions and Maf1 association with Pol III, supporting the hypothesis presented by Gajda et al. (32).
Here we examine the relationship between the Maf1-mediated repression and Pol III control by CK2. We demonstrate that phosphorylation of Maf1 by CK2 is critical for the release of Maf1 from tRNA genes and for the high rate of Pol III transcription after a shift from repressing (glycerol) to optimal growth conditions (glucose). In repressing conditions, Maf1 is physically associated with the Pol III complex and with tDNA chromatin (6). Inhibition of CK2 hampers the dissociation of Maf1 from the Pol III complex and its export out of the nucleus as normally observed upon the shift from respiration to fermentation. Moreover, the dissociation of CK2 catalytic subunit from Pol III chromatin is less efficient in the deletion strain maf1-Δ. These results suggest that Maf1 assists the arrangement of CK2 complex thus preventing CK2-mediated phosphorylation of TBP during repression.
Because CK2 activity is required for cell cycle progression at G1/S and G2/M (28), we asked whether the observed defect of Maf1 phosphorylation in CK2-deficient cells is an indirect effect of cell cycle blockage or is a direct result of the lowered CK2 activity. The relationship between checkpoint signaling and regulation of tRNA synthesis was analyzed extensively by a recent study showing that neither normal progression through S phase nor alpha-factor–mediated G1 arrest or G2/M arrest by nocodazole are associated with alterations in tRNA transcription (11). These results indicate that the Maf1-mediated effect on tRNA transcription correlated with CK2 depletion cannot be attributed to cell-cycle defects.
We believe that CK2 controls the dynamic association of Maf1 with chromatin by assessing Pol III during reinitiation. Regardless of the growth conditions, Maf1 could be recruited to the Pol III complex with each transcription cycle, during elongation or termination. Our previous study shows that a small amount of dephosphosphorylated Maf1 is Pol III bound even in cells growing exponentially on glucose (6). We propose that this dephosphorylated nuclear Maf1 is associated with the reinitiating complex. Following termination, before the new transcription cycle of reinitiating Pol III, Maf1 encounters CK2 bound to TFIIIB in the promoter region. Under favorable growth conditions, CK2 phosphorylates Maf1, thus releasing it from Pol III and enabling reinitiation. At the same time CK2 activates Pol III transcription by binding and phosphorylating TBP. Subsequent export of the phosphorylated Maf1 to the cytoplasm decreases its concentration in the nucleus, lowering the probability of association with the Pol III complex. Under stress, the catalytic subunits of CK2 dissociate from TBP and CK2 becomes inactive. The Pol III-associated Maf1 cannot be rephosphorylated, remains with Pol III, and reinitiation is not possible. Simultaneously, TBP cannot be phosphorylated by CK2. Moreover, dephosphorylated Maf1 is imported from the cytoplasm to the nucleus to strengthen the Pol III repression. Altogether, the presented idea fits with, and even strengthens the model of Pol III regulation by CK2, proposed previously by Ghavidel and Schultz (23).
Maf1 is regulated by a variety of pathways to adjust Pol III transcriptional rate to changing conditions. Besides CK2, three other kinases, PKA, Sch9, and TORC1, contribute to Maf1 phosphorylation in yeast. A question arises about the sequence of their action and priority in Maf1-dependent Pol III transcription regulation. We suggest that CK2 plays a major role by directly controlling Maf1–Pol III association, whereas phosphorylation by other kinases mainly contributes to cellular localization of Maf1. According to our model presented above, the nuclear abundance of Maf1 supports the control of Pol III activity by CK2. Maf1 phosphorylation by PKA and Sch9 contributes to excluding Maf1 from the nucleus, although inactivation of all potential Sch9 phosphorylation sites resulted in constitutive Maf1 association with Pol III (17). Phosphorylation of Maf1 by TORC1 is thought to take place on chromatin in nucleoli and to contribute to Maf1 regulation by excluding it from chromatin (19).
In cells lacking Maf1, CK2 still binds to tRNA genes and this association is even increased upon the shift to repressive (glycerol) conditions. We propose that dissociation of the CK2 catalytic subunits from the TBP subunit of TFIIIB is precluded in the absence of Maf1. Therefore, one hypothesis is that Maf1 is required to block TFIIIB activation by CK2, thereby effecting Pol III repression. This raises the question of whether Maf1 may influence the CK2 holoenzyme at the heart of the Pol III preinitiation complex.
One unanswered but fundamental question is the mechanism of CK2 regulation. The presence of many discrete CK2 subpopulations is crucial in resolving the current conflicting views on whether CK2 is constitutively active or is modulated in response to specific stimuli. Each local CK2 population may be regulated in a distinct manner to carry out its precise function(s). Most likely the CK2 activity is controlled through interactions of its regulatory subunits with other proteins. Apart from the transient formation of the holoenzyme with catalytic subunits of CK2 in humans, regulatory subunits can interact with at least 30 different proteins but are rapidly degraded when unassembled (for review see ref. 20). In humans, CK2-mediated phosphorylation of p53 protein is regulated by the FACT complex (hSpt16 and SSRP1) (33), and Pin-1 protein inhibits phosphorylation of topoisomerase IIα by CK2 (34). We propose here a model in which Maf1 regulates Pol III directly and also modulates CK2 activity toward TFIIIB. To confirm this conjecture, further studies are needed on how Maf1 influences the association of CK2 subunits with the TBP component of TFIIIB.
CK2 is also present in the human Pol III complex and phosphorylates the TFIIIB subunits Brf1 (24) and Bdp1 (26). Our results indicate that human Maf1 can also be phosphorylated by CK2 in vitro, although we have not tested this in vivo. Such evolutionary conservation suggests that phosphorylation of Maf1 by CK2 acting as a master switch of Pol III transcription is of fundamental importance.
Materials and Methods
Yeast Strains and Plasmids.
All strains and plasmids used in this study are described in the SI Materials and Methods (see also Tables S2 and S3).
Cell Growth Conditions and TBBt Treatment.
Rich media contained 1% yeast extract, 2% peptone, and 2% glucose (YPD) or 2% glycerol (YPGly). Minimal medium (SC) contained 2% glucose and 0.67% yeast nitrogen base without amino acids. SC −leu contained supplements required for growth (at 20 μg/mL each except leucine). Solid media contained 2% agar. All reagents were from Difco. TBBt stock solution (5 mM) was prepared in DMSO and added to YPD medium to a final concentration indicated.
Protein Extraction, Immunoblotting, and Immunofluorescence.
Western blot and immunofluorescence were performed as described previously (13).
Supplementary Material
Acknowledgments
We thank Maja Łebska for kindly providing purified human catalytic subunit of CK2, Christine Conesa for the Maf1-13myc strain, Kamil Gewartowski and Rafał Tomecki for help in CK2 purification, Andrew Oler for his stimulating comments, and anonymous reviewers for helpful comments. This work was supported by the Polish Ministry of Science and Education (N301 164035), the Foundation for Polish Science (START Program), and EMBO (Short-Term Fellowship Program).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010010108/-/DCSupplemental.
References
- 1.Harismendy O, et al. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J Mol Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 3.Dieci G, Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 4.Cabart P, Lee J, Willis IM. Facilitated recycling protects human RNA polymerase III from repression by Maf1 in vitro. J Biol Chem. 2008;283:36108–36117. doi: 10.1074/jbc.M807538200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pluta K, et al. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5031–5040. doi: 10.1128/MCB.21.15.5031-5040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oficjalska-Pham D, et al. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell. 2006;22:623–632. doi: 10.1016/j.molcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Vannini A, et al. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10:1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 9.Cieśla M, et al. Maf1 is involved in coupling carbon metabolism to RNA polymerase III transcription. Mol Cell Biol. 2007;27:7693–7702. doi: 10.1128/MCB.01051-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boisnard S, et al. H2O2 activates the nuclear localization of Msn2 and Maf1 through thioredoxins in Saccharomyces cerevisiae. Eukaryot Cell. 2009;8:1429–1438. doi: 10.1128/EC.00106-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen VC, et al. Replication stress checkpoint signaling controls tRNA gene transcription. Nat Struct Mol Biol. 2010;17:976–981. doi: 10.1038/nsmb.1857. [DOI] [PubMed] [Google Scholar]
- 12.Roberts DN, Wilson B, Huff JT, Stewart AJ, Cairns BR. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol Cell. 2006;22:633–644. doi: 10.1016/j.molcel.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Towpik J, Graczyk D, Gajda A, Lefebvre O, Boguta M. Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J Biol Chem. 2008;283:17168–17174. doi: 10.1074/jbc.M709157200. [DOI] [PubMed] [Google Scholar]
- 14.Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moir RD, et al. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc Natl Acad Sci USA. 2006;103:15044–15049. doi: 10.1073/pnas.0607129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Moir RD, Willis IM. Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J Biol Chem. 2009;284:12604–12608. doi: 10.1074/jbc.C900020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber A, et al. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Y, Zheng XF. Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis. Cell Cycle. 2009;8:4085–4090. doi: 10.4161/cc.8.24.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei Y, Tsang CK, Zheng XF. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J. 2009;28:2220–2230. doi: 10.1038/emboj.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsten ME, Litchfield DW. Order or chaos? An evaluation of the regulation of protein kinase CK2. Biochem Cell Biol. 2004;82:681–693. doi: 10.1139/o04-116. [DOI] [PubMed] [Google Scholar]
- 21.French AC, Luscher B, Litchfield DW. Development of a stabilized form of the regulatory CK2beta subunit that inhibits cell proliferation. J Biol Chem. 2007;282:29667–29677. doi: 10.1074/jbc.M706457200. [DOI] [PubMed] [Google Scholar]
- 22.Poole A, et al. A global view of CK2 function and regulation. Mol Cell Biochem. 2005;274:163–170. doi: 10.1007/s11010-005-2945-z. [DOI] [PubMed] [Google Scholar]
- 23.Ghavidel A, Schultz MC. TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell. 2001;106:575–584. doi: 10.1016/s0092-8674(01)00473-1. [DOI] [PubMed] [Google Scholar]
- 24.Johnston IM, et al. CK2 forms a stable complex with TFIIIB and activates RNA polymerase III transcription in human cells. Mol Cell Biol. 2002;22:3757–3768. doi: 10.1128/MCB.22.11.3757-3768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hockman DJ, Schultz MC. Casein kinase II is required for efficient transcription by RNA polymerase III. Mol Cell Biol. 1996;16:892–898. doi: 10.1128/mcb.16.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu P, Wu S, Hernandez N. A minimal RNA polymerase III transcription system from human cells reveals positive and negative regulatory roles for CK2. Mol Cell. 2003;12:699–709. doi: 10.1016/j.molcel.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 27.Hu P, Samudre K, Wu S, Sun Y, Hernandez N. CK2 phosphorylation of Bdp1 executes cell cycle-specific RNA polymerase III transcription repression. Mol Cell. 2004;16:81–92. doi: 10.1016/j.molcel.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Hanna DE, Rethinaswamy A, Glover CV. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J Biol Chem. 1995;270:25905–25914. doi: 10.1074/jbc.270.43.25905. [DOI] [PubMed] [Google Scholar]
- 29.Pagano MA, et al. The selectivity of inhibitors of protein kinase CK2: An update. Biochem J. 2008;415:353–365. doi: 10.1042/BJ20080309. [DOI] [PubMed] [Google Scholar]
- 30.Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci USA. 2010;107:11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michels AA, et al. mTORC1 directly phosphorylates and regulates human MAF1. Mol Cell Biol. 2010;30:3749–3757. doi: 10.1128/MCB.00319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gajda A, et al. Full repression of RNA polymerase III transcription requires interaction between two domains of its negative regulator Maf1. J Biol Chem. 2010;285:35719–35727. doi: 10.1074/jbc.M110.125286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller DM, et al. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol Cell. 2001;7:283–292. doi: 10.1016/s1097-2765(01)00176-9. [DOI] [PubMed] [Google Scholar]
- 34.Messenger MM, et al. Interactions between protein kinase CK2 and Pin1. Evidence for phosphorylation-dependent interactions. J Biol Chem. 2002;277:23054–23064. doi: 10.1074/jbc.M200111200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.