Abstract
Golgi-localized nucleotide sugar transporters (NSTs) are considered essential for the biosynthesis of wall polysaccharides and glycoproteins based on their characteristic transport of a large number of nucleotide sugars to the Golgi lumen. The lack of NST mutants in plants has prevented evaluation of this hypothesis in plants. A previously undescribed Golgi NST mutant, brittle culm14 (bc14), displays reduced mechanical strength caused by decreased cellulose content and altered wall structure, and exhibits abnormalities in plant development. Map-based cloning revealed that all of the observed mutant phenotypes result from a missense mutation in a putative NST gene, Oryza sativa Nucleotide Sugar Transporter1 (OsNST1). OsNST1 was identified as a Golgi-localized transporter by analysis of a fluorescence-tagged OsNST1 expressed in rice protoplast cells and demonstration of UDP-glucose transport activity via uptake assays in yeast. Compositional sugar analyses in total and fractionated wall residues of wild-type and bc14 culms showed a deficiency in the synthesis of glucoconjugated polysaccharides in bc14, indicating that OsNST1 supplies the glucosyl substrate for the formation of matrix polysaccharides, and thereby modulates cellulose biosynthesis. OsNST1 is ubiquitously expressed, with high expression in mechanical tissues. The inferior mechanical strength and abnormal development of bc14 plants suggest that OsNST1 has pleiotropic effects on cell wall biosynthesis and plant growth. Identification of OsNST1 has improved our understanding of how cell wall polysaccharide synthesis is regulated by Golgi NSTs in plants.
Keywords: UDP-glucose transporter, cell-wall regulation, rice brittleness mutant
In plants, most of the carbon that is fixed by photosynthesis is incorporated into carbohydrates, glycoproteins, and glycolipids, which are predominantly synthesized inside organelles [e.g., the endoplasmic reticulum (ER) and Golgi lumen]. However, the substrates (nucleotide sugars) used in these processes are mainly produced in the cytosol (1–3). The use of distinct sites for synthesis of substrates and glycoconjugated products requires mechanisms for the transport of metabolites across biological membranes. Therefore, organisms have evolved nucleotide sugar transporter (NST) proteins encoded by a large gene family for translocation of substrates from the cytoplasm to the Golgi/ER lumen (4, 5).
NSTs, which are present in all eukaryotes, are composed of 300 to 350 amino acids with 6 to 10 transmembrane domains (TMDs). This important gene family is closely related to triose phosphate translocators (5). Since the first NST gene was cloned from yeast in 1996 (6), database searches in organisms with known genome sequences have identified a large number of putative NSTs. The Arabidopsis genome contains more than 40 NSTs (7). Such abundance correlates with the diversity of carbohydrates and the substrate specificity of individual NSTs that generally act as antiporters (8). The relationship between sequence identity and substrate specificity is weak, probably because the sequence motifs required for substrate recognition in NSTs are not well defined. This lack of knowledge is a significant obstacle to the functional characterization of NSTs (4, 5).
Although many plant genes have features of an NST sequence, few have been functionally characterized. The first biochemical evidence for the existence of NSTs in plants came from activity analyses using pea Golgi vesicles (9–11). Later, GONST1 was cloned from Arabidopsis and shown to encode a Golgi-localized GDP-mannose transporter by complementing a yeast mutant, vrg4-2, that is unable to form mannosylated proteins (12). Four additional proteins, GONST2 through GONST5, were identified as transporters that perform the same activity as GONST1 in Arabidopsis (13). AtUTr1, -2, and -3 are UDP-glucose/UDP-galactose transporters (14–16). Ultimately, more UDP-galactose transporters were found (7, 17), indicating that plants may use multiple substrate channels in the glycosylation processes (18). However, most of these studies were performed in vitro at the biochemical or molecular level. Our knowledge of the biological functions of NSTs in planta is extremely limited. Recently, AtUTr1 and AtUTr3, which cooperate in transporting UDP-glucose into the ER, have been reported to function in late pollen development and embryo sac progression through the characterization of atutr1 atutr3 double mutants (16).
The plant Golgi apparatus is an important organelle for cell wall matrix polysaccharide and glycoprotein production (19, 20). At this site, nucleotide sugars are added to specific polysaccharide acceptors by the corresponding glycosyltransferases (GTs). Many studies have revealed the importance of substrate availability in polysaccharide biosynthesis. For example, mur1, which is deficient in the synthesis of GDP-fucose, has a compromised xyloglucan structure because of the lack of a terminal fucose (2, 21). A mutation in RHM2/MUM4 perturbs the biosynthesis of UDP-rhamnose and decreases the rhamnogalacturonan I contents in the mucilage of mutant plants (22–24). NSTs are believed to supply the substrates for Golgi GTs. NSTs are thus a potential key point for the control of wall composition and structure (5). However, there is currently no evidence to support this hypothesis because Golgi NST mutants have not been reported previously.
The Golgi NST mutant brittle culm14 (bc14) harbors a mutation in Oryza sativa Nucleotide Sugar Transporter1 (OsNST1), which was cloned by a positional cloning approach and found to encode a Golgi-localized NST. OsNST1 was shown to possess UDP-glucose transport activity. A deficiency of glucosyl residues in the matrix polysaccharides of the mutant lends support to this recently defined transport function of OsNST1. In addition, the reduced cellulose content and impaired mechanical strength in bc14 plants suggest the importance of OsNST1 for cell wall biosynthesis. As bc14 is a previously undescribed Golgi NST mutant, our findings provide unique biological evidence for the functions of Golgi NSTs in cell wall formation and plant growth.
Results
Mutant bc14 Has Reduced Mechanical Strength and Abnormal Plant Growth.
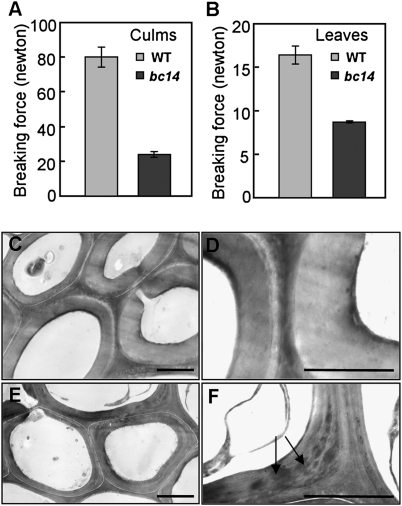
The mutant bc14 is one of our series of rice bc mutants that was isolated from a japonica cultivar, NE17. Easily broken culms and leaves are a major phenotype of mature bc14 plants. As shown in Fig. 1 A and B, the break force of culms and leaves in bc14 was reduced to ~30% and ~54% of the wild type, respectively, indicating that bc14 might have impaired cell wall structure or composition. Anatomical analysis of sclerenchyma cells in bc14 and wild-type culms revealed that the wall thickness of those cells was reduced in bc14 (Fig. S1 A and B). The cell size and length were not significantly different in the mutant culms (Fig. S1). The precise deficiency in sclerenchymatous walls was further analyzed by transmission electron microscopy. In addition to the reduction in wall thickness, bc14 had increased electron-dense materials deposited in the secondary walls (Fig. 1 E and F). In contrast, the wild-type walls were electron-translucent (Fig. 1 C and D). Taken together, these results suggest that the inferior mechanical strength observed in bc14 is the result of the reduced cell wall thickness and abnormal secondary cell wall.
Fig. 1.
Phenotypic observations in wild-type and bc14 plants. (A and B) Measurements of the breaking forces (newton) of culms (A) and leaves (B) (n = 3) ± SEM. (C–F) Transmission electron microscopy micrographs of wild-type (C and D) and bc14 (E and F) sclerenchyma cell walls. The increased electron-staining materials are indicated by arrows. (Scale bars, 0.5 μm.)
The bc14 mutants showed additional morphological abnormalities, including small stature that resulted from a weak growth tendency observed from the seedling to mature stages (Fig. S2 A to C), reduced fertility, and small seeds (Fig. S2D). The reduced seed size in bc14 caused the thousand-seeds-weight, a key unit for crop yield, to decrease by 50% (Fig. S2E). Therefore, the corresponding wild-type gene also participates in some basic processes of plant growth.
Map-Based Cloning of BC14/OsNST1.
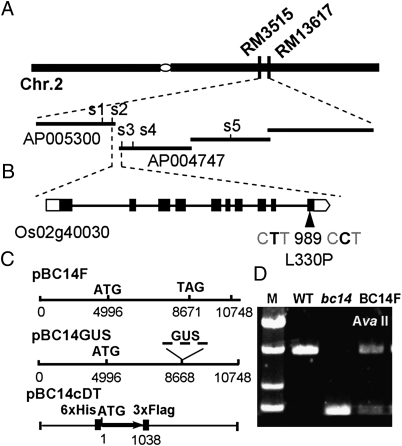
We used a map-based cloning approach to identify the gene responsible for the mutant phenotypes of bc14. Using 192 F2 mutant plants from a population generated by crossing bc14 with Kasalath (a wild-type polymorphic indica variety), the bc14 locus was mapped to an 815-kb region between two simple sequence repeat markers (RM3515 and RM13617) on chromosome 2q (Fig. 2A). The gene was further pinpointed within a 57-kb segment with simple sequence repeat markers (Table S1). This segment is situated at the ends of two contiguous BAC clones, AP005300 and AP004747. The 57-kb region contains six putative ORFs. After sequencing the six ORFs, a point mutation was found in the Os02g40030 ORF (Rice Genome Annotation Project, http://rice.plantbiology.msu.edu/) or the Os02g0614100 ORF [National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/] that led to a missense mutation at the 330th amino acid residue. This mutation changes leucine to a proline (Fig. 2B). The leucine at this position was found to be conserved in seven rice varieties (Fig. S3A). To identify BC14, an ~11-kb DNA fragment (pBC14F) that includes the entire ORF was introduced into bc14 mutant plants for a complementation assay (Fig. 2C). The genetic backgrounds of the transgenic plants were confirmed to be bc14 by the presence of a cleaved amplified polymorphic sequence (Fig. 2D). All of the transgenic plants had shown a wild-type appearance (Fig. S3 B and C). The mutant phenotypes, including brittleness (Fig. S3F), wall thickness (Fig. S3 G and H), and wall compositions (Tables 1 and 2), were fully rescued. We therefore concluded that we had cloned the corresponding wild-type gene.
Fig. 2.
Map-based cloning of the BC14 gene. (A) The bc14 locus was mapped to a 57-kb region on chromosome 2. (B) Structure of the BC14 gene. Lines represent introns and black boxes represent exons. (C) Constructs for complementation tests. (D) A cleaved amplified polymorphic sequence marker (digested with AvaII) used to distinguish between the backgrounds of the wild-type, bc14, and transgenic plants. M, DNA marker.
Table 1.
Monosaccharide compositional analysis of wall residues from the culms of wild-type and bc14 plants (μg mg−1 AIR)
| Residues | Rhamnose | Fucose | Arabinose | Xylose | Mannose | Galactose | Glucose | GalUA | GlcUA | Cellulose |
| WT | 3.5 ± 0.0* | 0.9 ± 0.0 | 86.2 ± 1.1* | 280.6 ± 1.8* | 0.4 ± 0.0* | 9.9 ± 0.3* | 45.2 ± 1.1* | 10.1 ± 0.3* | 6.5 ± 0.4* | 374.2 ± 5.6* |
| bc14 | 5.4 ± 0.3* | 1.0 ± 0.1 | 157.0 ± 8.0* | 400.4 ± 23.4* | 2.1 ± 0.1* | 15.1 ± 0.6* | 31.0 ± 1.1* | 16.6 ± 0.3* | 11.0 ± 1.2* | 234.9 ± 9.8* |
| pBC14F | 2.9 ± 0.1* | 0.8 ± 0.0 | 63.1 ± 0.9* | 249.3 ± 2.1* | 0.4 ± 0.0* | 10.3 ± 0.3* | 74.1 ± 1.6* | 10.8 ± 0.2* | 5.3 ± 0.0* | 361.5 ± 18.7* |
The mounts of monosaccharides are as determined by GC-MS analysis of trimethylsilane derivatives. Cellulose is as determined by Updegraff quantification after TFA hydrolysis.
*Significantly different (t test at P < 0.05) compared with wild type (n = 4) ± SD.
Table 2.
Monosaccharide compositional analysis of wall fractions derived from the culms of wild-type and bc14 plants (μg mg−1 AIR)
| Residue | Sample | Rhamnose | Fucose | Arabinose | Xylose | Mannose | Galactose | Glucose | Mass |
| Ammonium oxalate | WT | 0.1 ± 0.0 | ND | 0.3 ± 0.0 | 0.2 ± 0.0 | 0.1 ± 0.0 | 0.3 ± 0.0 | 2.0 ± 0.1* | 35.0 ± 2.0 |
| bc14 | 0.1 ± 0.0 | ND | 0.3 ± 0.0 | 0.4 ± 0.0 | 0.1 ± 0.0 | 0.3 ± 0.0 | 0.7 ± 0.0* | 37.4 ± 1.4 | |
| pBC14F | 0.1 ± 0.0 | ND | 0.3 ± 0.0 | 0.2 ± 0.0 | 0.1 ± 0.0 | 0.3 ± 0.0 | 3.0 ± 0.2* | 34.2 ± 3.1 | |
| 1 N KOH soluble | WT | 0.4 ± 0.0 | 0.2 ± 0.0 | 16.2 ± 0.6* | 120.7 ± 3.9* | 0.2 ± 0.0 | 4.6 ± 0.2 | 17.0 ± 0.6* | 289.8 ± 53.6 |
| bc14 | 0.5 ± 0.0 | 0.2 ± 0.0 | 30.6 ± 1.0* | 171.3 ± 5.5* | 0.5 ± 0.0 | 7.5 ± 0.3 | 6.9 ± 0.3* | 361.3 ± 7.6 | |
| pBC14F | 0.4 ± 0.0 | 0.2 ± 0.0 | 17.6 ± 0.3* | 124.4 ± 1.9* | 0.3 ± 0.0 | 5.0 ± 0.1 | 36.7 ± 0.6* | 273.2 ± 57.2 | |
| 4 N KOH soluble | WT | 0.2 ± 0.0 | 0.1 ± 0.0 | 4.5 ± 0.5* | 34.5 ± 3.5* | 0.1 ± 0.0 | 2.2 ± 0.2 | 9.3 ± 1.1* | 64.0 ± 4.4 |
| bc14 | 0.2 ± 0.0 | ND | 3.8 ± 0.3* | 20.0 ± 1.6* | 0.1 ± 0.0 | 1.7 ± 0.1 | 5.0 ± 0.3* | 50.4 ± 3.1 | |
| pBC14F | 0.2 ± 0.0 | 0.1 ± 0.0 | 3.5 ± 0.3* | 23.0 ± 1.9 | 0.1 ± 0.0 | 1.9 ± 0.2 | 11.4 ± 1.0* | 58.0 ± 4.0 | |
| 4 N KOH insoluble | WT | ND | ND | 0.4 ± 0.0 | 1.9 ± 0.1 | ND | 0.2 ± 0.0 | 9.9 ± 0.5* | ND |
| bc14 | 0.1 ± 0.0 | ND | 0.7 ± 0.0 | 2.1 ± 0.1 | 0.1 ± 0.0 | 0.4 ± 0.0 | 7.7 ± 0.2* | ND | |
| pBC14F | ND | ND | 0.4 ± 0.0 | 1.8 ± 0.1 | 0.0 ± 0.0 | 0.3 ± 0.0 | 10.2 ± 0.3* | ND |
ND, not detected. The amounts of monosaccharides are determined by GC-MS analysis of alditol acetate derivatives.
*Significantly different (t test at P < 0.05) compared with WT (n = 4) ± SD.
A Pfam search showed that this gene encodes a protein belonging to the triose phosphate translocator family (pfam03151) with an E-value of 5.8e-11. BC14 was defined as a putative NST by a search in the Conserved Domain Database of NCBI. Its full-length cDNA spans 1,438 nt, with a coding sequence (CDS) of 1,041 nt, as determined by the Knowledge-based Oryza Molecular Biological Encyclopedia (http://cdna01.dna.affrc.go.jp/cDNA/). The deduced BC14 protein has 346 amino acids and eight TMDs (Fig. S4), which is consistent with the common features of NSTs (i.e., 300–350 amino acids in length and 6–10 TMDs). We also fused the CDS of BC14 to FLAG and polyhistidine tags and transformed the bc14 mutant plants with the resulting construct (Fig. 2C). The completely rescued mutant phenotypes indicate that this CDS is functional (Fig. S3 D and I). A BLASTP search in the NCBI identified several BC14 homologs in plants (~60% sequence identity) and animals (~30% sequence identity). Given that phylogenetic analysis can provide information about a gene's evolution or function, a few activity-identified plant NSTs were selected to build an unrooted tree with BC14 and its homologs. BC14 was joined together with its homologs, including some activity-known animal NSTs, but it was separated into a different clade from the activity-identified plant NSTs (Fig. S5A), suggesting that BC14 is a putative NST and that its activity or evolution route may be different from that of the identified plant NSTs. BC14 was thus named Oryza sativa Nucleotide Sugar Transporter1 (OsNST1). Moreover, the point mutation of OsNST1 occurs at the end of the last putative transmembrane domain (Fig. S4). We analyzed the residue of this site in OsNST1 and its homologs from different plants and animals and found that an amino acid residue with a bulky hydrophobic side chain was present in each instance (Fig. S5B), suggesting that this type of side chain is important for the function of these NSTs.
OsNST1 Is Golgi-Localized and Transports UDP-Glucose.
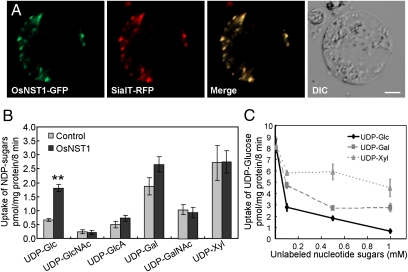
The subcellular localization of NSTs is highly associated with their biological function. No signal peptide was identified in OsNST1 by either the TargetP or the SignalP prediction servers, but OsNST1 was predicted to be a nonclassic, secreted protein by SecretomeP (http://www.cbs.dtu.dk/services/). To ascertain its subcellular location, we fused the GFP to the C terminus of OsNST1 and coexpressed this fusion protein with red fluorescent protein-tagged Golgi marker (sialytransferase-RFP) in rice protoplast cells. The OsNST1 fusion showed a dot-like pattern, which was identical to the RFP signals of this Golgi marker (Fig. 3A). A similar colocalization pattern was detected by coexpressing N-terminal GFP tagged OsNST1 with another Golgi marker, Man49-mCherry, in rice protoplast cells (Fig. S6). These data suggest that OsNST1 is a Golgi-localized protein.
Fig. 3.
OsNST1 is a Golgi-localized UDP-glucose transporter. (A) Rice protoplast cells coexpressing OsNST1-GFP (C-terminal tagging) with sialyltransferase (SialT)-RFP. (Scale bar, 5 μm.) (B) The nucleotide sugar uptake assay carried out in the Golgi-rich fraction of yeast expressing OsNST1 or empty vector (control) (n ≥ 2) ± SE. **Significant difference (t test at P ≤ 0.01). (C) The uptake competitive assay carried out in the Golgi-rich fraction of yeast expressing OsNST1 (n ≥ 2) ± SE.
Next, we sought to determine the substrates that are transported by OsNST1. Yeast mutant complementation assays were carried out in three yeast NST mutants: Δyea4 [UDP-GlcNAc transporter mutant (25)], vrg4-2 [GDP-mannose transporter mutant (26)], and Δhut1 [UDP-galactose transporter mutant (27)]. Expressing OsNST1 in the three yeast NST mutants, respectively (Fig. S7A), OsNST1 did not noticeably rescue the growth of all of the mutant strains to a wild-type level in the pharmaceutical assay (Fig. S8). We further performed uptake assays in yeast bearing the inducible pYES2 vector. The Golgi-rich vesicle fractions prepared from yeasts expressing His/Flag-OsNST1 or the empty vector (control) were used in the uptake assay (Fig. S7B). Vesicle preparations from individual transformed yeast were found to be up to 95% latent (Table S2), indicating that most of these vesicles had the proper membrane topographical orientation and were suitable for the transport assay. After incubation with several isotope-labeled nucleotide sugars, the abundance of UDP-glucose incorporated into the yeast fraction expressing OsNST1 was nearly twofold greater than that of the control (Fig. 3B). With the exception of an approximate 42% greater uptake activity for UDP-galactose, the activities for other substrates were indistinguishable from those of the control (Fig. 3B). Moreover, the uptake of radiolabeled UDP-glucose was greatly inhibited when the unlabeled UDP-glucose concentrations exceeded 10 μM (Fig. 3C). Such great inhibition was not observed in the presence of excess unlabeled UDP-xylose or UDP-galactose (Fig. 3C). These data indicate that OsNST1 transports UDP-glucose.
Mutation in OsNST1 Causes a Deficiency in the Synthesis of Glucoconjugated Polysaccharides and Cellulose.
Given the evidence indicating that OsNST1 transports UDP-glucose, a deficiency should be detected in cell wall residues of bc14 culms. We examined the wall composition of bc14 and wild-type culms by GC-MS analyses. The contents of glucose and cellulose were decreased by ~31% and ~37% in bc14, respectively, whereas the remaining monosaccharides were significantly increased in bc14 (Table 1). To determine whether the glucose deficiency could be ascribed to a specific wall polymer, we solubilized the wall residues into pectic and hemicellulosic fractions and determined the monosaccharide compositions. Interestingly, the reduction in glucose content was found in all fractions (Table 2), indicating that glucoconjugated polysaccharide synthesis is widely impaired in bc14. The levels of xylose and arabinose, the two major sugars of arabinoxylan, as well as galactose and mannose, were significantly higher in the 1 N KOH fraction of bc14 (Table 2).
These data indicate that mutation in OsNST1 causes a decrease in the glucose content of glucoconjugated polymers and cellulose, providing direct support for its role in transporting UDP-glucose.
OsNST1 Is Broadly Expressed.
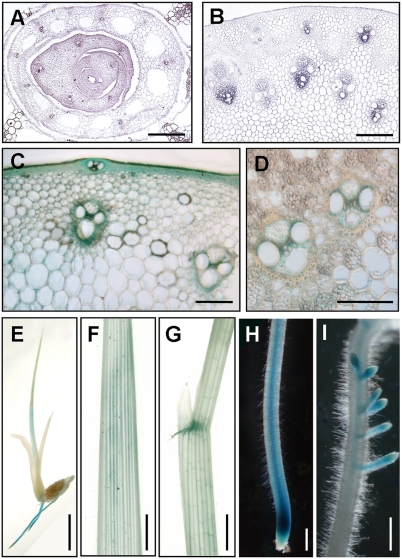
The expression pattern of OsNST1 at the cellular level was investigated by RNA in situ hybridization. Expression signals were detected in the vascular bundles of leaves and leaf sheaths (Fig. 4A) and young stems (Fig. 4B), in agreement with the inferior mechanical properties described above for bc14 mutants. To investigate its expression at the protein level, the GUS gene was inserted into the OsNST1 ORF, just before the stop codon (BC14-GUS), and this construct (pBC14GUS) (Fig. 2C) was introduced into bc14 mutant plants. The fully rescued mutant phenotypes of the resulting transgenic plants indicate that BC14-GUS retains the function of native OsNST1 protein (Fig. S3 E and J). Consistent with the expression pattern described above, GUS staining signals were observed in the sclerenchyma cells (Fig. 4C) and vascular bundles (Fig. 4 C and D) of T0 transgenic plants and were also present in the T1 transgenic plants, as evinced in the seedlings (Fig. 4E), leaves (Fig. 4F), leaf lamina (Fig. 4G), and veins of primary and lateral roots (Fig. 4 H and I). Therefore, OsNST1 was found to be expressed in many organs.
Fig. 4.
OsNST1 expression analysis. (A and B) RNA in situ hybridization of the leaf sheaths (A) and young stems (B). (Scale bars, 70 μm.) (C and D) GUS staining of the culms of 3-mo-old T0 transgenic plants. (Scale bars, 60 μm.) (E–I) GUS staining of 10-d-old T1 transgenic seedlings, showing GUS signals in a whole seedling (E), a leaf (F), a leaf lamina (G), and veins of primary and lateral roots (H and I). (Scale bars, 2 mm.)
Discussion
NSTs form a large family of proteins that are represented in all plants; however, only a few members have been characterized, and most studies have been carried out in vitro at the biochemical level (12–16). An important function of NSTs is to supply substrates for GTs in the synthesis of wall matrix polysaccharides and glycoproteins. Without available mutants, the effects of NSTs on cell wall formation had remained unclear. Here, we provide genetic and biochemical evidence for the function of a Golgi NST in the regulation of cell wall biosynthesis and plant growth through characterization of the bc14 mutant and its corresponding gene.
OsNST1 was cloned by a map-based cloning approach and encodes a hydrophobic protein composed of 346 amino acids with eight TMDs that was identified as a putative NST. Further studies verified that OsNST1 is targeted to the Golgi apparatus and that it transports UDP-glucose. In plants, a few UDP-glucose transporters have been reported, but they perform diverse functions. This functional discrepancy depends on the target organelle and their mechanistic role. AtUTr1 and AtUTr3, which transport UDP-glucose into the ER lumen, affect protein folding and pollen and embryo sac development (16). Although AtUTr3 also localizes to the Golgi apparatus, it is considered a short-time resident; ER localization is its steady-state pattern (16). A bacterial-type ABC transporter formed by STAR1 and STAR2 also transports UDP-glucose and contributes to aluminum tolerance in rice plants, but this transporter is localized in an unknown vesicular compartment (28). OsNST1, which was identified here, resides in the Golgi apparatus and has the same transport activity according to our in vitro biochemical assay. Cell wall compositional analyses indicate that it is involved in glucoconjugated polysaccharide synthesis.
Although the uptake assay clearly revealed that OsNST1 transports UDP-glucose, the in vitro transport assay showed weak uptake activity for UDP-galactose. The amount of inhibition of UDP-glucose uptake by UDP-galactose was between that observed for UDP-glucose, which is transported by OsNST1, and UDP-xylose, which is not transported by OsNST1. These results indicate that OsNST1 is possibly involved in transporting UDP-galactose. Several UDP-galactose transporters have been identified in plants (7, 14, 15, 17). AtUTr1 has both UDP-galactose and UDP-glucose transport activity (14). We suggest that the main function of OsNST1 is to transport UDP-glucose because its weak UDP-galactose transport activity seems insufficient to complement the growth of the yeast mutant, Δhut1, which is deficient in a UDP-galactose transporter. This hypothesis was further supported by the increase of galactose in bc14 wall residues. Given that plants have more than one transporter participating in UDP-galactose supply (7, 14, 15, 17), it seems that the increased amount of galactose observed in bc14 mutant walls results from the activity of these UDP-galactose transporters. It is also possible that the weak activity of OsNST1 in the transport of UDP-galactose is an artifact of the in vitro experiment because the structures of UDP-glucose and UDP-galactose are similar or because of other unknown reasons. Importantly, this activity was not supported by the yeast mutant complementation assay or the wall compositional analyses.
NST mutants are quite important for the evaluation of NST functions in vivo. GONST1, a GDP-mannose transporter, was the first Golgi-localized NST reported in plants (12). However, without a relevant mutant, its effect on glycoconjugates remains unclear. Thus, bc14 harbors a missense mutation in OsNST1 that had provided us with a tremendous opportunity to investigate the intrinsic functions of a Golgi NST in cell wall biosynthesis and plant development. NSTs generally work together with themselves or with GTs. An interesting question should be addressed in future is whether the replacement of leucine by a proline, the most rigid amino acid, in bc14 interferes with its transport activity or the interactions with other proteins. Consistent with the transport activity of OsNST1, the wall compositional analyses of total and fractionated wall residues revealed a glucose deficiency in bc14. Although the glucose value analyzed here cannot be completely interpreted as noncellulosic glucose, many glucosyl residues were derived from matrix polysaccharides, as indicated by the fractionation composition assay. Therefore, as a Golgi localized transporter, OsNST1 appears to form an important channel for the supply of UDP-glucose to the Golgi lumen during the synthesis of glucoconjugated polysaccharides. Glycan synthesis is an ordered process in the Golgi apparatus (19). OsNST1 may supply UDP-glucose for several GTs because glucosyl residues are common in many matrix polysaccharides (19). This finding was further substantiated by the decreased glucose content observed in several wall polymers of the mutant plants and the expression pattern of OsNST1. The reduced cellulose content is another important defect resulting from the bc14 mutation. Cellulose biosynthesis occurs in the plasma membrane using cytosolic UDP-glucose as a substrate (29). Therefore, the reduction of cellulose abundance in bc14 may be an indirect effect caused by the inferior synthesis of glucoconjugated matrix polysaccharides. Many studies have revealed that a deficiency in the synthesis of matrix polysaccharides or glycoproteins often results in aberrant assembly of nascent cellulose microfibrils, which could, in turn, impede cellulose synthesis (30–34). Another possibility to explain the reduced cellulose content in bc14 might be that OsNST1 is required for the synthesis of unique oligosaccharide sequences that may serve as primer for cellulose formation (35). However, this hypothesis should be backed up in further studies. In mutant plants, the reduced cellulose content was accompanied by the increases in the content of other cell wall components, including nonglucosidic neutral sugars and uronic acids. All of these changes cause abnormal wall structure and reduced mechanical strength in rice plants. As shown by several abnormal growth and developmental phenotypes, OsNST1 has pleiotropic effects on cell wall structure and plant development, suggesting its important roles in the monocot plant, rice.
Materials and Methods
Details of the plant material, growth conditions, testing of the breaking force of culms and leaves, microscopy, and bioinformatics are described in SI Materials and Methods.
Map-Based Cloning.
The bc14 locus was mapped and cloned using 192 mutant F2 plants obtained from the mapping population with the molecular markers listed in Table S1. The complementation assays were performed as described in the SI Materials and Methods.
Subcellular Localization and Transport Assay.
Cotransformation was performed in rice protoplasts as previously described (33) (see SI Materials and Methods). The complementation assay of yeast NST mutants, latency determination of yeast Golgi vesicles, and nucleotide sugar uptake and competitive assays in yeast are described in SI Materials and Methods.
Chemical Analysis of Wall Composition.
The second internodes of wild-type and mutant plants were used to prepare destarched alcohol insoluble residues (AIRs) of cell walls (33). The alditol acetate and trimethylsilane derivatives were prepared from AIRs (36), and then analyzed by GC-MS to measure the neutral sugars and uronic acids (for details, see SI Materials and Methods). Sequential fractionation of the destarched AIR and the analysis of monosaccharide composition were carried out as described in SI Materials and Methods. Crystalline cellulose was measured with a modified Updegraff method, as previously described (37) (for details, see SI Materials and Methods).
Gene Expression.
RNA in situ hybridization was performed as previously described (38). The GUS activity assays were conducted in T0 and T1 transgenic plants expressing pBC14GUS, as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Markus Pauly for the suggestions and helps on wall chemical analyses, Taihua Zhang for measurement of the breaking force of rice plants, and Neta Dean for kindly providing the yeast NDY5 strain. This work was supported by Grant 2008ZX08009-003 from the Ministry of Agriculture of China for transgenic research and Grant KSCX2-YW-G-033 from the Knowledge Innovation Program of the Chinese Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016144108/-/DCSupplemental.
References
- 1.Coates SW, Gurney T, Jr., Sommers LW, Yeh M, Hirschberg CB. Subcellular localization of sugar nucleotide synthetases. J Biol Chem. 1980;255:9225–9229. [PubMed] [Google Scholar]
- 2.Bonin CP, Potter I, Vanzin GF, Reiter WD. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc Natl Acad Sci USA. 1997;94:2085–2090. doi: 10.1073/pnas.94.5.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seifert GJ. Nucleotide sugar interconversions and cell wall biosynthesis: How to bring the inside to the outside. Curr Opin Plant Biol. 2004;7:277–284. doi: 10.1016/j.pbi.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Handford M, Rodriguez-Furlán C, Orellana A. Nucleotide-sugar transporters: Structure, function and roles in vivo. Braz J Med Biol Res. 2006;39:1149–1158. doi: 10.1590/s0100-879x2006000900002. [DOI] [PubMed] [Google Scholar]
- 5.Reyes F, Orellana A. Golgi transporters: Opening the gate to cell wall polysaccharide biosynthesis. Curr Opin Plant Biol. 2008;11:244–251. doi: 10.1016/j.pbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Abeijon C, Robbins PW, Hirschberg CB. Molecular cloning of the Golgi apparatus uridine diphosphate-N-acetylglucosamine transporter from Kluyveromyces lactis. Proc Natl Acad Sci USA. 1996;93:5963–5968. doi: 10.1073/pnas.93.12.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakker H, et al. Molecular cloning of two Arabidopsis UDP-galactose transporters by complementation of a deficient Chinese hamster ovary cell line. Glycobiology. 2005;15:193–201. doi: 10.1093/glycob/cwh159. [DOI] [PubMed] [Google Scholar]
- 8.Capasso JM, Hirschberg CB. Mechanisms of glycosylation and sulfation in the Golgi apparatus: Evidence for nucleotide sugar/nucleoside monophosphate and nucleotide sulfate/nucleoside monophosphate antiports in the Golgi apparatus membrane. Proc Natl Acad Sci USA. 1984;81:7051–7055. doi: 10.1073/pnas.81.22.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz P, Norambuena L, Orellana A. Evidence for a UDP-glucose transporter in Golgi apparatus-derived vesicles from pea and its possible role in polysaccharide biosynthesis. Plant Physiol. 1996;112:1585–1594. doi: 10.1104/pp.112.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neckelmann G, Orellana A. Metabolism of uridine 5′-diphosphate-glucose in golgi vesicles from pea stems. Plant Physiol. 1998;117:1007–1014. doi: 10.1104/pp.117.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wulff C, Norambuena L, Orellana A. GDP-fucose uptake into the Golgi apparatus during xyloglucan biosynthesis requires the activity of a transporter-like protein other than the UDP-glucose transporter. Plant Physiol. 2000;122:867–877. doi: 10.1104/pp.122.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baldwin TC, Handford MG, Yuseff MI, Orellana A, Dupree P. Identification and characterization of GONST1, a golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell. 2001;13:2283–2295. doi: 10.1105/tpc.010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handford MG, Sicilia F, Brandizzi F, Chung JH, Dupree P. Arabidopsis thaliana expresses multiple Golgi-localised nucleotide-sugar transporters related to GONST1. Mol Genet Genomics. 2004;272:397–410. doi: 10.1007/s00438-004-1071-z. [DOI] [PubMed] [Google Scholar]
- 14.Norambuena L, et al. Transport of UDP-galactose in plants. Identification and functional characterization of AtUTr1, an Arabidopsis thaliana UDP-galactos/UDP-glucose transporter. J Biol Chem. 2002;277:32923–32929. doi: 10.1074/jbc.M204081200. [DOI] [PubMed] [Google Scholar]
- 15.Norambuena L, et al. AtUTr2 is an Arabidopsis thaliana nucleotide sugar transporter located in the Golgi apparatus capable of transporting UDP-galactose. Planta. 2005;222:521–529. doi: 10.1007/s00425-005-1557-x. [DOI] [PubMed] [Google Scholar]
- 16.Reyes F, et al. The nucleotide sugar transporters AtUTr1 and AtUTr3 are required for the incorporation of UDP-glucose into the endoplasmic reticulum, are essential for pollen development and are needed for embryo sac progress in Arabidopsis thaliana. Plant J. 2010;61:423–435. doi: 10.1111/j.1365-313X.2009.04066.x. [DOI] [PubMed] [Google Scholar]
- 17.Rollwitz I, Santaella M, Hille D, Flügge UI, Fischer K. Characterization of AtNST-KT1, a novel UDP-galactose transporter from Arabidopsis thaliana. FEBS Lett. 2006;580:4246–4251. doi: 10.1016/j.febslet.2006.06.082. [DOI] [PubMed] [Google Scholar]
- 18.Seifert GJ, Barber C, Wells B, Dolan L, Roberts K. Galactose biosynthesis in Arabidopsis: Genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Curr Biol. 2002;12:1840–1845. doi: 10.1016/s0960-9822(02)01260-5. [DOI] [PubMed] [Google Scholar]
- 19.Lerouxel O, Cavalier DM, Liepman AH, Keegstra K. Biosynthesis of plant cell wall polysaccharides—A complex process. Curr Opin Plant Biol. 2006;9:621–630. doi: 10.1016/j.pbi.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Mohnen D. Pectin structure and biosynthesis. Curr Opin Plant Biol. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Zablackis E, et al. Substitution of L-fucose by L-galactose in cell walls of Arabidopsis mur1. Science. 1996;272:1808–1810. doi: 10.1126/science.272.5269.1808. [DOI] [PubMed] [Google Scholar]
- 22.Diet A, et al. The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell. 2006;18:1630–1641. doi: 10.1105/tpc.105.038653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Usadel B, Kuschinsky AM, Rosso MG, Eckermann N, Pauly M. RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol. 2004;134:286–295. doi: 10.1104/pp.103.034314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oka T, Nemoto T, Jigami Y. Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J Biol Chem. 2007;282:5389–5403. doi: 10.1074/jbc.M610196200. [DOI] [PubMed] [Google Scholar]
- 25.Roy SK, Chiba Y, Takeuchi M, Jigami Y. Characterization of yeast Yea4p, a uridine diphosphate-N-acetylglucosamine transporter localized in the endoplasmic reticulum and required for chitin synthesis. J Biol Chem. 2000;275:13580–13587. doi: 10.1074/jbc.275.18.13580. [DOI] [PubMed] [Google Scholar]
- 26.Dean N, Zhang YB, Poster JB. The VRG4 gene is required for GDP-mannose transport into the lumen of the Golgi in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1997;272:31908–31914. doi: 10.1074/jbc.272.50.31908. [DOI] [PubMed] [Google Scholar]
- 27.Fujimura H. The Saccharomyces cerevisiae MLF6/YPL244C gene, which encodes a possible yeast homologue of mammalian UDP-galactose transporter, confers resistance to the immunosuppressive drug, leflunomide. Curr Microbiol. 2001;43:93–95. doi: 10.1007/s002840010267. [DOI] [PubMed] [Google Scholar]
- 28.Huang CF, et al. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell. 2009;21:655–667. doi: 10.1105/tpc.108.064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 30.His I, Driouich A, Nicol F, Jauneau A, Höfte H. Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1,4-β-glucanase. Planta. 2001;212:348–358. doi: 10.1007/s004250000437. [DOI] [PubMed] [Google Scholar]
- 31.Persson S, et al. The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell. 2007;19:237–255. doi: 10.1105/tpc.106.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peña MJ, et al. Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell. 2007;19:549–563. doi: 10.1105/tpc.106.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, et al. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009;60:1055–1069. doi: 10.1111/j.1365-313X.2009.04022.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y, et al. BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.) Plant J. 2009;57:446–462. doi: 10.1111/j.1365-313X.2008.03703.x. [DOI] [PubMed] [Google Scholar]
- 35.Peng L, Kawagoe Y, Hogan P, Delmer D. Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science. 2002;295:147–150. doi: 10.1126/science.1064281. [DOI] [PubMed] [Google Scholar]
- 36.York WS, et al. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1985;118:3–40. [Google Scholar]
- 37.Updegraff DM. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, et al. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell. 2003;15:2020–2031. doi: 10.1105/tpc.011775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.