Abstract
The macrocyclic polyketides FK506, FK520, and rapamycin are potent immunosuppressants that prevent T-cell proliferation through initial binding to the immunophilin FKBP12. Analogs of these molecules are of considerable interest as therapeutics in both metastatic and inflammatory disease. For these polyketides the starter unit for chain assembly is (4R,5R)-4,5-dihydroxycyclohex-1-enecarboxylic acid derived from the shikimate pathway. We show here that the first committed step in its formation is hydrolysis of chorismate to form (4R,5R)-4,5-dihydroxycyclohexa-1,5-dienecarboxylic acid. This chorismatase activity is encoded by fkbO in the FK506 and FK520 biosynthetic gene clusters, and by rapK in the rapamycin gene cluster of Streptomyces hygroscopicus. Purified recombinant FkbO (from FK520) efficiently catalyzed the chorismatase reaction in vitro, as judged by HPLC-MS and NMR analysis. Complementation using fkbO from either the FK506 or the FK520 gene cluster of a strain of S. hygroscopicus specifically deleted in rapK (BIOT-4010) restored rapamycin production, as did supplementation with (4R,5R)-4,5-dihydroxycyclohexa-1,5-dienecarboxylic acid. Although BIOT-4010 produced no rapamycin, it did produce low levels of BC325, a rapamycin analog containing a 3-hydroxybenzoate starter unit. This led us to identify the rapK homolog hyg5 as encoding a chorismatase/3-hydroxybenzoate synthase. Similar enzymes in other bacteria include the product of the bra8 gene from the pathway to the terpenoid natural product brasilicardin. Expression of either hyg5 or bra8 in BIOT-4010 led to increased levels of BC325. Also, purified Hyg5 catalyzed the predicted conversion of chorismate into 3-hydroxybenzoate. FkbO, RapK, Hyg5, and Bra8 are thus founder members of a previously unrecognized family of enzymes acting on chorismate.
Keywords: calcineurin, mTOR
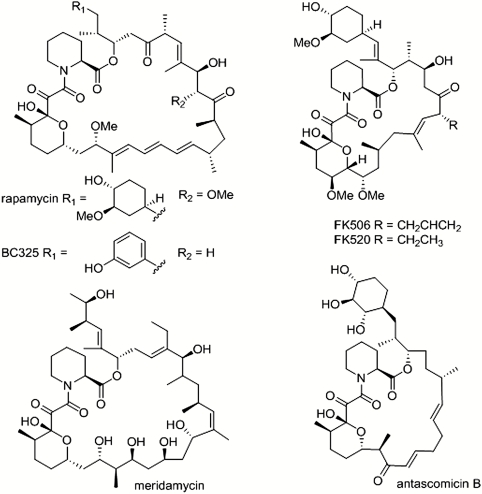
FK506, FK520 (ascomycin), and rapamycin are macrocyclic amino acid-linked polyketides isolated, respectively, from Streptomyces tsukubaensis, Streptomyces hygroscopicus subsp. ascomyceticus, and Streptomyces hygroscopicus. Both FK506 and rapamycin are important therapeutics in immunosuppression and in combating inflammatory disease. Since their discovery, additional related compounds [meridamycin (1) and antascomicin (2), which are not immunosuppressive] have been reported having in common the unusual masked tricarbonyl pipecolic acid moiety that terminates the polyketide chain (Fig. 1). This is the key structural determinant for binding to the immunophilins such as FKBP12 (3), which triggers inhibition of specific signaling pathways. They also contain (except for meridamycin) a highly substituted cyclohexyl group known to originate from the shikimic acid pathway (4). The immunosuppressive mechanisms of rapamycin and FK506/FK520 are distinct. The FK506/FK520-FKBP12 complex inhibits calcineurin. Calcineurin is a phosphatase required for the dephosphorylation of nuclear factor of activated T cells (NFAT), which then translocates to the nucleus and stimulates the release of IL-2 and other downstream cytokines. The rapamycin-FKBP12 complex inhibits the kinase function of mTOR (mammalian target of rapamycin), specifically only as part of mTOR complex 1 (mTORC1), blocking its ability to phosphorylate the substrates S6 kinase 1 (S6K1) and eIF4E-binding protein 1 (4E-BP1) (5). This leads to the inhibition of ribosome biogenesis and mRNA translation initiation and progression among other effects. Rapamycin and analogs have also found clinical application as the pharmacological component of drug-eluting stents in order to prevent restenosis (6). The selective inhibition of mTOR by the rapamycin-FKBP12 complex in cancer cells underlies the current clinical use of semisynthetic analogs of rapamycin against several malignancies including advanced renal cancer (5, 7). FK506 and rapamycin analogs also show potential therapeutic value against stroke (8), nerve degeneration (9), Parkinson disease (10), and Alzheimer’s disease (11, 12). Rapamycin has recently been shown to extend the average life span of genetically heterogeneous mice, even when fed late in life (13, 14).
Fig. 1.
Structures of immunophilin-binding polyketides.
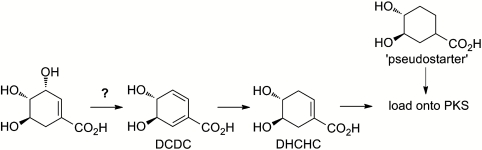
DNA sequencing of the biosynthetic gene cluster for rapamycin (15–17) provided important evidence for the biosynthetic origin of its key structural features: A nonribosomal peptide synthetase-like multienzyme (18) acts together with a modular polyketide synthase to produce the tricarbonyl pipecolate and to close the macrocycle, whereas (1R,3R,4R)-3,4-dihydroxycyclohexanecarboxylic acid appears to serve as the starter unit for polyketide chain synthesis (19). The true precursor of the starter unit was later shown, using isotope labeling and competition feeding experiments, to be (4R,5R)-4,5-dihydroxycyclohex-1-enecarboxylic acid (DHCHC) (Fig. 2), which is activated by an ATP-dependent adenylation domain in the loading module of the polyketide synthase, transferred to the acyl carrier protein domain of this module, and then reduced in situ by an adjacent enoylreductase domain (20). The route to DHCHC from shikimate remains unknown, but it has been suggested, based on extensive isotope labeling and heterologous gene expression analysis of related pathways in other bacteria (21–25), to involve the pathway shown in Fig. 2. Shikimic acid is first converted to (4R,5R)-4,5-dihydroxycyclohexa-1,5-dienecarboxylic acid (DCDC) by 1,4-conjugate elimination of water, after which the α,β-double bond is reduced in a syn fashion before undergoing a suprafacial 1,3-allylic rearrangement to give DHCHC.
Fig. 2.
Previously proposed pathway from shikimate to the rapamycin/FK506/FK520 starter unit DHCHC.
Initial comparison of the biosynthetic gene clusters for rapamycin (15–17), FK506 (26, 27), and FK520 (28) did not reveal obvious candidate genes encoding the enzymes of such a pathway. However, detailed genetic analysis has since revealed an essential role for rapK in the production of DHCHC during rapamycin biosynthesis (29). RapK was initially annotated as a putative pteridine-dependent dioxygenase involved in late-stage oxidation at C-9 of rapamycin (15, 16). However, deletion of rapK wholly abolished rapamycin production, and production was restored either by genetic complementation via ectopic expression with rapK (29) or by a close homolog of rapK, the fkbO gene from the FK520 gene cluster (29). Production was also restored by supplementation with the racemic pseudostarter unit (1R∗,3R∗,4R∗)-3,4-dihydroxycyclohexanecarboxylic acid (Fig. 2) (30), which presumably bypasses the normal in situ enoylreduction on the polyketide synthase loading module. These findings gave a strong hint that RapK/FkbO was an enzyme involved in the starter unit pathway, but left open the possibility that RapK might be a regulatory protein controlling this pathway (30).
We now provide evidence that FkbO and RapK are actually chorismate hydrolases, yielding DCDC (Fig. 2). We have also found that other previously obscure FkbO- and RapK-related enzymes catalyze a related unique reaction in which chorismate is converted into 3-hydroxybenzoate (3HBA), for subsequent incorporation into diverse natural products. All these enzymes are founder members of a previously undescribed family of enzymes acting on chorismate. They are also structurally related, in their C-terminal domain, to the large and enigmatic YjgF/YER057c/UK1114 protein superfamily, none of whose members has previously been ascribed an enzymatic function (31).
Results and Discussion
Chemical Complementation of a rapK-Deficient Mutant Strain of S. hygroscopicus.
BIOT-3410 is a higher-producing derivative of the rapamycin-producing strain of S. hygroscopicus NRRL5491, and BIOT-4010 is a mutant of BIOT-3410 in which rapK has been specifically deleted. Chemical analysis showed that BIOT-4010 produced no rapamycin, but upon addition of racemic synthetic pseudostarter unit (1R∗,3R∗,4R∗)-3,4-dihydroxycyclohexanecarboxylic acid (30) or the analog cyclohexanecarboxylic acid (30, 32) production of rapamycin or rapamycin analog production was fully restored (Fig. S1). DCDC has been shown to be an intermediate of DHCHC biosynthesis (20). To test whether its formation is before or after the action of RapK, DCDC was added (2 mM) to growing cultures of BIOT-4010, 24 h after their inoculation. Under these fermentation conditions, rapamycin production was restored to the mutant at titers [203.6 ± 44.5 mg/L (n = 3)] essentially identical to those from the control strain BIOT-3410 [251.8 ± 13.0 mg/L (n = 3)] (Fig. S1). These results clearly established DCDC as a competent precursor of the rapamycin starter unit and strongly suggested that RapK catalyzes the first committed step of DHCHC biosynthesis.
The C-terminal 125 amino acid residues of RapK or FkbO are strongly predicted, using the PHYRE (Protein Homology/analogY Recognition Engine) protein fold recognition program (www.phyre.com) (33), to adopt the distinctive protein fold of members of the YjgF/YER057c/UK1114 family, a fold known to be shared by chorismate mutase. This hint of a structural link to an enzyme utilizing chorismate prompted us to examine the possibility that RapK and FkbO catalyze a chorismatase reaction, releasing pyruvate from chorismate to yield DCDC in a single step.
Genetic Complementation of a rapK-Deficient Mutant Strain.
The fkbO gene from either the FK506-producing strain S. tsukubaensis or the FK520-producing strain S. hygroscopicus subsp. ascomyceticus was cloned into the integrative vector pGP9 and expressed ectopically in BIOT-4010 (SI Text), and recombinant clones were tested for rapamycin production. In both cases, rapamycin production was restored (Fig. S2), showing that FkbO and RapK have an identical function in their respective pathways. An initial experimental test of the idea that the proximal source of DCDC might be chorismate was suggested by the known ability of the isochorismatase EntB from Escherichia coli [which normally catalyzes synthesis of 2,3-dihydroxybenzoic acid during enterobactin biosynthesis (34)] to act as a slow chorismatase when supplied instead with chorismate (35). We utilized instead of entB the gene mxcF from Stigmatella aurantiaca Sga15, which encodes both an acyl carrier protein domain (not relevant here) and an EntB-like isochorismatase activity required for myxochelin biosynthesis (36, 37). The MxcF expression vector pSGK217 was introduced into BIOT-4010, and multiple resulting isolates were screened for production of rapamycin (SI Text). In all these recombinant clones, rapamycin was produced at low but significant levels (4.7 ± 1.5 mg/L (n = 15), the highest titer observed being 6.7 mg/L) (Fig. S2). These results are consistent with the idea that DCDC synthesis had been reinstated as a result of chorismatase activity. Rapamycin production was not fully restored to the titers of the parent strain, but this can be rationalized on the basis of previous work, which has shown convincingly that for EntB to be effective as a chorismatase in vivo, competing chorismate-utilizing pathways must be deleted (38, 39).
In Vitro Assays of FkbO as a Chorismatase Whose Product Is DCDC.
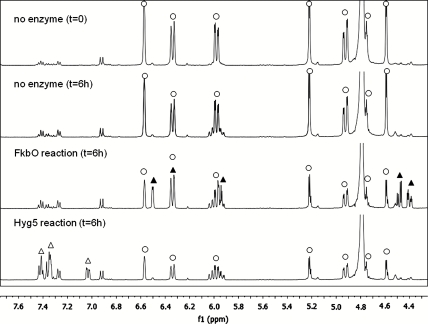
To test directly the ability of FkbO and similar proteins to act as chorismatases, the fkbO gene was cloned from the FK520 gene cluster of S. hygroscopicus subsp. ascomyceticus (fkbO520), and the protein was overexpressed in E. coli and purified (SI Text and Fig. S3). Chorismate was incubated in a buffer with purified FkbO520. Liquid chromatography (LC)-MS of reaction mixtures and comparison with standards showed that the product was DCDC (SI Text and Fig. S4). To confirm the identity of the product, 1H NMR was used to monitor the appearance of peaks corresponding to DCDC (SI Text and Fig. 3). After 6 h, the spectrum contained new sets of signals identical to those found for authentic DCDC.
Fig. 3.
NMR spectra of metabolites produced by the action of FkbO and Hyg5 on chorismate. Peaks were assigned to chorismate (O), DCDC (▴), or to 3-hydroxybenzoate (∆). See SI Text and Table S1 for detailed assignments. Other minor peaks arise from known degradation products of chorismate.
For determination of kinetic parameters, the FkbO520-catalyzed reaction was monitored spectrophotometrically by coupling pyruvate production to its NADH-linked reduction to lactate (35) (SI Text and Fig. S5). The kcat (17.6 ± 0.7 s-1) and the Km value (0.2 ± 0.03 mM) for chorismate for this enzyme compare very favorably to the kcat (approximately 21 s-1) and the Km (≥37 mM) previously reported for the chorismatase side activity of EntB isochorismatase (35).
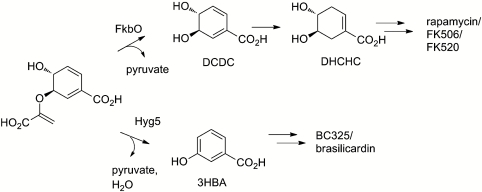
These results make it very likely that FkbO and the closely similar RapK proteins are true chorismatases and that DCDC for macrocyclic immunosuppressant biosynthesis is generated as shown in Fig. 4, rather than by the route shown in Fig. 2.
Fig. 4.
The role of FkbO and Hyg5 in polyketide starter unit production from chorismate.
A Rapamycin Analog (BC325) Derived from an Aromatic Starter Unit.
Although no detectable rapamycins derived from the DHCHC starter acid were produced by BIOT-4010, we consistently observed the low level production of a previously unnoticed rapamycin analog [3 ± 1 mg/L (n = 21); data accumulated from seven independent experiments] (Fig. S6). The previously undescribed compound (BC325, Fig. 1) was isolated and characterized by spectroscopic methods (SI Text and Tables S2 and S3). Surprisingly, the structure of BC325 suggested that it was derived from a 3HBA starter unit. In support of this, addition of 3HBA to growing BIOT-4010 (1 mM at 24 h after inoculation) increased the production of BC325 more than 10-fold [45.3 ± 2.3 mg/L (n = 3) vs. 2.9 ± 0.1 mg/L (n = 3)] (Fig. S6). Addition of benzoic acid, in contrast, had no effect [3.5 ± 0.1 mg/L (n = 3) vs. 3.5 ± 0.2 mg/L (n = 3)]. To probe the origin of 3HBA, d5-sodium benzoate and [1,7-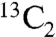 ]shikimic acid were separately fed to BIOT-4010. LC-MS analysis of the rapamycin produced showed efficient (> 50%) incorporation of the 1,7-
]shikimic acid were separately fed to BIOT-4010. LC-MS analysis of the rapamycin produced showed efficient (> 50%) incorporation of the 1,7- label, leading to an enhanced +2 amu peak, and intact incorporation of the 1,7-13C unit at C36 and C37 of rapamycin was subsequently verified by 13C-NMR (SI Text and Fig. S7). In contrast, very poor incorporation of label was observed with d5-sodium benzoate. These data argue in favor of direct production of 3HBA by a variant of the shikimic acid pathway, instead of via decarboxylation of phenylalanine to benzoate and subsequent 3-hydroxylation, because by the latter route the 7-13C label would have been lost during the decarboxylation of prephenate.
label, leading to an enhanced +2 amu peak, and intact incorporation of the 1,7-13C unit at C36 and C37 of rapamycin was subsequently verified by 13C-NMR (SI Text and Fig. S7). In contrast, very poor incorporation of label was observed with d5-sodium benzoate. These data argue in favor of direct production of 3HBA by a variant of the shikimic acid pathway, instead of via decarboxylation of phenylalanine to benzoate and subsequent 3-hydroxylation, because by the latter route the 7-13C label would have been lost during the decarboxylation of prephenate.
Phylogenetic Analysis of FkbO/RapK Reveals a Superfamily of Related Proteins Potentially Acting as Chorismatases.
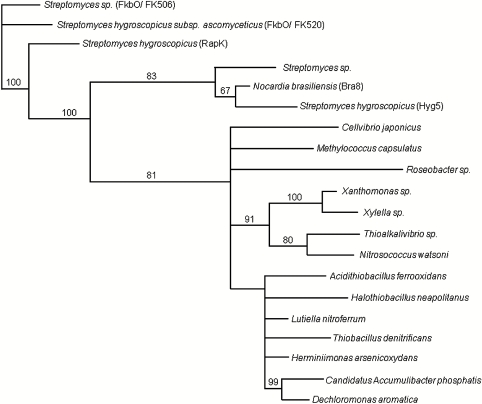
Comparison using BLAST (40) and ClustalW (41) of the sequences of FkbO and RapK with those of proteins in public databases showed that they belong to a large group of bacterial proteins of unknown function (Fig. 5). This analysis identified as their closest relatives a group of three mutually very similar proteins: an unidentified predicted protein from a Streptomyces species; another predicted protein (Hyg5) from the rapamycin-producing strain (42), and, most tellingly, a protein (Bra8) from Nocardia brasiliensis IFM 0406 encoded within the biosynthetic gene cluster for the glycosylated diterpene natural product brasiliocardin (43). Brasilicardin contains a 3HBA moiety. Bra8 was provisionally assigned an oxidative function by these authors (43). However, our recognition that RapK and FkbO are chorismatases suggests an alternative role for Bra8 in generating 3HBA directly from chorismate (Fig. 4).
Fig. 5.
Phylogenetic analysis of FkbO and RapK: Similar sequences (BLAST algorithm) were aligned with ClustalW, and the tree shown was generated using the Geneious software (53).
The existence of Bra8 focused our attention on Hyg5, encoded by a gene in an uncharacterized biosynthetic gene cluster (hyg) of the rapamycin-producing strain (42). Because the rapK mutant strain BIOT-4010 makes no rapamycins, it had been assumed that Hyg5 was either not expressed, or nonfunctional. Prompted by our discovery of BC325, and the identification of bra8, we hypothesized that Hyg5 and Bra8 represent another branch of the chorismatase family, comprising enzymes hydrolyzing chorismate to form 3HBA.
Hyg5 and Bra8 from the FkbO/RapK Family Are 3HBA Synthases Acting on Chorismate.
The gene encoding Hyg5 was cloned from S. hygroscopicus and the protein was overexpressed in E. coli and purified (SI Text and Fig. S3). Chorismate was incubated with purified Hyg5. LC-MS analysis of reaction mixtures and comparison with authentic standards showed that the product was 3HBA, not DCDC (SI Text and Fig. S8). To confirm the identity of the product, 1H NMR was used to monitor the appearance of peaks corresponding to 3HBA (SI Text and Fig. 3). After 6 h, the spectrum, when compared to the control without Hyg5 contained a set of signals identical to those for authentic 3HBA. The spectrum showed no evidence for the formation of DCDC.
For determination of kinetic parameters, the Hyg5-catalyzed reaction was monitored spectrophotometrically as for FkbO (35) (SI Text and Fig. S5). The kcat (22.4 ± 0.8 s-1) and the Km value (0.53 ± 0.1 mM) for chorismate show that Hyg5 is as efficient as FkbO in its action on chorismate. The purified Hyg5 (and to a lesser extent FkbO) used in these studies contained several minor protein contaminants, as revealed by SDS-PAGE analysis (SI Text and Fig. S3), which may mean that the kcat values reported here are slight underestimates. Nevertheless, taken together, these results using purified Hyg5 fully confirm the hypothesis that Hyg5 (and by extension Bra8) are chorismatases/3HBA synthases. They also bolster the conclusion that DCDC for macrocyclic immunosuppressant biosynthesis is produced by the route shown in Fig. 4, rather than by the previously proposed 1,4-conjugate elimination of water from shikimate (Fig. 2). In light of the present findings, it would be worthwhile to investigate the in vitro activity of those enzymes thought to catalyze the latter reaction (20–25) to obtain definitive evidence for or against their acting as chorismatases, rather than shikimate dehydratases.
The function of those proteins more distantly phylogenetically related to FkbO/RapK (Fig. 5) remains to be determined, but on the available evidence they are also likely to act as chorismatases/3HBA synthases. For example, the FkbO/RapK homolog in Xanthomonas species (the causative agent of black rot in plants) resides in a gene cluster (44) for the biosynthesis of xanthomonadins, yellow brominated aryl pigments apparently derived via the polyketide pathway from a 3HBA starter unit (45). A block in aroE in the early part of the shikimate pathway abolishes xanthomonadin biosynthesis (46). Deletion of the gene for the FkbO/RapK-like protein abolishes both pigment production and extracellular polysaccharide production dependent on a diffusible factor (47). Because these authors had relied on early annotation of FkbO/RapK-like proteins as pteridine-dependent dioxygenases, the recognition of these proteins as potential 3HBA synthases should lead to a useful reevaluation of their role in Xanthomonas and related phytopathogenic bacteria (48).
Ectopic Expression of Putative Chorismatases Hyg5 and Bra8 Increases BC325 Production.
Even if Hyg5 is normally expressed in the rapamycin producer, its failure to complement the rapK deletion in BIOT-4010 can be rationalized: It converts chorismate into 3HBA, not DCDC. However, 3HBA is a (poor) substrate for the loading module of the rapamycin polyketide synthase, giving rise to BC325. To test this, we cloned hyg5 and bra8 separately into the pGP9 expression vector and introduced the resulting plasmids (pSGK260 and pSGK259, respectively) into BIOT-4010 to give BIOT-4545 and BIOT-4538, respectively (SI Text). Multiple isolates were then selected, grown, and analyzed for production of rapamycin and BC325. None of these clones produced rapamycin, but several showed increased levels of BC325 production (up to 14.3 mg/L for hyg5, and 5.2 mg/L for bra8) (Fig. S6). When the analysis was repeated on the best-performing isolates, it showed an 8.3 ± 0.3-fold (n = 8) increase for Hyg5 and a 3.1 ± 0.7-fold (n = 8) increase for Bra8 in BC325 production, compared to BIOT-4010 (n = 4). These data clearly show that ectopic expression of Hyg5 or Bra8 leads to the enhanced production of BC325 in BIOT-4010, supporting their function as 3HBA synthases.
Conclusions
Chorismate has an impressive number of known alternative metabolic fates in bacteria (23, 49, 50): Chorismate mutase produces prephenate for tyrosine and phenylalanine formation and for secondary metabolite biosynthesis; anthranilate synthase produces 2-aminobenzoate for tryptophan and phenazine biosynthesis; salicylate synthase produces 2-hydroxybenzoate for yersiniabactin and mycobactin synthesis; 4-amino-4-deoxychorismate synthase produces 4-amino-4-deoxychorismate leading to secondary metabolites and to 4-aminobenzoic acid for folate biosynthesis; 2-amino-2-deoxychorismate synthase produces 2-amino-2-deoxychorismate for the benzoxazinolate moiety of the enediyne C-1027; isochorismate synthase produces isochorismate for both menaquinone and siderophore biosynthesis; and chorismate pyruvate-lyase produces 4-hydroxybenzoic acid, an essential intermediate in ubiquinone biosynthesis. The results presented here add two more reactions to this list, for the chorismatases we have identified catalyze one of two distinct reactions: either the simple hydrolysis of chorismate to form DCDC (FkbO, RapK) or the hydrolysis and concomitant dehydration of chorismate to form 3HBA (Hyg5, Bra8). The mechanistic details of these conversions, and the active site structural features that dictate which of these two pathways is followed, remain to be elucidated and will be the subject of further investigation. Meanwhile, the phylogenetic analysis has indicated the likely enzymatic activity of a large number of bacterial proteins of previously unknown function. The C-terminal domain of enzymes of the FkbO/RapK protein family also bears a strong structural resemblance to the YjgF/YER057c/UK1114 protein superfamily, highly conserved among eubacteria, archaea, and eukaryotes. X-ray crystal structures have been solved for several of these proteins (31), but their biochemical function remains ill-defined. The assignment of FkbO and RapK as chorismatases, and of Hyg5 and Bra8 as chorismatase/3HBA synthases, should encourage the functional characterization of other members of this superfamily.
Materials and Methods
A summary of experimental techniques is given here, with full methods presented in SI Text.
Construction of Plasmids for Heterologous Expression of FkbO520 and Hyg5.
Genes for FkbO520 (28) and Hyg5 (42) were amplified by PCR from genomic DNA isolated from S. hygroscopicus var. ascomyceticus and S. hygroscopicus, respectively. The products were digested with the appropriate restriction endonucleases and ligated into pET28a. The resulting plasmids (pET28a-FkbO and pET28a-Hyg5) were verified by sequencing.
Heterologous Expression and Purification of FkbO520 and Hyg5.
Cells were grown in inoculate LB medium supplemented with kanamycin and chloramphenicol at 37 °C/300 rpm. These cells were used to inoculate 2× tryptone-yeast extract medium (1 L) containing the same supplements and grown at 30 °C/200 rpm to an A600 of 0.4–0.6., then induced with IPTG (0.1 mM) and grown for 5 h before harvesting by centrifugation. Cells were broken by sonication on ice using a Mysonix Incorporated Sonicator ® Ultrasonic Processor XL2020 sonicator, clarified, and the resulting extract was purified over a Ni-NTA column (Fig. S3).
FkbO520 and Hyg5 in Vitro Assays.
NMR assays (1 mL total volume): deuterated potassium phosphate buffer (20 mM, pD 7) containing chorismate (10 mg/mL, 44.2 mM) and enzyme (0.5 mg/mL) at 20 °C. The samples were analyzed immediately and after 6 h by 1H NMR. An experiment without enzyme served as control to monitor chorismate degradation. LC-MS assays (1 mL total volume): potassium phosphate buffer (100 mM, pH 7), chorismate (1 mM), and enzyme (100 μg/mL) at 20 °C for 2 h. Spectrophotometric assay: the resulting pyruvate was determined spectrophotometrically based on a LDH (lactate dehydrogenase) coupled assay described elsewhere (35). Various amounts of chorismate (0–4 mM) were preincubated in potassium phosphate buffer (100 mM) containing lactate dehydrogenase (LDH) (2.5 U/mL; Sigma) and NADH (0.5 mM). Kinetic data (Fig. S5) were fitted according to the Michaelis–Menten model using Origin software (ORIGIN® 7G, OriginLab Corporation).
Genetic Manipulation, Fermentation, and Analysis of S. hygroscopicus Mutants.
Plasmid DNA was introduced into S. hygroscopicus by conjugal transfer as described previously (51). pGP9 is an E. coli-actinomycete shuttle plasmid shown to be effective for gene expression in S. hygroscopicus (52). Fermentation was performed in 50 mL Falcon tubes (8 mL working volume) or Braun 22 L Bioreactors (15 L working volume) as described previously (51). Feeding experiments were performed routinely in 50 mL Falcon tubes, with the required carboxylic acid feed being added after 24 h growth in production media.
Fermentation, Isolation, and Structural Characterization of BC325.
BIOT-4010 was grown in a Braun 22 L bioreactor (×2) and subsequent processing was performed as described previously (51). The resulting material (13.0 g) was suspended in a methanol∶water mix (80∶20; 300 mL) and extracted twice with hexane (300 mL). The methanol∶water fraction was concentrated to yield a crude oil (8.2 g; 17 mg BC325). This was purified using standard chromatographic methods, and its structure was solved by MS and NMR.
Supplementary Material
Acknowledgments.
We thank Professors Heinz G. Floss (University of Washington, Seattle, WA), Donald Hilvert [Eidgenössische Technische Hochschule (ETH) Zürich, Zurich, Switzerland], Peter Kast (ETH Zürich, Zurich, Switzerland), Taifo Mahmud (Oregon State University, Corvallis, OR), and Rolf Müller (Saarland University, Saarbrüken, Germany) for the kind provision of materials. We also thank Drs. Frank Hahn and Fanglu Huang for helpful discussions. This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/C519597/1 (to P.F.L.), a German Academic Exchange Service postdoctoral fellowship (to J.N.A.), and an ERASMUS studentship (to A.-S.Z.).
Footnotes
Conflict of interest statement: S.G.K., M.A.G., C.J.M., S.J.M., P.F.L., and B.W. hold Biotica shares. F.E.K., J.S.S., and G.T.C. hold Pfizer shares.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015773108/-/DCSupplemental.
References
- 1.Salituro GM, et al. Meridamycin: A novel nonimmunosuppresive FKBP12 ligand from Streptomyces hygroscopicus. Tetrahedron Lett. 1995;36:997–1000. [Google Scholar]
- 2.Fehr T, et al. Antascomicins A, B, C, D and E. Novel FKBP12 binding compounds from a Micromonospora strain. J Antibiot. 1996;49:230–233. doi: 10.7164/antibiotics.49.230. [DOI] [PubMed] [Google Scholar]
- 3.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 4.Paiva NL, Roberts MF, Demain AL. The cyclohexane moiety of rapamycin is derived from shikimic acid in Streptomyces hygroscopicus. J Ind Microbiol. 1993;12:423–428. [Google Scholar]
- 5.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slavin L, Chhabra A, Tobis JM. Drug-eluting stents: Preventing restenosis. Cardiol Rev. 2007;15:1–12. doi: 10.1097/01.crd.0000200844.16899.fc. [DOI] [PubMed] [Google Scholar]
- 7.Vignot S, Faivre S, Aguire D, Raymond E. mTOR-targeted therapy of cancer with rapamycin derivatives. Ann Oncol. 2005;16:525–537. doi: 10.1093/annonc/mdi113. [DOI] [PubMed] [Google Scholar]
- 8.Ruan B, et al. Binding of rapamycin analogs to calcium channels and FKBP52 contributes to their neuroprotective activities. Proc Natl Acad Sci USA. 2008;105:33–38. doi: 10.1073/pnas.0710424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klettner A, Herdegen T. FK506 and its analogs—Therapeutic potential for neurological disorders. Curr Drug Targets CNS Neurol Disord. 2003;2:153–162. doi: 10.2174/1568007033482878. [DOI] [PubMed] [Google Scholar]
- 10.Tain LS, et al. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci. 2009;12:1129–1135. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spilman P, et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS One. 2010;5(4):e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caccamo A, Majumder S, Richardson A, Strong R, Oddo S. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and tau: Effects on cognitive impairments. J Biol Chem. 2010;285:13107–13120. doi: 10.1074/jbc.M110.100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller RA, et al. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerentol A Biol Sci Med Sci. 2011;66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwecke T, et al. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aparicio JF, et al. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: Analysis of the enzymatic domains in the modular polyketide synthase. Gene. 1996;169:9–16. doi: 10.1016/0378-1119(95)00800-4. [DOI] [PubMed] [Google Scholar]
- 17.Molnár I, et al. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: Analysis of genes flanking the polyketide synthase. Gene. 1996;169:1–7. doi: 10.1016/0378-1119(95)00799-7. [DOI] [PubMed] [Google Scholar]
- 18.König A, et al. The pipecolate-incorporating enzyme for the biosynthesis of the immunosuppressant rapamycin: Nucleotide sequence analysis, disruption and heterologous expression of rapP from Streptomyces hygroscopicus. Eur J Biochem. 1997;247:526–534. doi: 10.1111/j.1432-1033.1997.00526.x. [DOI] [PubMed] [Google Scholar]
- 19.Lowden PAS, Böhm GA, Staunton J, Leadlay PF. The nature of the starter unit for the rapamycin polyketide synthase. Angew Chem Int Edit. 1996;35:2249–2251. [PubMed] [Google Scholar]
- 20.Lowden PAS, et al. Origin and true nature of the starter unit for the rapamycin polyketide synthase. Angew Chem Int Edit. 2001;40:777–779. [PubMed] [Google Scholar]
- 21.Wallace KK, et al. Biosynthetic studies of ascomycin (FK520): Formation of the (1R,3R,4R)-3,4-dihydroxycyclohexanecarboxylic acid-derived moiety. J Am Chem Soc. 1994;116:11600–11601. [Google Scholar]
- 22.Reynolds KA, et al. Biosynthesis of the shikimate-derived starter unit of the immunosuppressant ascomycin: Stereochemistry of the 1,4-conjugate elimination. J Antibiot. 1997;50:701–703. doi: 10.7164/antibiotics.50.701. [DOI] [PubMed] [Google Scholar]
- 23.Floss HG. Natural products derived from unusual variants of the shikimate pathway. Nat Prod Rep. 1997;14:433–452. doi: 10.1039/np9971400433. [DOI] [PubMed] [Google Scholar]
- 24.Chen S, et al. Biosynthesis of ansatrienin (mycotrienin) and napthomycin. Identification and analysis of two separate biosynthetic gene clusters in Streptomyces collinus Tü1892. Eur J Biochem. 1999;261:98–107. doi: 10.1046/j.1432-1327.1999.00244.x. [DOI] [PubMed] [Google Scholar]
- 25.Cropp TA, Wilson DJ, Reynolds KA. Identification of a cyclohexylcarbonyl CoA biosynthetic gene cluster and application in the production of doramectin. Nat Biotechnol. 2000;18:980–983. doi: 10.1038/79479. [DOI] [PubMed] [Google Scholar]
- 26.Motamedi H, Cai SJ, Shafiee A, Elliston KO. Structural organization of a multifunctional polyketide synthase involved in the biosynthesis of the macrolide immunosuppressant FK506. Eur J Biochem. 1997;244:74–80. doi: 10.1111/j.1432-1033.1997.00074.x. [DOI] [PubMed] [Google Scholar]
- 27.Motamedi H, Shafiee A. The biosynthetic gene cluster for the macrolactone ring of the immunosuppressant FK506. Eur J Biochem. 1998;256:528–534. doi: 10.1046/j.1432-1327.1998.2560528.x. [DOI] [PubMed] [Google Scholar]
- 28.Wu K, Chung L, Revill WP, Katz L, Reeves CD. The FK520 gene cluster of Streptomyces hygroscopicus var ascomyceticus (ATCC 4891) contains genes for biosynthesis of unusual polyketide extender units. Gene. 2000;251:81–90. doi: 10.1016/s0378-1119(00)00171-2. [DOI] [PubMed] [Google Scholar]
- 29.Gregory MA, et al. Isolation and characterization of pre-rapamycin, the first macrocyclic intermediate in the biosynthesis of the immunosuppressant rapamycin by S. hygroscopicus. Angew Chem Int Edit. 2004;43:2551–2553. doi: 10.1002/anie.200453764. [DOI] [PubMed] [Google Scholar]
- 30.Gregory MA, et al. Mutasynthesis of rapamycin analogues through the manipulation of a gene governing starter unit biosynthesis. Angew Chem Int Edit. 2005;44:4757–4760. doi: 10.1002/anie.200462784. [DOI] [PubMed] [Google Scholar]
- 31.Burman JD, Stevenson CE, Sawers RG, Lawson DM. The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057/UK114 family. BMS Struct Biol. 2007;7:30. doi: 10.1186/1472-6807-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowden PAS, Böhn GA, Metcalfe S, Staunton J, Leadlay PF. New rapamycins by precursor-directed biosynthesis. ChemBioChem. 2004;5:535–538. doi: 10.1002/cbic.200300758. [DOI] [PubMed] [Google Scholar]
- 33.Kelley LA, Sternberg MJE. Protein structure prediction on the web: A case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 34.Walsh CT, Liu J, Rusnak F, Sakaitani M. Molecular studies on enzymes in chorismate metabolism and the enterobactin biosynthetic pathway. Chem Rev. 1990;90:1105–1129. [Google Scholar]
- 35.Rusnak F, Liu J, Quinn N, Berchtold GA, Walsh CT. Subcloning of the enterobactin biosynthetic gene entB: Expression, purification, characterization, and substrate specificity of isochorismatase. Biochemistry. 1990;29:1425–1435. doi: 10.1021/bi00458a013. [DOI] [PubMed] [Google Scholar]
- 36.Silakowski B, et al. The myxochelin iron transport regulon of the myxobacterium Stigmatella aurantiaca Sg a15. Eur J Biochem. 2000;267:6476–6485. doi: 10.1046/j.1432-1327.2000.01740.x. [DOI] [PubMed] [Google Scholar]
- 37.Gaitatzis N, Kunze B, Müller R. In vitro reconstitution of the myxochelin biosynthetic machinery of Stigmatella aurantiaca Sg a15: Biochemical characterization of a reductive release mechanism from nonribosomal peptide synthetases. Proc Natl Acad Sci USA. 2001;98:11136–11141. doi: 10.1073/pnas.201167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller R, Breuer M, Wagener A, Schmidt K, Leistner E. Bacterial production of trans-dihydroxycyclohexadiene carboxylates by metabolic pathway engineering. Microbiology. 1996;142:1005–1012. doi: 10.1099/00221287-142-4-1005. [DOI] [PubMed] [Google Scholar]
- 39.Franke D, et al. (S,S)-2,3-Dihydroxy-2,3-dihydrobenzoic acid: Microbial access with engineered cells of Escherichia coli and application as starting material in natural-product synthesis. Chemistry. 2003;9:4188–4196. doi: 10.1002/chem.200204265. [DOI] [PubMed] [Google Scholar]
- 40.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson JD, Higgins W, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruan X, Stassi D, Lax SA, Katz L. A second type-I PKS gene cluster isolated from Streptomyces hygroscopicus ATCC 29253, a rapamycin-producing strain. Gene. 1997;203:1–9. doi: 10.1016/s0378-1119(97)00450-2. [DOI] [PubMed] [Google Scholar]
- 43.Hayashi Y, et al. Cloning of the gene cluster responsible for the biosynthesis of brasilicardin A, a unique diterpenoid. J Antibiot. 2008;61:164–174. doi: 10.1038/ja.2008.126. [DOI] [PubMed] [Google Scholar]
- 44.Poplawsky AR, Urban SC, Chun W. Biological role of xanthomonadin pigments in Xanthomonas campestris pv. campestris. Appl Environ Microb. 2000;66:5123–5127. doi: 10.1128/aem.66.12.5123-5127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starr MP, Jenkins CL, Bussey LB, Andrewes AG. Chemotaxonomic significance of the xanthomonadins, novel brominated aryl-polyene pigments produced by bacteria of the genus Xanthomonas. Arch Microbiol. 1977;113:1–9. doi: 10.1007/BF00428572. [DOI] [PubMed] [Google Scholar]
- 46.Goel AK, Rajagopal L, Sonti RV. Pigment and virulence deficiencies associated with mutations in the aroE gene of Xanthomonas oryzae pv. oryzae. Appl Environ Microbiol. 2001;67:245–250. doi: 10.1128/AEM.67.1.245-250.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poplawsky AR, Walters DM, Rouviere PE, Chun W. A gene for a dioxygenase-like protein determines the production of the DF signal in Xanthomonas campestris pv. Campestris. Mol Plant Pathol. 2005;6:653–657. doi: 10.1111/j.1364-3703.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 48.He Y-W, Zhang L-H. Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol Rev. 2008;32:842–857. doi: 10.1111/j.1574-6976.2008.00120.x. [DOI] [PubMed] [Google Scholar]
- 49.Dosselaere F, Vanderleyden J. A metabolic node in action: Chorismate-utilizing enzymes in microorganisms. Crit Rev Microbiol. 2001;27:75–131. doi: 10.1080/20014091096710. [DOI] [PubMed] [Google Scholar]
- 50.Van Lanen SG, Lin S, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027 involves a new branching point in chorismate metabolism. Proc Natl Acad Sci USA. 2008;105:494–499. doi: 10.1073/pnas.0708750105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gregory MA, et al. Rapamycin biosynthesis: Elucidation of gene product function. Org Biomol Chem. 2006;4:3565–3568. doi: 10.1039/b608813a. [DOI] [PubMed] [Google Scholar]
- 52.Kuščer E, et al. Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J Bacteriol. 2007;189:4756–4763. doi: 10.1128/JB.00129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drummond AJ, et al. Geneious v5.1. 2010. available at http://www.geneious.com.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.