Abstract
MicroRNA-122 (miR-122) is believed to stimulate hepatitis C virus (HCV) replication through interaction with two adjacent sites downstream of stem loop I (SLI) within the HCV 5′ untranslated region (5′ UTR). Recently, it was demonstrated that locked nucleic acid SPC3649-induced miR-122 antagonism suppressed HCV genotype 1a and 1b infection in vivo. However, virus-producing culture systems with 5′ UTR of different HCV genotypes have not been available for testing 5′ UTR-based treatment approaches. Using JFH1-based Core-NS2 genotype recombinants, we developed 5′ UTR-NS2 recombinants of HCV genotypes 1a, 1b, 2a, 2b, 3a, 4a, 5a, and 6a with efficient growth in Huh7.5 cells. Deletion mutagenesis studies demonstrated that the 5′ UTR SLI was essential for genotypes 1–6 infection. However, lack of SLI could be compensated for by insertion of other structured HCV or host RNA sequences, including U3 small nucleolar RNA. We demonstrated that SPC3649-induced miR-122 antagonism had a potent antiviral effect against HCV genotypes 1–6 5′ UTR-NS2 viruses. Strikingly, HCV recombinant virus with substitution of SLI and miR-122 binding site 1 (S1) by the U3 RNA sequence was not affected by miR-122 antagonism; this was attributed to the lack of an intact S1 by reverse genetics studies. Therefore, we engineered the corresponding U3 RNA sequences into S1 and demonstrated that HCV recombinants with wild-type SLI and single or combined mutations at four of eight nucleotides of S1 were viable in Huh7.5 cells. These mutations reduced the efficacy of SPC3649 treatment, indicating that escape variants to miR-122 antagonism-based HCV therapy could potentially occur.
Keywords: cell culture infection, locked nucleic acid therapy, miRNA, RNA recombination
Hepatitis C virus (HCV) infects ~180 million people worldwide, often causing fatal chronic liver diseases. HCV is a ~9.6-kb positive-sense RNA virus belonging to the genus Hepacivirus in the Flaviviridae family (1), which has been classified into seven major genotypes and numerous subtypes (2, 3). The viral genome consists of a single ORF flanked by 5′ and 3′ UTRs. The 5′ UTR consists of ~341 nucleotides forming four major domains. Domain I [nucleotides 1–43, numbering according to reference isolate H77 (GenBank accession no. AF009606)] contains one stem-loop structure (SLI), essential for RNA replication (1), and two microRNA-122 (miR-122) binding sites (Fig. S1), through which liver-abundant miR-122 stimulates HCV replication and translation (4–6). Although the 5′ UTR is a conserved region, a number of genotype-specific nucleotides naturally exist, which have been used for HCV genotyping (3). To date, genotype-specific functional analysis of the HCV 5′ UTR has been hampered by lack of suitable culture systems.
Current therapy with pegylated IFN-α and ribavirin has severe side effects and the outcome is dependent on the infecting HCV genotype. Overall, ~50% of patients completing treatment are cured. Thus, therapeutic drugs that are more effective against diverse HCV genotypes are urgently needed. Recently, miR-122 antagonism induced by 2′O-ribose–methylated miR-122 antisense oligonucleotides was found to inhibit HCV genotype 1a RNA replication and genotype 2a infection in vitro (5–7). Treatment with miR-122 antisense locked nucleic acid (LNA) SPC3649 efficiently suppressed HCV genotype 1a and 1b infections in chimpanzees, with no evidence of viral resistance or side effects (8), thus strengthening the therapeutic potential of miR-122 antagonism. Because the miR-122 binding sites are highly conserved across HCV genotypes, we aimed at testing the efficacy of miR-122 antagonism for HCV genotypes 1–6 infection.
Following development of the HCV genotype 2a JFH1 culture system (9) and a JFH1-based Core-NS2 recombinant with HCV genotype 2a (strain J6) (10), our group developed JFH1-based HCV genotypes 1–7 Core-NS2 recombinants with robust growth in Huh7.5 cells (2). However, these recombinants contained the 5′ UTR from JFH1. Here we developed efficient JFH1-based culture systems containing genotype-specific 5′ UTR-NS2 for HCV genotypes 1a, 1b, 2a, 2b, 3a, 4a, 5a, and 6a, and used these systems for functional analysis of the 5′ UTR domain I and for testing the antiviral effect of miR-122 antagonism.
Results
JFH1-Based HCV Culture Systems with Genotypes 1–6 Specific 5′ UTR, Core, E1, E2, p7, and NS2 (5′ UTR-NS2).
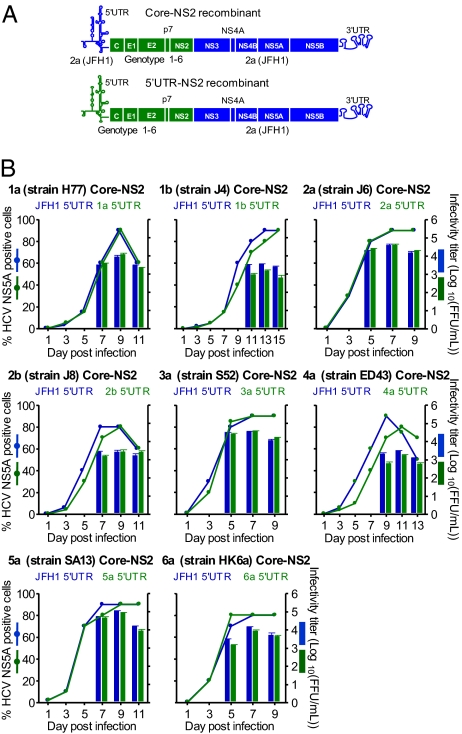
We previously found that JFH1-based genotypes 1–6 Core-NS2 recombinants with (genotypes 1a, 1b, 3a, 4a, 5a, and 6a) and without (2a and 2b) adaptive mutations efficiently produced infectious virus particles in Huh7.5 cells (2). Here, by replacing the JFH1 5′ UTR of these recombinants with the strain-specific 5′ UTR, we have developed unique HCV recombinants with genotypes 1–6 specific 5′ UTR-NS2 (Fig. 1A). After transfection of Huh7.5 cells with RNA transcripts, the 5′ UTR-NS2 recombinants spread efficiently in culture and produced HCV infectivity titers comparable to the corresponding virus with JFH1 5′ UTR. Comparative growth kinetic studies of transfection-derived viruses further showed that spread kinetics, as well as peak intra- and extracellular HCV infectivity and HCV RNA titers, of the 5′ UTR-NS2 recombinant viruses were comparable to that of the corresponding viruses with JFH1 5′ UTR (Fig. 1B and Table S1). Thus, these JFH1-based HCV recombinants with genotypes 1–6 specific 5′ UTR-NS2 apparently were fully functional in Huh7.5 cells.
Fig. 1.
Robust JFH1-based HCV culture systems with genotypes 1–6 specific 5′ UTR-NS2. (A) The JFH1 5′ UTR of previously developed genotypes 1–6 Core-NS2 recombinants (2) was replaced with strain-specific 5′ UTR to generate genotypes 1–6 specific 5′ UTR-NS2 recombinants. (B) Naïve Huh7.5 cells were infected with transfection-derived 5′ UTR-NS2 viruses (green) and the corresponding Core-NS2 viruses (blue); multiplicity of infection was 0.003 focus-forming units (FFU) per cell. The percentage of HCV-infected cells (Left y axis) and the peak HCV infectivity titers (mean ± SEM of triplicates; Right y axis) are shown.
Sequence analysis of the full-length genome of the genotypes 1–6 5′ UTR-NS2 viruses recovered from cultures of the growth kinetic studies (Fig. 1B) revealed that no mutations were acquired in the ORF and 3′ UTR (Table S2). The 5′ UTR sequences were also maintained, except the 5′-terminal G of genotypes 1a, 1b, 2b, 3a, and 6a 5′ UTR-NS2 viruses that in all cases changed to A. The 5′-terminal A of the 2a, 4a, and 5a 5′ UTR-NS2 viruses was maintained, whereas the G inserted immediately upstream of the 5′-terminal A to facilitate in vitro transcription was deleted (Table S2). Thus, all recovered JFH1-based 5′ UTR-NS2 genotype viruses had A at the 5′ terminus.
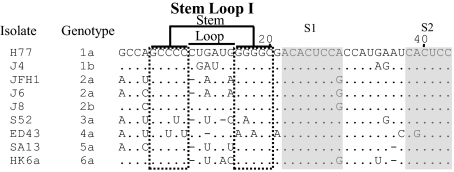
Because only the 5′-terminal sequence of the 1a (strain H77) 5′ UTR was previously determined (11), here we determined the 5′-terminal sequences of the prototype genotype strains used in development of respective 5′ UTR-NS2 recombinants, by sequence analysis of viruses recovered from plasma pools of experimentally infected chimpanzees (12) (Fig. 2 and Fig. S1). We found that genotypes 1b, 2a, 2b, 4a, 5a, and 6a 5′ UTR sequences were identical to those used in respective 5′ UTR-NS2 recombinants. However, the prototype 3a strain S52 had A at the 5′ terminus, whereas G was used in the 3a 5′ UTR-NS2 recombinant. Thus, the change from G to A in the recovered 3a 5′ UTR-NS2 virus might represent a reversion.
Fig. 2.
Identification of the 5′-terminal sequence of HCV prototype isolates of genotypes 1–6 from chimpanzee plasma pools. The entire 5′ UTR sequence of HCV prototype genotypes 1b, 2a, 2b, 3a, 4a, 5a, and 6a isolates was determined in this study (Fig. S1 and SI Materials and Methods). The sequence of domain I of the 5′ UTR (nucleotides 1–43) is shown and numbered according to H77 isolate (GenBank accession no. AF009606) (11). Dots indicate sequences identical to H77; dashes indicate gaps. The sequences comprising the SLI structures are indicated; S1 and S2 are shaded. The JFH1 5′ UTR (AB047639) (9) was included for comparison.
Next, we examined whether A at the 5′ terminus was also required for in vivo infection following intrahepatic transfection of chimpanzees with transcribed authentic HCV genomes. We thus analyzed the 5′ UTR sequence of HCV infectious clones of genotypes 1a (strain H77) (13) and 4a (ED43) (14) using acute phase sera of transfected chimpanzees. In contrast to JFH1-based 5′ UTR-NS2 genotype viruses recovered in vitro, the 5′-terminal G was maintained in the authentic 1a virus in vivo. The G inserted immediately upstream of the 5′-terminal A of the authentic 4a genome was deleted, in line with in vitro observations.
5′ UTR SLI Structure Was Essential for Production of HCV Genotypes 1–6 in Huh7.5 Cells.
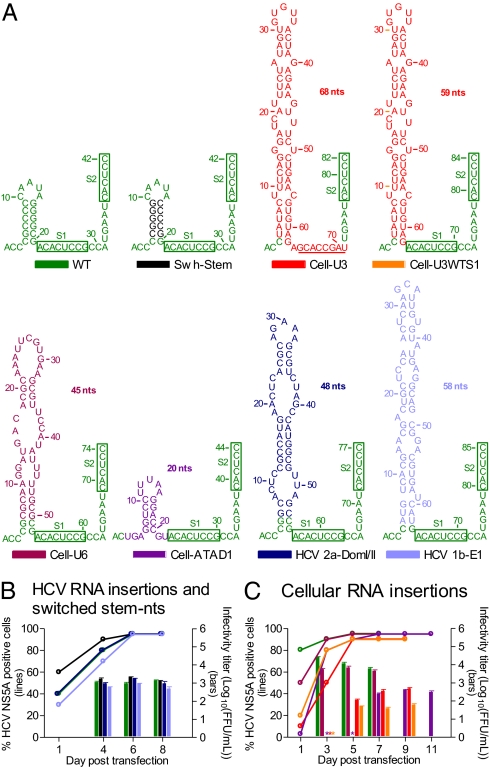
The HCV genotypes 1–6 specific 5′ UTR-NS2 culture systems permitted detailed functional analysis of the 5′ UTR in the context of the complete HCV life cycle. Here we focused on the 5′ UTR SLI. We deleted the entire SLI (ΔSLI), the 5 bp of stem-forming nucleotides (ΔStem) or the loop (ΔLoop) of genotypes 1–6 5′ UTR-NS2 recombinants (Fig. 2); 1a and 2a recombinants with partial SLI deletion [ΔSLI(nt7–17)] were also constructed. In addition, a 2a recombinant, in which 5 bp of stem-forming nucleotides were switched, was constructed (Swh-Stem; Fig. 3A). RNA transfection of Huh7.5 cells showed that all 26 mutants were either nonviable (NS5A staining negative through day 29) or highly attenuated (Table S3). However, the Swh-Stem spread as efficiently as the corresponding virus with wild-type SLI sequence and produced virus particles with comparable infectivity titers (Fig. 3B). The 5′ UTR and ORF sequences of first-passage viruses were maintained. Thus, these data indicate that the structure, and not the specific nucleotide sequence, of SLI was an essential element for viability of HCV in vitro.
Fig. 3.
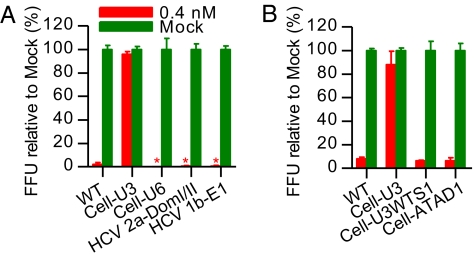
Structured host cellular RNAs and HCV downstream sequences inserted in domain I of the HCV 5′ UTR could compensate for the lack of SLI. (A) RNA insertion sequences from U3 snoRNA, U6 snRNA, ATAD1, HCV domain I/II of 2a 5′ UTR, and HCV 1b E1 were engineered into the corresponding position of the 2a 5′ UTR-NS2 recombinant to obtain Cell-U3, Cell-U6, Cell-ATAD1, HCV 2a-DomI/II, and HCV 1b-E1, respectively. A modified Cell-U3 containing wild-type S1 and downstream nucleotides CCA was also constructed (Cell-U3WTS1). A 2a 5′ UTR-NS2 recombinant with switch of the 5-bp stem-forming nucleotides was included (Swh-Stem). G was not inserted immediately upstream of the 5′-terminal A of the 2a 5′ UTR; all had efficient in vitro transcription. The secondary structure of domain I was predicted by Mfold (15). S1 and S2 are boxed. In Cell-U3, the sequences corresponding to S1 are underlined. (B and C) Huh7.5 cells were transfected with HCV RNA transcripts of recombinants shown in A. Percentage of HCV-infected cells (Left y axis) and selected HCV infectivity titers after viral spread (Right y axis) were determined. *, FFU counts were not determined. Mean of triplicates ± SEM is shown.
Identification of Structured RNA Sequences from Host Cells or from Downstream HCV Genome Regions Compensating for the Lack of SLI.
Because the highly attenuated SLI deletion recombinants showed evidence of very low levels of HCV infection or late emerging infection, we followed these cultures until viruses eventually spread to most cells (Table S3). The transfection-derived viruses could be passaged to naïve Huh7.5 cells. Peak infectivity titers of first-passage 1aΔLoop, 1bΔStem, and 1bΔLoop were similar to the corresponding virus with wild-type 5′ UTR; however, infectivity titers of 2aΔSLI, 2aΔSLI(nt7–17), 2aΔStem, 3aΔStem, 3aΔLoop, 5aΔSLI, and 5aΔLoop were 0.3–2.2 log10 lower (Table S3). Sequence analysis of first-passage viruses revealed that 2aΔSLI(nt7–17) and 5aΔSLI acquired RNA insertions from host noncoding RNAs: U6 small nuclear RNA (snRNA) and U3 small nucleolar RNA (snoRNA), respectively. 3aΔStem acquired an RNA insertion from the 5′ UTR of ATPase family AAA domain-containing 1 (ATAD1). Some of the other deletion mutants acquired RNA sequences from downstream HCV genome regions (Table S3). In general, these RNA sequences were inserted at the SLI position of the SLI deletion mutants, with intact 5′-terminal nucleotides ACC (ACU in ATAD1), and the unique 5′ UTR domain I sequences with host or viral RNA insertions were predicted to form single or multiple SL structures by Mfold (15). In addition, in most recovered viruses, point mutations were identified in other genome regions (Table S3 and S4).
RNA insertion fragments from host U3, U6, and ATAD1 RNAs, as well as from HCV genotype 2a 5′ UTR domain I and II and genotype 1b E1 gene, were engineered into the corresponding SLI position of the 2a 5′ UTR-NS2 recombinant to obtain mutants Cell-U3, Cell-U6, Cell-ATAD1, HCV 2a-DomI/II, and HCV 1b-E1, respectively (Fig. 3A). In RNA transfections of Huh7.5 cells, these recombinants spread efficiently, and the peak viral infectivity titers were similar to SLI wild-type virus, with exception of Cell-U3 and Cell-ATAD1 with titers being ~20-fold lower (Fig. 3 B and C). Sequence analysis of first-passage viruses showed that 5′ UTR and ORF sequences of these SLI insertion mutants were maintained. Hence, the RNA insertion sequences of host and viral origin with SL structure(s) could apparently compensate for the lack of HCV SLI.
Antagonism of miR-122 by LNA SPC3649 Efficiently Suppressed Infection with HCV Genotypes 1–6 5′ UTR-NS2 Recombinants.
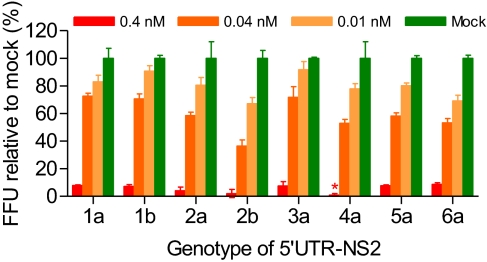
Because suitable culture systems were not available, the antiviral effect of miR-122 antagonism was previously demonstrated only for HCV genotype 1a in vitro replication (5, 6); 1a and 1b in vivo infection (8); and 2a in vitro infection (7). The 5′ UTR-NS2 culture systems developed here provided a unique opportunity for testing the efficacy of miR-122 antagonism against HCV genotypes 1–6 infections. We transfected Huh7.5 cells with miR-122 antisense LNA SPC3649 and subsequently infected with first-passage–derived stock viruses. In cells transfected with 0.4 nM SPC3649 (cytotoxicity observed at ≥1.2 nM), infection with 5′ UTR-NS2 recombinants of HCV genotypes 1–6 was efficiently suppressed, with <5% and <9% infection in two independent experiments, compared with respective infection in SPC3649-free mock-transfected controls (100%). The inhibitory effect of SPC3649 transfection was dose-dependent for all genotype recombinant viruses (Fig. 4). Hence, miR-122 antagonism had potent antiviral effect against HCV genotypes 1–6 infections.
Fig. 4.
Antagonism of miR-122 by LNA SPC3649 suppressed infection of JFH1-based genotypes 1–6 5′ UTR-NS2 recombinant viruses. Huh7.5 cells were transfected with the indicated concentrations of SPC3649 (Materials and Methods). No cytotoxic effect was observed. The treated cells were infected with culture-derived 5′ UTR-NS2 viruses (Fig. 1). Percentage of virus infection, by FFU counts (SI Materials and Methods), was expressed relative to the respective infection in SPC3649-free mock-transfected controls, which was set at 100%. Asterisk (*) indicates <2%. Mean of triplicates ± SEM is shown. In an independent experiment, transfection of 0.4 nM SPC3649 resulted in <5% infection for all HCV recombinants.
HCV Recombinant with S1 Substituted by U3 snoRNA Sequence Was Not Affected by miR-122 Antagonism.
The insertion sequence of U3 snoRNA replaced the S1 (Fig. 3A and Table S3), which is believed to be critical for miR-122–mediated stimulation of HCV replication and translation (4, 5). We thus tested the effect of miR-122 antagonism on the infection with U3 insertion mutant Cell-U3 (Fig. 5 A and B). In five independent experiments, Huh7.5 cells transfected with 0.4 nM SPC3649 showed 94%, 96%, 101%, 80%, and 88% Cell-U3 infection compared with SPC3649-free mock-transfected controls (100%), whereas the wild-type virus showed 0%, 2%, 0%, 3%, and 8% infection. Hence, infection with the Cell-U3 recombinant virus, lacking authentic S1, was unaffected by miR-122 antagonism.
Fig. 5.
An HCV recombinant with S1 substituted by U3 snoRNA sequence was not affected by miR-122 antagonism. For details on SPC3649 transfection, virus infection and determination of infection rate, see Materials and Methods and legend to Fig. 4. Viruses used were described in Fig. 3. A and B show two different experiments. Asterisk (*) indicates <2%. Mean of triplicates ± SEM is shown.
To elucidate whether lack of an intact S1 or the complex SL structure conferred the resistance of Cell-U3 virus to miR-122 antagonism, we engineered the wild-type S1 into the Cell-U3 recombinant, but maintained the heterologous SL structure, to obtain Cell-U3WTS1 (Fig. 3A). After RNA tranfection, the Cell-U3WTS1 spread efficiently in Huh7.5 cells and produced infectious virus. The infectivity titers produced were ~eightfold lower than for Cell-U3 (Fig. 3C). Sequence analysis of the 5′ UTR and ORF of first-passage virus confirmed that the mutated genome sequences were maintained. In Huh7.5 cells transfected with 0.4 nM SPC3649, infection of Cell-U3WTS1 was efficiently suppressed, with 6% infected cells, which was comparable to that of 2a wild-type virus (8%); the Cell-U3 recombinant showed 88% infection in this particular experiment (Fig. 5B). All infection rates were relative to the respective infection in mock-transfected controls (100%). Similarly, other insertion mutants (Cell-U6, HCV 2a-DomI/II, HCV 1b-E1, and Cell-ATAD1) that contained diverse heterologous SL structures, but had intact S1, were suppressed by miR-122 antagonism (Fig. 5 A and B). Thus, the resistance of the Cell-U3 recombinant virus to miR-122 antagonism was attributed to the lack of an intact S1; the SL structure apparently had no influence on this effect.
The Cell-U3 recombinant virus contained an intact S2, which has previously been demonstrated to regulate HCV replication in cooperation with S1 (6). The fact that Cell-U3 infection was apparently not affected by miR-122 antagonism suggested that interaction of S2 with miR-122 might not be essential for the HCV life cycle. We thus tested the efficacy of miR-122 antagonism against a fully viable 2a 5′ UTR-NS2 recombinant with A-to-G substitution at position 6 of S2 (S2-A6G). Mutation at position 6 of S2 was recently found to impair the miR-122 binding, although to a lesser extent than a equivalent mutation in S1 (16). The S2-A6G mutant had peak infectivity titers equivalent to wild-type 2a recombinant virus (104.2 FFU/mL). In Huh7.5 cells transfected with 0.4 nM SPC3649, S2-A6G infection was 6%, similar to the 2a recombinant virus with wild-type S2 (5%), compared with respective infection in mock-transfected controls (100%). The infection was SPC3649 dose-dependent (Fig. S2). This is in line with the effect of miR-122 antagonism observed for the genotype 4a 5′ UTR-NS2 recombinant, which also contains G at position 6 of S2 (Fig. 4). Thus, miR-122 antagonism could efficiently inhibit the infection of HCV recombinant virus with a point mutation in S2, suggesting that the interaction of miR-122 and the S2 might not be essential for the infection of JFH1-based HCV recombinants.
HCV Recombinants with Point Mutations within the S1 Were Fully Viable and Only Partially Inhibited by miR-122 Antagonism.
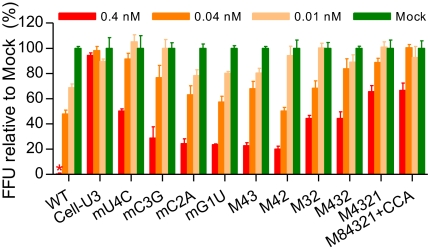
In the viable mutant Cell-U3, an U3 RNA stretch (GCACCGAU, defined as position 8–1) substituted the HCV 5′ UTR S1 (ACACUCCG, defined as position 8–1) and downstream nucleotides CCA (Fig. 3A), which rendered the Cell-U3 unaffected by miR-122 antagonism (Fig. 5 A and B). The nucleotides at positions 8, 4, 3, 2, and 1 of the HCV 5′ UTR S1 were different from the corresponding nucleotides of the U3 stretch, whereas the nucleotides at positions 7, 6, and 5 were identical. However, heterogeneities of positions 7, 6, and 5 were observed in reported HCV patient sequences (HCV databases) (8). We thus mutated positions 8–1 of the 2a 5′ UTR-NS2 recombinant to the corresponding nucleotide appearing in the U3 RNA stretch or in reported sequences, to obtain mutants with single changes or with combinations of these changes (Table S5). A recombinant with mutations at positions 8, 4, 3, 2, and 1, and deletion of the downstream CCA, was also constructed (Table S5). Transfection of Huh7.5 cells with RNAs from 14 HCV 5′ UTR S1 mutants revealed that mA8G, mC7G, M84321, and M84321+CCA were highly attenuated (≤1% NS5A-positive cells at day 1); mA6G, mC5U, mU4C, mC3G, mC2A, M43, M42, M32, and M432 were impaired (5–40%); and mG1U was equivalent compared with wild-type virus (80%; Table S5). The transfection supernatant from peak infection was passaged to naïve Huh7.5 cells. Sequence analysis of the complete 5′ UTR of first-passage viruses demonstrated that mA8G, mC7G, mA6G, and mC5U had reverted to the wild-type sequence, whereas mU4C, mC3G, mC2A, mG1U, M43, M42, M32, and M432 maintained the mutations in the 5′ UTR. For M84321, the mutation at position 8 had reverted, and mutations at positions 4, 3, 2, and 1 were maintained (designated M4321 for subsequent use). In M84321+CCA, all mutations were maintained; however, changed sequences were observed upstream of SLI (Table S5). No mutations were acquired in the ORF sequence of recovered viruses containing 5′ UTR mutation(s). The infectivity titers of recovered viruses with 5′ UTR mutation(s) were ~3- to 10-fold lower than that of wild-type viruses, with exception of mG1U, whereas titers of the viruses with reversion of mutated sequences were comparable to that of wild-type viruses (Table S5). Thus, mutations at positions 4, 3, 2, and 1 of HCV 5′ UTR S1, either single or in combination, were permitted in vitro.
Next, we examined the antiviral effect of miR-122 antagonism on infection with HCV 5′ UTR S1 mutant viruses (Fig. 6). In parallel, wild-type S1 HCV and the Cell-U3 viruses were tested. In Huh7.5 cells transfected with 0.4 nM SPC3649, the percentages of infection compared with mock-transfected controls in two independent experiments were as follows: wild-type virus (2% and 0%), Cell-U3 (94% and 96%), mU4C (46% and 50%), mC3G (28% and 28%), mC2A (30% and 24%), M432 (46% and 44%), M4321 (68% and 65%), and M84321+CCA (73% and 66%). In two other experiments, the values for additional mutants were wild-type virus (6% and 5%), Cell-U3 (88% and 103%), M43 (22% and 31%), M42 (20% and 25%), and M32 (44% and 50%), respectively. In yet another experiment, the values for wild-type, Cell-U3, and mG1U viruses were 11%, 85%, and 23%, respectively. The inhibitory effect of miR-122 antagonism was SPC3649 dose-dependent for all viruses, except for Cell-U3, which showed no suppression at any dose tested (Fig. 6). Therefore, combinations of or even single mutations in the 5′ UTR S1 reduced the antiviral effect of miR-122 antagonism.
Fig. 6.
Mutations in the 5′ UTR S1 impaired the response of HCV recombinants to miR-122 antagonism. For details on SPC3649 transfection, virus infection, and determination of infection rate, see Materials and Methods and legend to Fig. 4. Viruses used were described in Fig. 3 and Table S5. Two independent experiments were performed for each mutant (5% and 6% infection for wild-type virus), except for mG1U, which was tested in a single experiment (11% infection for wild-type virus), and the data from independent experiments were similar. The data from one experiment are shown. Asterisk (*) indicates <2%. Mean of triplicates ± SEM is shown.
Discussion
We developed robust JFH1-based culture systems containing HCV genotypes 1–6 specific 5′ UTR-NS2. To date, only the full-length genome of JFH1 (genotype 2a) was reported to efficiently produce virus particles in Huh7 and derived cell lines (9, 17), whereas other genomes—for example, H77-S (1a) (18)—had only low-level virus production. Most HCV strains are apparently nonviable in vitro. Thus, the efficient 5′ UTR-NS2 genotypes 1–6 systems represent a significant advance in the development of HCV culture systems, and will facilitate HCV basic research and provide a useful tool for development and preclinical testing of novel HCV drugs. A genotype 1a (strain H77) 5′ UTR-NS2 recombinant was previously reported to grow in culture with different adaptive mutations from those used in this study (19, 20). Using these systems, we here demonstrated the universal antiviral effect of miR-122 antagonism against HCV genotypes 1–6 infections. Importantly, we discovered that RNA sequences from the cellular U3 snoRNA could substitute for SLI and S1 of the HCV 5′ UTR, and that they conferred resistance to miR-122 antagonism. Our observation led to the development of viable HCV recombinants with point mutations in S1, being only partially inhibited by miR-122 antagonism. This discovery indicates that in chronic HCV patients the therapeutic effect of miR-122 antagonism-based therapy could potentially be reduced by development of resistance mutants.
MicroRNAs (miRNAs) have important roles in various cellular processes and viral infections, and miRNA-based therapies are currently being tested in animal models and clinical trials. The potential therapeutic effect of miR-122 antagonism by antisense LNA SPC3649 was recently demonstrated in chimpanzees infected with HCV genotypes 1a and 1b (8). Here, we demonstrated a universal antiviral effect of miR-122 antagonism against HCV genotypes 1–6 infections (Fig. 4), and the specificity of this treatment effect was validated by the resistant phenotype of several HCV mutants (Figs. 5 and 6). Thus, our data strengthen the potential of miR-122 antagonism as a therapeutic approach for patients infected with HCV of diverse genotypes.
Virus resistance is a major concern for antiviral HCV drug development. Potential resistance mutants were not detected during administration of SPC3649 in HCV-infected chimpanzees (8); however, the treatment period was only 12 wk. Our finding that point mutations in S1 reduced the antiviral effect of miR-122 antagonism (Fig. 6) suggests that the effect of this therapy could potentially be reduced or eliminated by occurrence of variants in chronic HCV patients or in vivo infection models. Thus, it will be of interest to determine whether resistance mutants could emerge during long-term in vitro treatment and in HCV patients enrolled in ongoing clinical phase II trials. It will further be of interest to test the in vivo viability of culture-derived miR-122 binding-site mutants to further address the relevance of these introduced mutations. Nevertheless, SPC3649-mediated miR-122 antagonism could be applied in a combined therapy with other antivirals, such as NS3/4A protease inhibitors, to increase the barrier to the development of viral resistance mutations.
The S1 and S2 miR-122 binding sites were demonstrated to function cooperatively in regulation of HCV replication (6). Our data showed that the Cell-U3 virus, a 2a 5′ UTR-NS2 virus containing the U3 RNA substitution of the S1 (resulting in mutations at S1 positions 1, 2, 3, 4, and 8) and a wild-type S2 could grow in culture (Fig. 3) and was unaffected by miR-122 antagonism (Figs. 5 and 6). A 2a 5′ UTR-NS2 recombinant with mutation at position 6 of S2 (S2-A6G) and the 4a 5′ UTR-NS2 recombinant that naturally had G at this position were fully viable in Huh7.5 cells, and both were efficiently suppressed by miR-122 antagonism (Fig. 4 and Fig. S2). It was recently shown that mutating position 6 of S2 reduced miR-122 binding (16). Thus, our data on these binding-site mutants suggested that the most important interaction of miR-122 was that with S1, and that the interaction with S2 might not be essential for the HCV life cycle. This was also in line with a recent report showing that mutation of position 6 in S2 decreased, but to a smaller degree than a equivalent mutation in S1, production of infectious virus particles (16).
miR-122 might promote stability of HCV genomic RNA by binding to the miR-122 binding sites. The U3 RNA insertion and mutations in S1 may interfere, perhaps to different extents, with miR-122 binding, resulting in lower stability of mutated compared with wild-type HCV genomes in a miR-122 abundant environment. This might explain the observation that such mutants had relative low virus titers in naïve Huh7.5 cells (Fig. 3 and Table S5). However, interference mediated by mutations in S1 may be synergized when having a U3 RNA sequence with a long SL structure instead of SLI. Thus, the titer of Cell-U3 was lower than that of other S1 mutant viruses with wild-type SLI. As an exception, the mG1U mutant was not attenuated in virus production, which was in accordance with a previous report that mutation at this position did not affect 1a H77 genomic RNA accumulation (5). This might explain why mG1U appeared to be more sensitive to miR-122 antagonism than other mutants (Fig. 6).
It is possible that in an environment with low levels of miR-122 induced by LNA SPC3649 treatment, the U3 RNA insertion and the mutations in S1 rendered HCV RNA more stable than wild-type RNA. This would explain why infection of these mutant viruses was less affected by miR-122 antagonism (Fig. 6). Alternatively, the U3 RNA insertion sequence or point mutations might enable these viruses to grow using unknown factors replacing miR-122 function, such as other miRNAs, making them completely or partially independent of miR-122. Thus, in future studies it would be of interest to test whether these mutant viruses could infect cell types with low levels of miR-122, such as CD81-expressing HepG2 cells, reported to support early infection of wild-type J6/JFH1 at a low level (21), or humanized mouse cell lines, which support HCV entry (22). Because the RNA interference (RNAi) pathway (involved in miRNA biogenesis) and miR-122 are both required for HCV replication (7), and because Cell-U3 was not affected by miR-122 antagonism (Figs. 5 and 6), in future studies it would also be of interest to determine whether the RNAi machinery is still important for infection with this unique mutant virus.
The S1 contains eight nucleotides (Fig. 2). Previous studies showed that mutation at positions 6 (A to U) and double mutations at positions 4 and 3 (UC to AG) abolished RNA accumulation of the 1a H77 genome (5) and production of 1a Core-NS2 virus HJ3-5 (16). In addition, mutation at position 3 (C to G), but not at position 1 (A to G), was shown to abolish mutated H77 genomic RNA accumulation, observed 5 d after RNA electroporation (5). We observed reversion of single mutations introduced at positions 8, 7, 6, and 5 of S1, which either represented sequences found in the corresponding U3 host RNA sequence in the Cell-U3 virus (position 8) or in HCV isolates. However, single or combined mutations at positions 4, 3, 2, and 1, all observed in Cell-U3, resulted in genetically stable HCV recombinants (Table S5). The discrepancy regarding the data on mutation at position 3 might be caused by the different culture system used. The high replication capacity of JFH1-based systems (9, 10), compared with the lower capacity of the adapted H77-S genomes (18), may compensate for the effect of this mutation in an early replication phase.
To date, host cellular RNA insertions in the HCV genome were not reported. We found that in Huh7.5 cultures host cellular RNAs U3, U6, and ATAD1, with a defined stem-loop structure, could be inserted in the HCV 5′ UTR domain I and compensate for the lack of SLI (Fig. 3 and Table S3). The recombination event could either be a result of HCV polymerase template switching or an HCV-mediated repair mechanism of the 5′ end using random recombination with RNA fragments present in the cytoplasm. We speculate that the incorporated RNA sequences might be closely associated with the HCV RNA replication complex and that viable recombined genomes emerged by natural selection. ATAD1 belongs to the ATPase family, but its function is unclear. The nucleus resident RNAs U3 and U6 are transcribed by RNA polymerase II and III, respectively, and play essential roles in processing of ribosomal RNAs and catalysis of the spliceosome during RNA editing, respectively (23). They could potentially enter the cytoplasm by associating with nucleolin, or following stress-induced nucleolus disruption. Less likely, HCV RNA synthesis could have a not-yet-identified nuclear phase. The 3′-terminal U stretch of U6 could associate with La protein, a protein required for IRES-mediated translation of HCV RNA (23, 24). Thus, in future studies it would be of great interest to determine whether U3, U6, and ATAD1, as well as associated proteins, are involved in the HCV replication process. In addition, it would be interesting to conduct additional studies under deletion selection pressure to determine whether a broad spectrum of RNAs is subjected to recombination; such studies would help clarify whether RNA is selected randomly or by active involvement in HCV replication.
Both A and G exist naturally as the 5′-terminal nucleotide of HCV isolates (Fig. 2) (25). In our 5′ UTR-NS2 culture systems, 5′-terminal G was in all cases changed to A despite early efficient replication. In contrast, the 5′-terminal G was maintained in vivo in chimpanzee infection with HCV full-length infectious clones. Thus, the G-to-A change at the 5′ terminus of JFH1-based 5′ UTR-NS2 recombinants could perhaps be attributed to the function of JFH1 NS3-3′ UTR. The prototype JFH1 genome naturally has A at the 5′ terminus (9). However, this change could also be an in vitro effect, because in the Con1 replicon system 5′-terminal G was changed to A upon multiple rounds of replication (25). Thus, future studies analyzing the 5′ terminus of JFH1-based 5′ UTR-NS2 HCV recombinants from in vivo transfection-derived infections would be of interest to further study this phenomenon.
In conclusion, we developed robust JFH1-based HCV cell culture systems containing genotypes 1–6 specific 5′ UTR-NS2, which allow genotype-specific functional analysis of the 5′ UTR, structural proteins, p7 and NS2, as well as broad testing of antivirals targeting these regions. These systems might also contribute to future development of full-length culture systems of different HCV genotypes. Importantly, we demonstrated the functional importance of the 5′ UTR SLI in the HCV life cycle, and that antagonism of miR-122 had a universal antiviral effect against HCV of diverse genotypes. Although the approach of host-factor miR-122 antagonism has the potential for HCV therapy and is being tested in clinical trials, the reduced antiviral effect by single mutations in S1 support the reevaluation of this approach as monotherapy for future HCV treatment. Identification of host cellular RNA insertions in the HCV genome might facilitate understanding of virus-host interactions and could contribute to the identification of cellular interacting partners as future drug targets.
Materials and Methods
The JFH1 5′ UTR of HCV genotypes 1–6 Core-NS2 cDNA clones developed previously (2) were replaced with genotype-specific 5′ UTR to obtain corresponding genotype 5′ UTR-NS2 recombinants (Fig. 1). Other recombinant mutants were subsequently constructed using standard cloning techniques. HCV RNA in vitro transcription, transfection, infection, and sequence analysis of recovered viruses, as well as HCV infectivity and RNA titers (intra- and extracellular), were performed as described in Gottwein et al. (2) and Scheel et al. (20). See SI Materials and Methods for detailed information.
For LNA SPC3649 treatment experiments, Huh7.5 cells were seeded in Nunc 96-well optical bottom plates (6 × 103 cells/well) for 16 h. The cells were transfected with synthesized SPC3649 (Exiqon; 5′-CcAttGTcaCaCtCC-3′; LNA in capitals, DNA in lowercase, complete phosphorothioate backbone, capital C denotes LNA methylcytosine) (8) using Lipofectamine 2000 (Invitrogen), incubated for 10 h, and then retransfected with SPC3649 and incubated for another 14 h. The cell viability was determined by CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega). The transfected cells were infected with 5′ UTR-NS2 viruses, incubated for 48 h, fixed and immunostained for HCV NS5A, and then analyzed for FFU counts (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank L. Mikkelsen, L. Ghanem, A. L. Sørensen, and B. Landt for technical assistance; S. Ladelund for statistical advice; J. O. Nielsen, O. Andersen, and K. Schønning for providing valuable support; and R. H. Purcell, C. M. Rice, and T. Wakita for providing reagents. This study was supported by PhD stipends from the Faculty of Health Sciences, University of Copenhagen (to T.K.S. and T.B.J.) and research grants from the Lundbeck Foundation (to J.B.), the Danish Cancer Society (to J.M.G. and J.B.), the Novo Nordisk Foundation (to J.M.G. and J.B.), and the Danish Council for Independent Research, Medical Science (to J.B.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JF343780–JF343793).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016606108/-/DCSupplemental.
References
- 1.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 2.Gottwein JM, et al. Development and characterization of hepatitis C virus genotype 1–7 cell culture systems: Role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 3.Simmonds P, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 4.Henke JI, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 6.Jopling CL, Schütz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randall G, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 11.Kolykhalov AA, et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 12.Bukh J, et al. Challenge pools of hepatitis C virus genotypes 1-6 prototype strains: Replication fitness and pathogenicity in chimpanzees and human liver-chimeric mouse models. J Infect Dis. 2010;201:1381–1389. doi: 10.1086/651579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottwein JM, et al. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): Genetic analyses and in vivo pathogenesis studies. J Virol. 2010;84:5277–5293. doi: 10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci USA. 2006;103:2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMullan LK, et al. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci USA. 2007;104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheel TK, et al. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc Natl Acad Sci USA. 2008;105:997–1002. doi: 10.1073/pnas.0711044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mee CJ, et al. Polarization restricts hepatitis C virus entry into HepG2 hepatoma cells. J Virol. 2009;83:6211–6221. doi: 10.1128/JVI.00246-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ploss A, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiss T. Biogenesis of small nuclear RNPs. J Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 24.Mondal T, et al. Structural determinant of human La protein critical for internal initiation of translation of hepatitis C virus RNA. J Virol. 2008;82:11927–11938. doi: 10.1128/JVI.00924-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai Z, Liang TJ, Luo G. Effects of mutations of the initiation nucleotides on hepatitis C virus RNA replication in the cell. J Virol. 2004;78:3633–3643. doi: 10.1128/JVI.78.7.3633-3643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.