Abstract
Neurofibromatosis type 2 (NF2) is a multiple neoplasia syndrome and is caused by a mutation of the NF2 tumor suppressor gene that encodes for the tumor suppressor protein merlin. Biallelic NF2 gene inactivation results in the development of central nervous system tumors, including schwannomas, meningiomas, ependymomas, and astrocytomas. Although a wide variety of missense germline mutations in the coding sequences of the NF2 gene can cause loss of merlin function, the mechanism of this functional loss is unknown. To gain insight into the mechanisms underlying loss of merlin function in NF2, we investigated mutated merlin homeostasis and function in NF2-associated tumors and cell lines. Quantitative protein and RT-PCR analysis revealed that whereas merlin protein expression was significantly reduced in NF2-associated tumors, mRNA expression levels were unchanged. Transfection of genetic constructs of common NF2 missense mutations into NF2 gene-deficient meningioma cell lines revealed that merlin loss of function is due to a reduction in mutant protein half-life and increased protein degradation. Transfection analysis also demonstrated that recovery of tumor suppressor protein function is possible, indicating that these mutants maintain intrinsic functional capacity. Further, increased expression of mutant protein is possible after treatment with specific proteostasis regulators, implicating protein quality control systems in the degradative fate of mutant tumor suppressor proteins. These findings provide direct insight into protein function and tumorigenesis in NF2 and indicate a unique treatment paradigm for this disorder.
Neurofibromatosis type 2 (NF2) is a multiple neoplasia syndrome with an incidence of 1 in 25,000 live births (1). It is the result of the mutation of the NF2 tumor suppressor gene that is on the long arm of chromosome 22 and can be inherited in an autosomal dominant manner. The NF2 gene encodes for the tumor suppressor protein merlin (69 kDa). Merlin indirectly regulates cellular pathways involved in tumorigenesis, including cell to cell adhesion, cytoskeletal architecture, and membrane protein organization (2–4). Decreased or absent merlin function in NF2 predisposes patients to develop a number of nervous system tumors, including bilateral vestibular schwannomas (over 90% of patients), other cranial nerve schwannomas (25–50%), meningiomas (50%), ependymomas (20–50%), and astrocytomas (5, 6).
Missense mutations in the coding sequences of the NF2 gene occur in NF2. After biallelic inactivation, these and other types of NF2 gene mutations result in loss of merlin function, which underlies NF2-associated tumor development. Although a wide variety of missense mutations of the NF2 gene cause loss of merlin function, the cause of this functional loss remains unknown. Specifically, the ability of various different single nucleotide substitutions by missense mutations in the NF2 gene coding regions to give rise to similar manifestations of merlin function loss indicates that there may be a convergent effect of mutations on merlin protein. To gain insight into the mechanisms underlying loss of merlin function in NF2, we investigated mutated merlin homeostasis and function in NF2-associated tumors and cell lines.
Results
Quantitative Tumor Suppressor Protein Levels and mRNA Expression.
To assess the levels of merlin expression in NF2 tumors, we quantitatively measured merlin expression in microdissected tumors from NF2 patients using Western blot analysis (Fig. 1A). Quantitative protein analysis revealed a significant reduction in merlin expression in NF2-associated meningiomas and schwannomas (95% reduction in merlin expression; seven tumors) compared with normal adjacent central nervous system tissue. Consistent with Western blot analysis of merlin expression in NF2-associated tumors, immunofluorescence in NF2-associated tumor specimens demonstrated an absence of merlin expression (Fig. 1B), with robust merlin expression in corresponding normal tissue in NF2 patients.
Fig. 1.
Quantification of merlin protein and RNA expression. (A) Western blot of merlin proteins from various microdissected NF2-associated tumor samples. (B) Immunofluorescence staining for merlin (red) in tumor specimens and normal tissue derived from patients with NF2. The nuclei of cells are counterstained with Hoechst (blue). (C) RT-PCR for NF2 mRNA expression from various NF2-associated tumors.
Because NF2 mutations could affect associated merlin levels through either decreased mRNA and/or associated total protein levels, we measured mRNA transcript levels for merlin in multiple tumors from NF2 patients (five tumors from five patients). RT-PCR analysis revealed that NF2 mRNA encoding for merlin is expressed at the same levels in NF2-associated tumors compared with meningioma control cells without genetic mutations in the NF2 gene (Fig. 1C). The reduced merlin expression in the setting of normal mRNA expression levels in NF2-associated tumors suggests that inactivation of tumor suppressor function occurs as a result of an absolute decrease in merlin levels rather than a decrease in NF2 gene-encoded mRNA.
Half-Life Reduction of Merlin in NF2 Tumor-Derived Mutants.
Because NF2-associated functional loss of merlin appeared to occur at the protein level, we hypothesized that increased protein degradation underlies the net loss of function seen in NF2-associated tumors. To test this hypothesis, we investigated the stability of mutant and wild-type merlin. Because missense mutations occur and are well-characterized in NF2, we identified hotspot missense mutations (L46R, L141P, A211D, K413E, Q324L, and L535P) from patients with NF2 and inserted these coding sequences into pCMV6-Entry vectors. After transfection of these vectors into meningioma cell lines deficient in NF2 gene expression, we investigated protein stability using the [35S]-methionine mediated pulse chase assay to quantitatively identify half-life changes in newly synthesized mutant merlin (Fig. 2A).
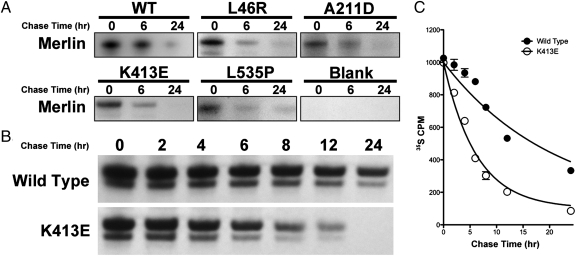
Fig. 2.
Half-life kinetic analysis of reduction of mutant merlin proteins. (A) Radioactive 35S pulse chase assay of wild-type merlin and different merlin missense mutants at 0, 6, and 24 h demonstrating increased turnover and degradation of mutant merlin compared with wild-type control. (B) Radioactive 35S pulse chase assay at several time points (in hours) demonstrating the degradation kinetics of merlin mutant K413E compared with wild-type control. (C) Liquid scintillation measurement of 35S-labeled merlin demonstrating an increased degradation rate of K413E merlin mutant compared with wild-type control.
Consistent with effective mutant and wild-type protein translation, pulse chase analysis revealed that newly synthesized mutant and wild-type merlin exist in their full length immediately after pulse labeling and that there was no difference between mutant and wild-type merlin expression levels after initial translation. Over time, there was a marked reduction in labeled merlin expression compared with wild type, consistent with increased mutant merlin turnover and a reduced mutant merlin half-life compared with wild type (Fig. 2 B and C). Autoradiography and 35S liquid scintillation revealed that there was minimal reduction in wild-type merlin expression 6 h after labeling, but 24 h after labeling, the majority of the wild-type merlin was degraded, indicative of merlin turnover under normal conditions. Pulse chase analysis of mutant merlin revealed rapid degradation and a significantly decreased half-life compared with wild-type merlin. For example, in merlin mutant K413E, [35S]-merlin expression was found to be <50% of wild-type merlin 4 h after labeling.
Kinetics of Merlin Degradation.
To precisely determine differences between mutant and wild-type merlin half-life, we assessed the rate of degradation of wild-type and mutant merlin. Using an established exponential decay regression (7), merlin mutant half-lives were significantly shorter (L46R t1/2, 1.8 h; L141P t1/2, 7.8 h; A211D t1/2, 2.2 h; Q324L t1/2, 3.3 h; K413E t1/2, 4.3 h) than that of wild-type merlin (t1/2, 14.6 h).
Mutant Merlin Protein Maintains Intrinsic Functional Capacity.
To determine if reduced merlin function in NF2 also results from impaired merlin functional capacity, we investigated whether mutant merlin maintains its functional capacity by reintroducing it into CH157 NM meningioma cells lacking analogous NF2 gene expression (8). Specifically, we measured the effects of mutant merlin transfection on cytoskeletal reorganization and growth control. Although merlin does not control cellular proliferation and differentiation directly, it affects these processes indirectly through interactions with cell membrane and cytoskeletal components. In particular, inactivation of NF2 protein causes abnormalities in intracellular filamentous actin (F-actin) organization and stress fiber formation, both of which are characteristics of meningiomas and schwannomas in NF2 patients, as well as the CH157 NM cell line (9, 10).
Based on the well-described effects of merlin on cytoskeletal function, we initially tested mutant merlin function through its effects on F-actin. Consistent with previous findings (11), strong, bundle-like F-actin stress fiber formation occurred in tumor cells deficient in merlin protein. With the introduction of wild-type merlin into these cells, cytoskeletal abnormalities were eliminated. The bundle-like F-actin was eliminated from the cytoplasm and became localized adjacent to the plasma membrane. Similarly, the introduction of merlin mutants significantly reduced F-actin stress fiber formation. With this intervention, bundle-like F-actin also decreased and accumulated in the cytoplasm and on the plasma membrane. In particular, merlin mutant K413E was found at ruffled areas of cell membrane, suggesting its role in the regulation of membrane dynamics and motility (Fig. 3A).
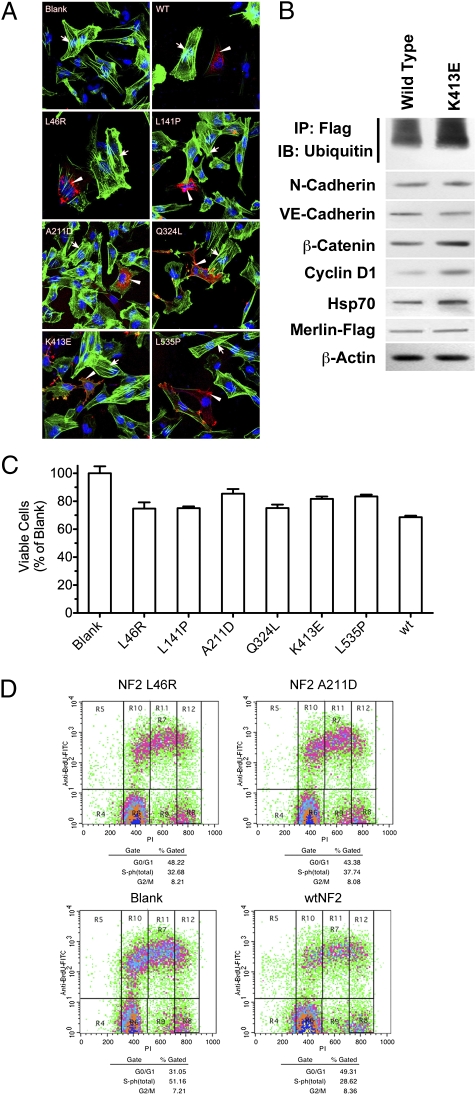
Fig. 3.
Mutant merlin protein maintains intrinsic functional capacity. (A) Immunofluorescence staining for F-actin (green) and merlin mutants (red) in CH157 NM cells transfected with wild-type merlin, merlin mutants, as well as empty vector (blank). The nuclei of cells were counterstained with Hoechst (blue). Arrows point to F-actin stress fibers and arrowheads point to mutant merlin. (B) Western blot of lysates from CH157 NM cells transfected with either wild-type or K413E mutant merlin protein. K413E-transfected cells demonstrate increased merlin ubiquitylation and up-regulation of β-catenin and cyclin D1 cytoplasmic expression. (C) MTT proliferation assay demonstrating suppression of CH157 NM cell growth with mutant merlin transfection compared with empty vector (blank). (D) Flow cytometric BrdU-PI cell cycle analysis demonstrating reduced proliferation in CH157 NM cells transfected with L46R and A211D merlin mutants compared with wild type and empty vector (blank).
Next, we examined the effect of introducing mutant or wild-type merlin on cell to cell contact inhibition in NF2-deficient cells. Functional loss of merlin is also known to affect cellular contact inhibition, which may occur through β-catenin-mediated signaling pathways (12). Specifically, loss of merlin function results in down-regulation of β-catenin expression via cyclin D1-regulated pathways, which occurs in NF2-associated meningiomas and schwannomas (11, 13). To determine if wild-type or mutant merlin can maintain normal β-catenin regulation, we tested whether β-catenin and cyclin D1 expression were changed in CH157 NM cells after reintroduction of wild-type and mutant merlin. We found that with such intervention, both mutant and wild-type merlin up-regulated catenin expression and cyclin D1 expression. Further, this effect appears to be specific, because expression of upstream cadherins, including N- and VE-cadherin, remained unaffected with the introduction of mutant and wild-type merlin (Fig. 3B).
Effect of Mutant Merlin on Tumor Growth.
To determine whether transfected mutant merlin can regulate growth of meningioma cells, we performed an MTT proliferation assay on CH157 NM meningioma cell lines transfected with various merlin missense mutations (14). In cell lines transfected with wild-type merlin, cell growth was reduced by 31.4% compared with cell lines transfected with an empty control vector. Although not as effective as wild-type merlin, introduction of merlin mutants reduced cell growth from 14.6 to 24.9%, a significant effect compared with control (Fig. 3C). The effect of mutant merlin on cell growth was further assessed by cell cycle analysis using a BrdU incorporation assay. A reduction of 22.5% was found in the S-phase population of cells when wild-type merlin was introduced. When a similar analysis was performed using merlin mutants, L46R and A211D mutants reduced the S-phase population by 18.5 and 13.4%, respectively (Fig. 3D). These results indicate that merlin mutants maintain intrinsic tumor suppressor function.
Mechanism of Merlin Degradation.
It has previously been shown that truncated merlin protein products are degraded through ubiquitin-mediated proteasomal pathways (15). However, it remains unclear whether missense mutations in merlin result in accelerated degradation by similar mechanisms. To investigate the mechanism of degradation of merlin mutants, immunoprecipitated mutant merlin was probed with an antibody for ubiquitin. Based on Western blot analysis, there was a 42% increase in ubiquitylated merlin mutants compared with wild-type merlin protein (Fig. 3B). This finding suggests that missense merlin mutants are degraded by a ubiquitin-mediated proteasomal pathway, similar to the known mechanism of wild-type merlin turnover under normal conditions. Thus, deficiency of mutant merlin function in NF2 patients may be due to protein maturation effects resulting from changes in protein structure and conformation, with minimal effects due to changes in intrinsic protein function (16).
Increased Merlin Expression after Treatment with Proteostasis Regulators.
After determining that mutant merlin retains intrinsic tumor suppressor function and that NF2-related loss of merlin function is due to rapid degradation, we sought to investigate a potential treatment strategy that could reduce mutant protein degradation and minimize the effects of decreased tumor suppressor protein function. Specifically, we screened small molecule compounds known to modulate protein degradation and proteostasis. Among these compounds, two flavanoid compounds, celastrol and quercetin, were found to be effective in delaying merlin degradation. Pulse chase analysis demonstrated that celastrol and quercetin treatments both result in increased mutant merlin half-life (Fig. 4A). This result was confirmed by liquid scintillation analysis of radiolabeled merlin, which revealed an increase in proteasomal mutant merlin from 79.7 to 108.7 and 136.9 cpm (*P < 0.05, **P < 0.01) after treatment with celastrol and quercetin, respectively (Fig. 4B). Because celastrol had a potent effect on prolonging mutant merlin half-life at lower doses compared with quercetin, we further quantified the effect of celastrol treatment on mutant merlin degradation. Using nonlinear regression, the half-life of merlin mutant K413E was found to increase from 3.7 to 5.5 h after celastrol treatment. These results indicate that modulation of proteostasis and proteosomal degradation pathways can increase mutant merlin half-life and decrease its degradation (Fig. 4 C and D).
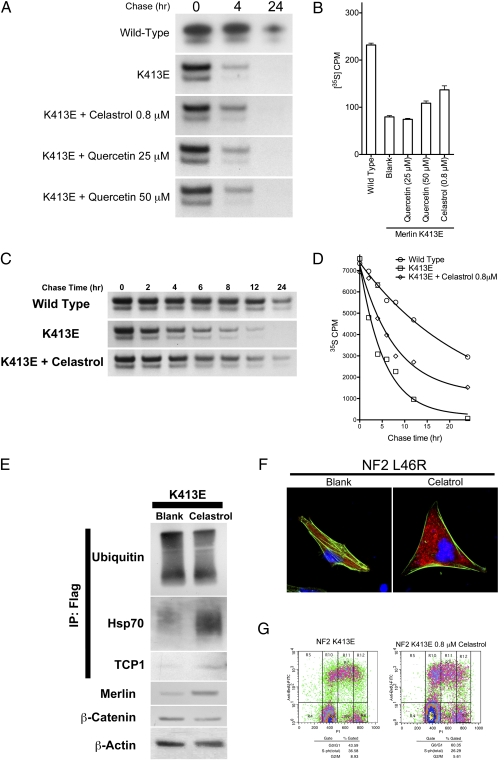
Fig. 4.
Increased cellular expression levels of mutant merlin protein after treatment with proteostasis regulators. (A) Radioactive 35S pulse chase assay demonstrating prolonged K413E merlin mutant protein half-life with celastrol and quercetin treatment. (B) 35S liquid scintillation measurement demonstrating a quantitative decrease in K413E mutant merlin degradation rate with celastrol and high dose quercetin treatment. (C) Radioactive 35S pulse chase assay at several time points (in hours) demonstrating the degradation kinetics of merlin mutant K413E before and after treatment with celastrol compared with wild type. (D) 35S liquid scintillation measurement at several time points (in hours) demonstrating the degradation kinetics of mutant merlin K413E before and after treatment with celastrol compared with wild-type control. (E) Western blot of lysates from CH157 NM cells transfected with merlin mutant K413E before and after celastrol treatment, demonstrating decreased ubiquitin binding, increased Hsp70 and TCP1 binding to immunoprecipitated merlin protein, as well as decreased β-catenin expression after celastrol treatment. (F) Flow cytometric analysis of CH157 NM cells transfected with K413 mutant merlin demonstrating decreased cell proliferation after treatment with celastrol. (G) Immunofluorescence staining for F-actin (green) and merlin K413E mutant protein (red) in CH157 NM cells before and after celastrol treatment. The nuclei of cells are counterstained with Hoechst (blue).
Posttranslational interactions of nascent proteins include those with important mediators of the protein folding and maturation process. To better define the molecular interactions of mutant merlin in the posttranslational period and to further elucidate pathways of merlin degradation, we assessed the levels of known mediators of such processes before and after celastrol treatment. Immunoprecipitated mutant merlin protein treated with celastrol was found to have decreased binding to ubiquitin and increased binding to heat shock protein 70 (Hsp70) and T-complex protein 1 (TCP1) (Fig. 4E), two well-accepted mediators of protein folding and maturation (17, 18). The effect of celastrol on tumor suppressor protein functional recovery is evidenced by an increase in mutant merlin expression levels and reduced cell proliferation, β-catenin expression, and F-actin stress fiber formation (Fig. 4 F and G). These results indicate that celastrol prolongs merlin mutant half-life, likely through its effects on mutant protein binding with important mediators of the protein folding and maturation process.
Discussion
Although Knudsen's two-hit model of tumorigenesis describes the genetic mechanism that underlies inherited neoplastic syndromes (19, 20), the specific pathogenic mechanisms underlying functional loss of gene-specific translated proteins are not known. Despite variable mutations in the NF2 tumor suppressor gene, biallelic loss of this gene results in the loss of merlin function (tumor suppressor protein encoded by the NF2 gene). The loss of merlin function in NF2 results in dysregulation of cellular growth control pathways and uncontrolled cellular proliferation, which gives rise to tumors of the nervous system. To gain insight into mechanisms of merlin functional loss in NF2, we investigated pathways that mediate mutant merlin function, quality control, and degradation. After determining that accelerated protein degradation underlies the loss of mutant merlin function, we identified potential modifiers of merlin degradation pathways that could serve as putative therapeutics in NF2.
To assess whether reduced merlin underlies its functional loss in NF2, we analyzed merlin expression levels in tumors (meningiomas and schwannomas) from NF2 patients. Western blot analysis revealed that there was a significant decrease (95% reduction in meningiomas and schwannomas) in the quantity of merlin across all samples compared with controls. Immunofluorescence underscored these quantitative results and demonstrated the absence of merlin in NF2-associated tumors. These findings are consistent with results from previous work examining the qualitative expression pattern of merlin in tumors from NF2 patients (21). Although these data indicate that reduced expression or increased degradation could underlie tumor formation in NF2, the potential mechanisms that underlie quantitative loss of merlin in NF2 were unknown.
To determine the cause of underlying quantitative decreases in expression and function of mutant merlin in NF2, we investigated possible mechanisms, including altered gene transcription, abnormal protein translation, and reduced stability of mutant merlin products. Consistent with intact NF2 gene transcription, RT-PCR analysis of NF2-associated tumors demonstrated that NF2 gene mRNA expression levels were similar to those in control cells. Evidence of effective translation was established by pulse chase analysis, which revealed that successful translation of both mutant and wild-type merlin occurs. Whereas evidence of effective transcription and translation of mutant merlin was demonstrated, pulse chase analysis revealed a reduction in mutant merlin half-life across all mutations, which was found to be 2–4 times shorter than that of wild-type merlin. Additionally, increased ubiquitylation of mutant merlin occurred, which suggests that reduced merlin expression in NF2-associated tumors is the result of enhanced degradation through previously described ubiquitin-mediated merlin degradation pathways (7).
To determine whether reduced total mutant merlin function results from impaired intrinsic function in addition to enhanced degradation, we introduced plasmid vectors containing common NF2 missense mutations (22, 23) into a meningioma cell line lacking the NF2 gene. Reintroduction of wild-type or mutant merlin genes in this cell line reversed previously described effects of NF2 deficiency (i.e., abnormalities in intracellular F-actin organization, stress fiber formation, reduced cell to cell contact inhibition, and increased cell proliferation). Specifically, introduction of mutant or wild-type NF2 genes resulted in normalization of the cytoskeleton, including reduction of F-actin stress-fiber formation (11) and restoration of cell to cell contact inhibition. Moreover, incorporation of mutant or wild-type NF2 genes led to a reduction in the S-phase population of cells, indicating a reduction in the proliferative and mitotic capacity. These changes in cell cycle were validated by a reduction in the proliferation of meningioma cells transduced with mutant and wild-type merlin vectors compared with empty vector controls.
Taken together, the preceding results suggest that missense mutations in the NF2 gene encode for mutant merlin that is functionally competent and that intrinsic mutant merlin functional loss does not underlie the pathogenesis of NF2 tumor formation. Rather, enhanced degradation of mutant forms of merlin may in fact underlie the pathogenesis of NF2. These findings provide potential insight into the clinical findings associated with NF2. Specifically, variations in missense mutations resulting in a spectrum of mutant merlin proteins with different degradation rates (as demonstrated by pulse chase and scintillation analyses) may underlie the variable severity of NF2 clinical manifestations and/or the variability of growth patterns associated with NF2-associated tumors. A similar mechanism of protein loss underlies the pathogenesis and disease severity described in Gaucher disease patients in which enzyme quantity, rather than function, was decreased in patients with this heritable neurological disorder (17).
The finding that amino acid substitutions in mutant merlin result in quantitative reductions in protein expression, rather than intrinsic functional changes, suggests that NF2 missense mutations result in altered protein stability and implicates a protein quality control pathway through which such mutant merlin is detected and degraded within cells. Acceleration of these degradative pathways with missense merlin mutants may occur as a result of protein misfolding, a concept supported by previous work examining the crystal structure of merlin; results from this study demonstrated accelerated degradation of missense merlin mutants, such as L64P, that have altered secondary and tertiary protein structure and may thus lead to improper protein folding (24). Also consistent with the idea of increased degradation of mutant merlin by protein quality control pathways, the application of proteostasis regulators (celastrol and quercetin) significantly increased merlin half-life.
The results of this study have implications for both diagnostics and therapeutics for NF2 and, potentially, other heritable tumor suppressor syndromes. Current molecular diagnostics are aimed at detecting gene mutations and loss of heterozygosity in tumor suppressor genes, which largely rely on the comparison of single-nucleotide polymorphisms of different alleles using a relatively large quantity of tumor sample as well as patient-matched normal control tissue. The current results suggest that a tumor suppressor protein quantification assay might be of greater utility in the diagnosis of the disease and in the prediction of disease severity. Further, because manipulation of protein quality control pathways using proteostasis regulators results in increased expression of functional mutant tumor suppressor protein, specific molecular mediators involved in these pathways may provide a unique treatment paradigm for NF2 and other similar disorders.
Materials and Methods
NF2 Tumor Samples and Tissue Dissection.
All tissue was collected at the Surgical Neurology Branch at National Institute of Neurological Disorders and Stroke. Tissue samples and clinical information were obtained as part of an Institute Review Board-approved study. Frozen tissue samples included three meningiomas, four schwannomas, one perineurioma, and two normal brain and skin samples. Tissue dissection was performed as previously described (25).
Western Blot.
Microdissected tissue and cell pellets were lysed in RIPA lysis buffer (Thermo), sonicated, and centrifuged. The quantity of protein was determined in the supernatant solution using a Bio-Rad Protein Assay kit. Proteins were separated by NuPAGE 4–12% Bis-Tris gel (Invitrogen) and transferred to PVDF membranes (Invitrogen). Membranes were blocked in 5% skim dried milk and immunoblotted with primary antibody. The following antibodies were used: Merlin (1:1,000, Sigma), Flag (1:2,000, Origene), Ubiquitin (1:1,000, Cell Signaling), N-Cadherin (1:1,000, Abcam), VE-Cadherin (1:1,000, Abcam), β-catenin (1:2,000, Cell Signaling), Cyclin D1 (1:1,000, Cell Signaling), Hsp70 (1:1,000, Sigma), Hsp90 (1:1,000, Cell Signaling), TCP1 (1:1,000, Sigma), and β-Actin (1:2,000, Sigma).
Immunoprecipitation.
Immunoprecipitation was performed as previously described (17) with minor modifications. Two hundred micrograms of whole-cell lysates was precipitated using DynaBeads Protein G immunoprecipitation kit (Invitrogen) with monoclonal antibodies against Flag (1:200, Origene). Precipitated protein was analyzed by Western blot.
Immunofluorescence Analysis.
Cells or tissue slices were fixed in Histochoice and labeled with primary antibodies overnight. Anti-Flag antibody (1:200, Origene) was used for visualization of mutant merlin. Tissue sections from NF2-associated tumors were labeled with merlin antibody (1:200, Sigma). F-actin was labeled with Alexa Fluor 488 phalloidin (Invitrogen) for 20 min. Cell nuclei were counterstained with Hoechst 33342 (Invitrogen). The specimens were visualized using a Zeiss LSM 510 confocal microscope.
RT-PCR.
Tumor samples were dissected and RNA extraction was performed using the RNeasy extraction kit (Qiagen). RNA was reverse-transcribed with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) for cDNA. NF2 expression was determined by PCR using gene-specific primers (Invitrogen).
Cell Culture and Transfection.
CH157 NM cells were maintained in DMEM (Invitrogen) containing 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were transfected with wild-type or mutant NF2 vectors by using Lipofectamine 2000 (Invitrogen).
DNA Cloning and Site-Directed Mutagenesis.
Mutagenesis was performed using a QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent). Missense mutations were selected based on frequent mutations in the NF2 syndrome (22). Mutations in the NF2 gene were generated in pCMV6-Entry vectors with the full-length wild-type NF2 gene (Origene). For each mutant, the sequence of mutant NF2 genes was verified with DNA sequencing of the entire coding region.
Metabolic 35S Labeling and Pulse Chase Assay.
A total of 7 × 105 CH157 cells was transfected with mutant merlin vectors 12 h before labeling. Radioisotopic protein labeling with radioisotopes was performed by methionine starvation for 15 min followed by growth in methionine-free DMEM supplemented with 0.2 mCi/mL [35S]-methionine (>1,000 Ci/mmol specific activity, Perkin-Elmer). The cells were then chased in DMEM with 10% FBS and 3 mg/mL methionine. Cells were lysed with RIPA lysis buffer containing a proteinase inhibitor mixture (Thermo). Merlin proteins were immunoprecipitated using 200 μg from whole cell lysates with monoclonal Flag antibody (Origene). Precipitated proteins were fractionated with NuPAGE Novex 4–12% Bis-Tris Gel (Invitrogen) and measured by liquid scintillation counting. The gel was fixed and incubated in EN3HANCE (Perkin-Elmer) for 30 min and dried for subsequent X-ray film exposure.
MTT Metabolic Assay.
CH157 NM cells were lipofected with mutant NF2 vectors and aliquoted in triplicate in 96-well plates with a concentration of 5 × 103 cells/100 μL of growth media. Cell proliferation was measured using a modified MTT assay kit (ATCC). Absorbance values of formazan were measured at 590 nm with an automatic plate reader.
BrdU Incorporation and Flow Cytometry.
To visualize cell cycle changes, CH157 NM cells were labeled with 20 μM BrdU (Sigma) for 2 h. Cells were fixed with 70% ethanol on ice. Cells were then washed with PBS and sequentially treated with 2 N HCl/Triton X-100 and 0.1 M Na2B4O7. Cells were blocked with Superblock (Thermo) and stained with anti-BrdU antibody (BD) followed by Alexa Fluor 488 conjugated secondary antibody. DNA was counterstained with 0.1 mg/mL propidium iodide. Cells were analyzed using a dual-laser FACSVantage SE flow cytometer (Becton Dickinson). Cell Quest Acquisition and Analysis software (Becton Dickinson) was used to acquire and quantify fluorescent signal intensities and to graph the data as bivariate dot density plots.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Neurologic Disorders and Stroke at the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
References
- 1.Martuza RL, Eldridge R. Neurofibromatosis 2 (bilateral acoustic neurofibromatosis) N Engl J Med. 1988;318:684–688. doi: 10.1056/NEJM198803173181106. [DOI] [PubMed] [Google Scholar]
- 2.Trofatter JA, et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;72:791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- 3.Lallemand D, Curto M, Saotome I, Giovannini M, McClatchey AI. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 2003;17:1090–1100. doi: 10.1101/gad.1054603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McClatchey AI, Giovannini M. Membrane organization and tumorigenesis–the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- 5.Asthagiri AR, et al. Neurofibromatosis type 2. Lancet. 2009;373:1974–1986. doi: 10.1016/S0140-6736(09)60259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asthagiri AR, Helm GA, Sheehan JP. Current concepts in management of meningiomas and schwannomas. Neurol Clin. 2007;25:1209–1230. doi: 10.1016/j.ncl.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Gautreau A, et al. Mutant products of the NF2 tumor suppressor gene are degraded by the ubiquitin-proteasome pathway. J Biol Chem. 2002;277:31279–31282. doi: 10.1074/jbc.C200125200. [DOI] [PubMed] [Google Scholar]
- 8.Ragel BT, et al. A comparison of the cell lines used in meningioma research. Surg Neurol. 2008;70:295–307. doi: 10.1016/j.surneu.2007.06.031. discussion 307. [DOI] [PubMed] [Google Scholar]
- 9.Mackay DJ, Esch F, Furthmayr H, Hall A. Rho- and rac-dependent assembly of focal adhesion complexes and actin filaments in permeabilized fibroblasts: an essential role for ezrin/radixin/moesin proteins. J Cell Biol. 1997;138:927–938. doi: 10.1083/jcb.138.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jay V, Edwards V. Filamentous aggregates in a meningioma. Ultrastruct Pathol. 1996;20:577–583. doi: 10.3109/01913129609016362. [DOI] [PubMed] [Google Scholar]
- 11.James MF, et al. Modeling NF2 with human arachnoidal and meningioma cell culture systems: NF2 silencing reflects the benign character of tumor growth. Neurobiol Dis. 2008;29:278–292. doi: 10.1016/j.nbd.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutmann DH, et al. Increased expression of the NF2 tumor suppressor gene product, merlin, impairs cell motility, adhesion and spreading. Hum Mol Genet. 1999;8:267–275. doi: 10.1093/hmg/8.2.267. [DOI] [PubMed] [Google Scholar]
- 13.Saydam O, et al. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol Cell Biol. 2009;29:5923–5940. doi: 10.1128/MCB.00332-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta V, Samuleson CG, Su S, Chen TC. Nelfinavir potentiation of imatinib cytotoxicity in meningioma cells via survivin inhibition. Neurosurg Focus. 2007;23:E9. doi: 10.3171/FOC-07/10/E9. [DOI] [PubMed] [Google Scholar]
- 15.Huang J, Chen J. VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene. 2008;27:4056–4064. doi: 10.1038/onc.2008.44. [DOI] [PubMed] [Google Scholar]
- 16.Wenzel T, Baumeister W. Conformational constraints in protein degradation by the 20S proteasome. Nat Struct Biol. 1995;2:199–204. doi: 10.1038/nsb0395-199. [DOI] [PubMed] [Google Scholar]
- 17.Lu J, et al. Decreased glucocerebrosidase activity in Gaucher disease parallels quantitative enzyme loss due to abnormal interaction with TCP1 and c-Cbl. Proc Natl Acad Sci USA. 2010;107:21665–21670. doi: 10.1073/pnas.1014376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 19.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 21.Hitotsumatsu T, et al. Expression of neurofibromatosis 2 protein in human brain tumors: an immunohistochemical study. Acta Neuropathol. 1997;93:225–232. doi: 10.1007/s004010050608. [DOI] [PubMed] [Google Scholar]
- 22.Ahronowitz I, et al. Mutational spectrum of the NF2 gene: a meta-analysis of 12 years of research and diagnostic laboratory findings. Hum Mutat. 2007;28:1–12. doi: 10.1002/humu.20393. [DOI] [PubMed] [Google Scholar]
- 23.Nordstrom-O'Brien M, et al. Genetic analysis of von Hippel-Lindau disease. Hum Mutat. 31:521–537. doi: 10.1002/humu.21219. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu T, et al. Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain. J Biol Chem. 2002:277, 10332–10336. doi: 10.1074/jbc.M109979200. [DOI] [PubMed] [Google Scholar]
- 25.Furuta M, et al. Protein patterns and proteins that identify subtypes of glioblastoma multiforme. Oncogene. 2004;23:6806–6814. doi: 10.1038/sj.onc.1207770. [DOI] [PubMed] [Google Scholar]