Abstract
To cleave DNA, the Type III RM (restriction–modification) enzymes must communicate the relative orientation of two recognition sequences, which may be separated by many thousands of base pairs. This long-range interaction requires ATP hydrolysis by a helicase domain, and both active (DNA translocation) and passive (DNA sliding) modes of motion along DNA have been proposed. Potential roles for ATP binding and hydrolysis by the helicase domains are discussed, with a focus on bipartite ATPases that act as molecular switches.
Keywords: ATP-binding cassette transporter (ABC transporter), DNA sliding, gyrase, heat-shock protein 90, histidine kinase and MutL ATPase (GHKL ATPase), helicase, molecular switch
Abbreviations: 3D, three-dimensional; ABC transporter, ATP-binding-cassette transporter; AFM, atomic force microscopy; CTD, C-terminal domain; dsDNA, double-stranded DNA; GHKL, gyrase, heat-shock protein 90, histidine kinase and MutL; Hsp90, 90 kDa heat-shock protein; MMR, mismatch repair; NBD, nucleotide-binding domain; NTD, N-terminal domain; RE, restriction endonuclease; RM, restriction–modification; SF, superfamily; TMD, transmembrane domain
Introduction
Many cellular transactions require enzymes that bind and hydrolyse nucleoside triphosphates. Many of these are ‘molecular motors proteins’: they couple chemical energy to mechanical events such as protein motion. The helicases represent one class of motor protein [1]. They are widespread in all domains of life and play roles in every aspect of genome biology. On the basis of amino acid sequence motifs, they are classified into six SFs (superfamilies). All are NTPases and the classically defined role of a ‘helicase’ is separation of two strands of DNA/RNA. A large body of evidence has coalesced into a basic mechanism for coupling NTP-binding and hydrolysis to stepwise motion along a polynucleotide (see below) [1–4]. However, there are also a growing number of examples of helicases that can unwind multiple base pairs without stepwise motion and with the consumption of relatively few ATP molecules [5]. In addition, there are numerous other examples where ATP-coupling appears to play an alternative role to strand separation, e.g. in dsDNA (double-stranded DNA) translocation [6]. This review considers one such non-classical system, the Type III RM (restriction–modification) enzymes [7,8]. Possible roles for nucleotide-binding are discussed by analogy to NTP-driven motors and molecular switches.
The ATP-dependent Type III RM enzymes
Type III RM enzymes cleave DNA following recognition of specific DNA sequences (e.g. 5′-CAGCAG-3′ for EcoP15I) and play an important role in bacteria and archaea by protecting against infection by parasitic nucleic acids. They form heterotetramers of two Res and two Mod subunits [9,10]: Mod contains motifs characteristic of an adenine MTase (methyltransferase), recognizes the target site and methylates one DNA strand to prevent host genome cleavage; Res contains motifs characteristic of SF2 DNA helicases [in the NTD (N-terminal domain)] and PD(D/E)XK nucleases [in the CTD (C-terminal domain)] [7,11]. DNA cleavage requires two Res2Mod2 complexes to bind two target recognitions sequences on the same DNA molecule in an indirectly repeated orientation, i.e. either HtH (head-to-head) or TtT (tail-to-tail) [12]. Type III enzymes thus show ‘site orientation selectivity’ [8]. ATP hydrolysis is absolutely required for DNA cleavage via long-range communication between the sites [13,14].
Despite the presence of dual helicase subunits, there is no known role for duplex unwinding by Type III enzymes. Instead, and by analogy to the related ATP-dependent Type I RM enzymes, it was first suggested that Type III enzymes couple ATP hydrolysis to unidirectional dsDNA loop translocation [8,15]. Evidence for loop translocation has been obtained using AFM (atomic force microscopy) [16,17]. Translocation without loop formation has also been suggested [18]. However, other studies did not find evidence for either long-lived DNA loops or directional translocation [12,19,20]. Alternatively, it was suggested that ATP is used to catalyse a conformational switch from DNA recognition to one-dimensional DNA diffusion (also known as ‘DNA sliding’) [8,19]. Communication is therefore driven by thermal energy and does not require ATP hydrolysis except during initiation.
To identify the true communication mechanism, it is important to resolve the role of ATP. In addressing this problem one needs to consider: (i) why two Res subunits (and thus two helicases) are present; and (ii) the apparently high ATP coupling efficiency compared with Type I RM enzymes [15,19,21].
Translocating: nucleotide hydrolysis coupled to stepping motion on nucleic acids
SF2 helicase structures reveal a protein architecture built around two linked RecA-like domains (N-core and C-core) that are involved in nucleotide binding, polynucleotide binding and mechanochemical coupling [1]. N- and C-cores form a nucleotide-binding pocket within which are arrayed conserved amino acid helicase motifs. Binding of ATP between the domains ‘zippers up’ the pocket, causing a conformation change that is coupled to the DNA or RNA. For the classical helicases, this results in the ‘inchworm’ model in which alternating protein contacts are used to walk along single-stranded polynucleotides with the consumption of one ATP molecule per nucleotide moved [1–4]. This underlying mechanochemical coupling appears universal and dsDNA translocases show a similar unitary coupling ratio [22]. dsDNA translocation also requires principal motor contacts to one duplex strand and can be said to have a polarity [6]. A modified inchworm mechanism for 3′–5′ dsDNA translocation is illustrated (Figure 1A).
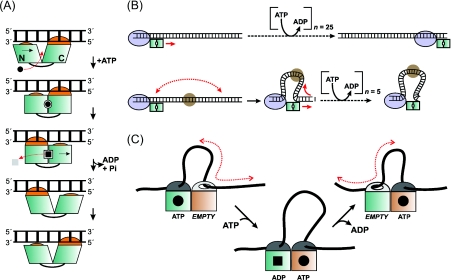
Figure 1. Models for ATP-coupling by Type III restriction enzymes.
(A) Modified inchworm model for dsDNA translocation [6,22]. ATP binding between the N- and C-core RecA domains of Res causes domain motions that are coupled to motion along one strand of intact dsDNA. In making 1 bp steps, the motor or DNA must rotate around the helical axis (not shown) [22]. (B) 3D DNA looping to shorten DNA translocation distances. The Mod complex is represented as a blue oval, and a non-specific bound protein as a brown circle. Each single base pair step along DNA consumes one ATP molecule. (C) Facilitated diffusion against a reflecting barrier [29]. Separate helicase subunits are shown as green and brown squares and DNA is shown as a black line.
Although inchworm translocation appears an attractive option given the similarities between the RM enzymes, an important limitation in applying the model to Type III enzymes is that they consume at least 1000-fold fewer ATP molecules than their Type I counterparts. Although a coupling ratio has not been measured directly, one can be inferred by comparing ATP and DNA hydrolysis rates. This gives a range of (at least) tens to hundreds of base pairs communicated per ATP molecule [19,21]. These values are incompatible with current structural views of helicases. Moreover, in contrast with the Type I enzymes [22], the ATPase kinetics of the Type III enzymes have not provided simple Michaelis–Menten relationships [13]. Finally, triplex displacement assays that have been successfully used with every other bona fide translocase have failed to provide evidence for Type III translocation [19].
Without invoking Brobdingnagian helicase step sizes, two models have been suggested that could account for unexpected bp/ATP coupling ratios.
Movement of the motor by passive 3D (three-dimensional) looping to a distant site
Based on the observation by AFM of stable DNA loops formed by Type III REs (restriction endonucleases) [16], it was suggested that 3D DNA looping could shorten the distance to a target, thus giving a larger apparent coupling ratio (Figure 1B). However, while DNA looping should allow Type III motors to by-pass downstream DNA-binding proteins, communication is actually inhibited by protein roadblocks [12,15]. Another important issue is that without special geometric constraints, passive 3D looping will not preserve relative site orientation and will also allow communication between sites on separate DNA strands [23–25]. Type II REs that use 3D looping can communicate between DNA catenane rings [26–28], whereas Type III REs cannot [20].
‘Facilitated diffusion against a reflecting barrier’
The ‘facilitated diffusion against a reflecting barrier’ scheme was originally suggested for the DNA MMR (mismatch repair) protein MutS to account for similar observations of low ATPase rates and DNA loops [29]. Two ATPase domains alternate between tight and loose DNA binding states. During the loose-bound state, DNA diffuses back-and-forth past the enzyme. This could produce loops, allow movement of >1 bp per ATP molecule, and retain relative binding orientation. It also requires a ‘two cylinder’ mechanism using dual ATPase domains, consistent with two Res subunits. Because DNA loop motion is passive, this model is essentially a modified sliding scheme. However, DNA cleavage by Type III REs is force-independent [19], suggesting that loops do not play a critical role in diffusive motion.
In favouring an inchworm model (Figure 1A), one must also question the role of dual Res subunits. Helicases display a variety of oligomerization states, but are principally active as hexamers or monomers [1]. Unlike the hexameric helicases where a composite ATPase active site is formed at the interface of two subunits, SF2 enzymes use contacts between N- and C-cores in one subunit. Where dimers do form, each helicase domain remains independent [30]. A hand-over-hand (or ‘rolling’) model for motion by helicase dimers has been superseded by the monomeric inchworm model. Dimerization may be an evolutionary accident that simply sequesters hydrophobic interfaces. Alternatively, it may activate helicase activity by sequestering autoinhibitory domains [31].
Switching and gating: NTPases that modulate protein conformations
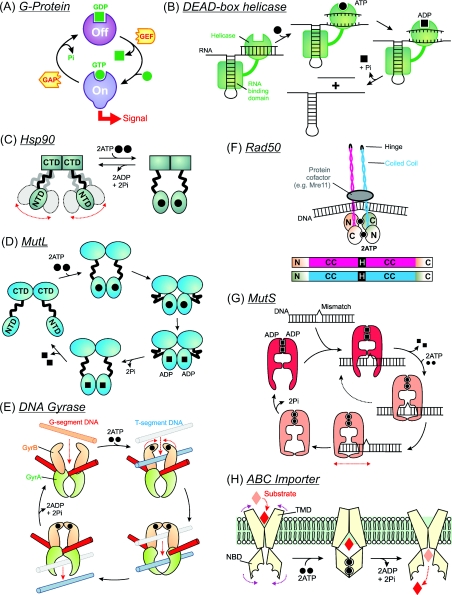
Rather than being coupled to a power stroke, NTP hydrolysis can also allow cycling between multiple conformational states, producing a so-called ‘molecular switch’ as first described for small monomeric GTPases (Figure 2A) [32]. In these G-proteins, signalling is activated by GTP binding and inactivated by GTP hydrolysis. Regulators guide the switching: GAPs (GTPase-activating protein) increase GTP hydrolysis, whereas GEFs (guanine-nucleotide-exchange factors) potentiate GDP release. Roles for similar NTP-driven molecular switches on DNA and RNA now appear widespread. One example are rRNA chaperones (Figure 2B), where monomeric helicase domains can unwind multiple base pairs through a conformational change upon binding a single ATP molecule [5]. Hydrolysis actually drives product release so that the enzyme can turn over. This is quite distinct to the inchworm model and illustrates that helicases can also act as one-step switches.
Figure 2. Schematic models for molecular switches and pumps.
Models illustrated are driven by the binding of one NTP (A and B) or two NTPs (C–H). See the text for further details.
Both the above examples require only a single NBD (nucleotide-binding domain). But there are also dimeric switches in which two nucleotides must bind and where nucleotide-binding stabilizes composite active sites. Two motor classes will be considered here. The first of these are members of the GHKL (gyrase, heat-shock protein 90, histidine kinase and MutL) ATPase SF [33], including the protein chaperone Hsp90 (90 kDa heat-shock protein), the DNA MMR protein MutL and the type II topoisomerases. The family shares a common NBD architecture (the Bergerat fold) and has a necessity for dimerization coupled to large-scale conformational changes as part of their ATP-binding cycles. Domain sharing during dimerization activates ATP hydrolysis because some catalytic residues must act in trans between subunits. ATP-binding and domain dimerization also initiates a cascade of protein–protein assembly events, a common feature of the dual ATP switches. Hydrolysis of ATP and ADP/Pi release then allows the proteins to release their cofactors and turnover. Measured ATPase rates are often relatively low (<1/min) [34], which may in part reflect their switch role.
Hsp90
Hsp90 comprises three domains, with a stable dimer interface between the CTDs and reversible dimerization of the NTDs driven by ATP binding [35]. In the absence of nucleotides, the NTDs are free and highly flexible (Figure 2C). Association of two ATP molecules causes a pincer movement that dimerizes the NBDs and entraps client proteins for stabilization or partial refolding. ATP hydrolysis and ADP/Pi opens the clamp and releases the client, resetting Hsp90 for another binding event. The ATPase cycle is also tightly linked to the binding of other co-chaperones.
MutL
Bacterial MutL and eukaryotic homologues play molecular ‘matchmaker’ roles in MMR and other repair pathways [36]. The bacterial enzymes are homodimers with a CTD dimer interface and N-terminal NBDs that both dimerizes upon nucleotide binding and alter in orientation relative to the CTDs (Figure 2D) [34]. As above, the ATPase cycles (and thus conformational changes) are intimately coupled to the recruitment of other proteins. It is not clear, however, if dimerization directly entraps other proteins and/or DNA. The eukaryotic homologues are heterodimers with different ATP hydrolysis rates in each subunit, leading to asymmetry in the reaction cycle [36].
Type II topoisomerases
These enzymes illustrate another property of switches: the co-ordinated opening and closing of multiple protein ‘gates’ (Figure 2E) [37]. For example, bacterial DNA gyrase is a heterotetramer of two GyrA subunits and two GyrB subunits (the latter containing the NBDs). Gates are formed from protein dimer interfaces. Changes in DNA topology occur by crossing two dsDNA, cutting one of the helices (the G-segment) and passing the other (the T-segment) through it, before resealing the break. ATP-induced dimerization of the NBDs forms the ATP gate that captures the T-segment and sets in motion the cascade of domain motions. The exact role of ATP hydrolysis has been much debated as many of the reactions of gyrase are energetically favourable [38]. Since the broken G-segment is potentially lethal, ATP binding and hydrolysis may be vital in tightly regulating gating.
The second family of dual ATP switches is the ABC transporter (ATP-binding cassette transporter) SF [39], which includes the membrane-bound ABC transporters, the DNA MMR protein MutS and the SMC (structural maintenance of chromosome)-related DNA repair enzyme Rad50. They share a RecA-like fold similar to the helicases and other ATP-dependent machines [40]. However, the NBDs associate as head-to-tail dimers [41], with ATP molecules bridging the interface to interact with Walker A and B motifs in one subunit and with an ABC-specific signature motif in the partner subunit. As above, ATPase rates can be relatively low [42].
Rad50
Rad50 is a required component of double strand break repair and comprises an NBD with a >600-residue heptad repeat insertion that forms a coiled-coil between the N- and C-terminal ATPase lobes (Figure 2F) [39,41]. Nucleotide-binding promotes dimerization of NBDs leading to formation of a globular head at the end of two large coiled-coil arms, with the latter forming assembly sites for DNA breaks and protein cofactors.
MutS
Homodimeric bacterial MutS and heterodimeric eukaryotic homologues recognize DNA mismatches and initiate MMR [42,43]. A MutS subunit consists of five domains: domain V contains the NBD/dimerization interface, domains I and IV bind to DNA and the other domains act as connector domains to relay conformational changes. Events at the NBDs and their effects on mismatch recognition have been thoroughly studied, although a consensus model is still to be accepted. In a striking parallel to the Type III enzymes, DNA translocation, DNA looping and DNA sliding models have all been proposed [43]. A highly-simplified view of the sliding scheme is illustrated in Figure 2(G). The ADP-bound form (which is a stable enzyme–product complex) binds to the mismatch, exchanges with ATP and forms a sliding clamp. On linear DNA substrates with free ends, ATP induces MutS dissociation, while on linear DNA with ends blocked with streptavidin, the ATP-induced dissociation is significantly reduced. Similarly, DNA cleavage by Type III REs is enhanced on linear DNA with streptavidin-blocked ends [19]. Upon release of MutS from the DNA, ATP hydrolysis resets the ADP-bound state for another repair cycle.
Asymmetry in nucleotide binding and hydrolysis between each MutS ATPase site produces different nucleotide bound states with different affinities for hetero- and homo-duplex DNA [42]. It is not clear how cellular nucleotide levels would influence the model. Since many of these studies used isolated MutS rather than the complete MMR machinery, ATP-induced MutL conformational changes (and other protein cofactors) could further alter the mechanism.
ABC membrane transporters
This large family of membrane-associated importers and exporters comprise four domains, two TMDs (transmembrane domains) and two NBDs, which can be found on one, two or four polypeptides [44]. A substrate import mechanism is shown in Figure 2(H). Substrate enters the TMD, partially crossing the bilayer as far as a membrane gate. NBD dimerization causes TMD rearrangements, closing the entry gate and opening the membrane gates. The substrate can now exit and ATP hydrolysis resets the system. A stoichiometry of two ATP molecules per transported cargo molecule has been measured [45], suggesting a coupled mechanism, although alternative models have also been discussed [46].
Roles for ATP in the Type III RM enzymes
Taking a straightforward view of the Res subunits as dsDNA translocases (Figure 1A), one possible role for ATP might be to catalyse stepping along a short stretch of dsDNA adjacent to the target site that pulls (or pushes) the Mod2 complex into a sliding configuration. This is akin to established models for nucleoprotein remodelling by helicases, for example displacement of stalled RNA polymerases [47]. Remodellers can also have relatively low ATPase rates [48]. Nonetheless, the dsDNA translocation activity of remodelling helicases can be measured using triplex displacement [31]. One reason for the lack of displacement activity with Type III REs [19] might be that the translocation only occurs at the target site, and that motion thereafter is by diffusion alone.
Alternatively, ATP could drive a switch, co-ordinating conformational changes in Res and/or Mod. Of particular relevance is the role of ATP in switching MutS from mismatch recognition to non-specific diffusion (Figure 2G). Nothing is known about the atomic structure of Res or Mod. Nevertheless it is tempting to speculate that the two Res monomers (perhaps in combination with the Mod2 dimer) form a sliding clamp similar to that suggested for MutS (Figure 2G). For the bipartite ATPases (Figure 2C–H), dimerization is a necessity as the ATPase motifs in a single subunit are physically separated and cannot interact. In contrast, SF2 helicases appear to form complete nucleotide-binding sites using one protein subunit and there is no indication that Type III RE have ABC- or GHKL-family motifs. A modified sliding model could be envisaged where separate (rather than bipartite) ATPase sites catalyse the domain motions required to form the protein clamp.
To clarify the role of ATP, it is important to confirm the Type III subunit stoichiometry, particularly the form that moves on DNA, and to determine the stoichiometry of the ATP hydrolysis cycle. The proposed sliding model could require as little as one or two ATP hydrolysis events to allow dissociation from the site [8]. However, if ATP binding by two Res subunits is required, uncoupled hydrolysis steps could alter the observed coupling ratios.
Acknowledgements
I thank Kara van Aelst, Júlia Tóth, Friedrich Schwarz, David Dryden, Steve Halford, Mark Dillingham, Nigel Savery and Ralf Seidel for discussions on Type III enzymes.
Funding
This work was funded by the Wellcome Trust.
References
- 1.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 2.Dillingham M.S., Wigley D.B., Webb M.R. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 3.Saikrishnan K., Powell B., Cook N.J., Webb M.R., Wigley D.B. Mechanistic basis of 5′–3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 4.Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 5.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanley L.K., Seidel R., Van Der Scheer C., Dekker N.H., Szczelkun M.D., Dekker C. When a helicase is not a helicase: dsDNA tracking by the motor protein EcoR124I. EMBO J. 2006;25:2230–2239. doi: 10.1038/sj.emboj.7601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sistla S., Rao D.N. S-adenosyl-L-methionine-dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:1–19. doi: 10.1080/10409230490440532. [DOI] [PubMed] [Google Scholar]
- 8.Szczelkun M.D., Friedhoff P., Seidel R. Maintaining a sense of direction during long-range communication on DNA. Biochem. Soc. Trans. 2010;38:404–409. doi: 10.1042/BST0380404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janscak P., Sandmeier U., Szczelkun M.D., Bickle T.A. Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J. Mol. Biol. 2001;306:417–431. doi: 10.1006/jmbi.2000.4411. [DOI] [PubMed] [Google Scholar]
- 10.Sears A., Szczelkun M.D. Subunit assembly modulates the activities of the Type III restriction–modification enzyme PstII in vitro. Nucleic Acids Res. 2005;33:4788–4796. doi: 10.1093/nar/gki788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClelland S.E., Szczelkun M.D. The type I and III restriction endonucleases: structural elements in the molecular motors that process DNA. Nucleic Acids Mol. Biol. 2004;14:111–135. [Google Scholar]
- 12.van Aelst K., Toth J., Ramanathan S.P., Schwarz F.W., Seidel R., Szczelkun M.D. Type III restriction enzymes cleave DNA by long-range interaction between sites in both head-to-head and tail-to-tail inverted repeat. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9123–9128. doi: 10.1073/pnas.1001637107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saha S., Rao D.N. ATP hydrolysis is required for DNA cleavage by EcoPI restriction enzyme. J. Mol. Biol. 1995;247:559–567. doi: 10.1016/s0022-2836(05)80137-8. [DOI] [PubMed] [Google Scholar]
- 14.Saha S., Rao D.N. Mutations in the Res subunit of the EcoPI restriction enzyme that affect ATP-dependent reactions. J. Mol. Biol. 1997;269:342–354. doi: 10.1006/jmbi.1997.1045. [DOI] [PubMed] [Google Scholar]
- 15.Meisel A., Mackeldanz P., Bickle T.A., Kruger D.H., Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995;14:2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crampton N., Roes S., Dryden D.T., Rao D.N., Edwardson J.M., Henderson R.M. DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes. EMBO J. 2007;26:3815–3825. doi: 10.1038/sj.emboj.7601807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crampton N., Yokokawa M., Dryden D.T., Edwardson J.M., Rao D.N., Takeyasu K., Yoshimura S.H., Henderson R.M. Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12755–12760. doi: 10.1073/pnas.0700483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raghavendra N.K., Rao D.N. Unidirectional translocation from recognition site and a necessary interaction with DNA end for cleavage by Type III restriction enzyme. Nucleic Acids Res. 2004;32:5703–5711. doi: 10.1093/nar/gkh899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramanathan S.P., van Aelst K., Sears A., Peakman L.J., Diffin F.M., Szczelkun M.D., Seidel R. Type III restriction enzymes communicate in 1D without looping between their target sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1748–1753. doi: 10.1073/pnas.0807193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peakman L.J., Szczelkun M.D. DNA communications by Type III restriction endonucleases–confirmation of 1D translocation over 3D looping. Nucleic Acids Res. 2004;32:4166–4174. doi: 10.1093/nar/gkh762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sears A., Peakman L.J., Wilson G.G., Szczelkun M.D. Characterization of the type III restriction endonuclease PstII from Providencia stuartii. Nucleic Acids Res. 2005;33:4775–4787. doi: 10.1093/nar/gki787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seidel R., Bloom J.G., Dekker C., Szczelkun M.D. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingston I.J., Gormley N.A., Halford S.E. DNA supercoiling enables the type IIS restriction enzyme BspMI to recognise the relative orientation of two DNA sequences. Nucleic Acids Res. 2003;31:5221–5228. doi: 10.1093/nar/gkg743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watson M.A., Gowers D.M., Halford S.E. Alternative geometries of DNA looping: an analysis using the SfiI endonuclease. J. Mol. Biol. 2000;298:461–475. doi: 10.1006/jmbi.2000.3676. [DOI] [PubMed] [Google Scholar]
- 25.Adzuma K., Mizuuchi K. Interaction of proteins located at a distance along DNA: mechanism of target immunity in the Mu DNA strand-transfer reaction. Cell. 1989;57:41–47. doi: 10.1016/0092-8674(89)90170-0. [DOI] [PubMed] [Google Scholar]
- 26.Gowers D.M., Bellamy S.R., Halford S.E. One recognition sequence, seven restriction enzymes, five reaction mechanisms. Nucleic Acids Res. 2004;32:3469–3479. doi: 10.1093/nar/gkh685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall J.J., Gowers D.M., Halford S.E. Restriction endonucleases that bridge and excise two recognition sites from DNA. J. Mol. Biol. 2007;367:419–431. doi: 10.1016/j.jmb.2006.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood K.M., Daniels L.E., Halford S.E. Long-range communications between DNA sites by the dimeric restriction endonuclease SgrAI. J. Mol. Biol. 2005;350:240–253. doi: 10.1016/j.jmb.2005.04.053. [DOI] [PubMed] [Google Scholar]
- 29.Blackwell L.J., Martik D., Bjornson K.P., Bjornson E.S., Modrich P. Nucleotide-promoted release of hMutSα from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem. 1998;273:32055–32062. doi: 10.1074/jbc.273.48.32055. [DOI] [PubMed] [Google Scholar]
- 30.Rudolph M.G., Klostermeier D. The Thermus thermophilus DEAD box helicase Hera contains a modified RNA recognition motif domain loosely connected to the helicase core. RNA. 2009;15:1993–2001. doi: 10.1261/rna.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith A.J., Szczelkun M.D., Savery N.J. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 2007;35:1802–1811. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wittinghofer A., Pai E.F. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem. Sci. 1991;16:382–387. doi: 10.1016/0968-0004(91)90156-p. [DOI] [PubMed] [Google Scholar]
- 33.Dutta R., Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 34.Ban C., Junop M., Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- 35.Pearl L.H., Prodromou C., Workman P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem. J. 2008;410:439–453. doi: 10.1042/BJ20071640. [DOI] [PubMed] [Google Scholar]
- 36.Polosina Y.Y., Cupples C.G. Wot the ‘L-Does MutL do? Mutat. Res. 2010;705:228–238. doi: 10.1016/j.mrrev.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Schoeffler A.J., Berger J.M. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 38.Bates A.D., Maxwell A. The role of ATP in the reactions of type II DNA topoisomerases. Biochem. Soc. Trans. 2010;38:438–442. doi: 10.1042/BST0380438. [DOI] [PubMed] [Google Scholar]
- 39.Hopfner K.P., Tainer J.A. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 40.Ye J., Osborne A.R., Groll M., Rapoport T.A. RecA-like motor ATPases: lessons from structures. Biochim. Biophys. Acta. 2004;1659:1–18. doi: 10.1016/j.bbabio.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 41.Hopfner K.P., Karcher A., Shin D.S., Craig L., Arthur L.M., Carney J.P., Tainer J.A. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 42.Kunkel T.A., Erie D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 43.Iyer R.R., Pluciennik A., Burdett V., Modrich P.L. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 44.Locher K.P. Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. London Ser. B. 2009;364:239–245. doi: 10.1098/rstb.2008.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patzlaff J.S., Van Der Heide T., Poolman B. The ATP/substrate stoichiometry of the ATP-binding cassette (ABC) transporter OpuA. J. Biol. Chem. 2003;278:29546–29551. doi: 10.1074/jbc.M304796200. [DOI] [PubMed] [Google Scholar]
- 46.Jones P.M., O'Mara M.L., Georg A.M. ABC transporters: a riddle wrapped in a mystery inside an enigma. Trends Biochem. Sci. 2009;34:520–531. doi: 10.1016/j.tibs.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Savery N.J. The molecular mechanism of transcription-coupled DNA repair. Trends Microbiol. 2007;15:326–333. doi: 10.1016/j.tim.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Selby C.P., Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]