Abstract
The development of new catalytic methods to functionalize carbon-hydrogen (C-H) bonds continues to progress at a rapid pace due to the significant economic and environmental benefits of these transformations over traditional synthetic methods. In nature, enzymes catalyze regio- and stereoselective C-H bond functionalization using transformations ranging from hydroxylation to hydroalkylation under ambient reaction conditions. The efficiency of these enzymes relative to analogous chemical processes has led to their increased use as biocatalysts in preparative and industrial applications. Furthermore, unlike small molecule catalysts, enzymes can be systematically optimized via directed evolution for a particular application and can be expressed in vivo to augment the biosynthetic capability of living organisms. While a variety of technical challenges must still be overcome for practical application of many enzymes for C-H bond functionalization, continued research on natural enzymes and on novel artificial metalloenzymes will lead to improved synthetic processes for efficient synthesis of complex molecules. In this critical review, we discuss the most prevalent mechanistic strategies used by enzymes to functionalize non-acidic C-H bonds, the application and evolution of these enzymes for chemical synthesis, and a number of potential biosynthetic capabilities uniquely enabled by these powerful catalysts.
I. Introduction
Although the field of chemical synthesis has matured to a discipline capable of generating compounds of amazing complexity, inefficiencies in the synthetic process often limit the application of these molecules toward societal problems.i Improving economies of atoms, chemical steps, and process operations incurred throughout syntheses has therefore been a major goal of chemists seeking to develop new chemical transformations. Ideally, these reactions should enable the synthesis of target molecules from simple building blocks without the need for functional group manipulations or protecting groups.ii Reactions involving catalytic functionalization of carbon-hydrogen (C-H) bonds provide one means to improve synthetic efficiency, and, as this thematic issue illustrates, this field has experienced impressive progress.
Catalytic functionalization of C-H bonds presents three fundamental challenges to chemists: activating an inert C-H bond, functionalizing the activated organic fragment in a manner that permits catalyst turnover, and accomplishing both of these tasks with high chemo-, site/regio-, and stereoselectivity.iii Activation of C-H bonds has been demonstrated on a wide range of substrates, particularly arenes, using numerous transition metal complexes, and many methods for catalytic functionalization have also emerged.iv However, obtaining suitable selectivity, particularly on functionally complex compounds, remains highly challenging. The most common approach has involved using structural and electronic properties of a substrate that render only a single C-H bond of that substrate (e.g. one proximal to a directing group) reactive toward a given catalyst. While these methods eliminate the need for prefunctionalization in some cases, their restricted substrate scope limits their utility, particularly in complex molecules with many C-H bonds of similar reactivity.
Researchers have also appended molecular recognition elements to catalysts capable of activating many substrate C-H bonds to orient the substrate such that only a single C-H bond is presented to the catalyst for reaction.v Unfortunately, adapting such systems for selective recognition of different substrates or different C-H bonds on a given substrate presents significant synthetic challenges. Of course, the synthetic utility of molecular recognition was long ago discovered in nature and provides a major component of the amazing rate enhancements of enzyme-catalyzed reactions, including C-H functionalization, over their non-catalyzed congeners. Unlike any synthetic structures explored, however, the activity and selectivity of enzymes can be systematically optimized for reaction of a substrate via directed evolution.vi
This review is intended to serve as an introduction to the exciting array of enzyme-catalyzed reactions in which non-acidic C-H bonds (i.e. not alpha to a ketone or imine) are functionalized. Only reactions in which C-H bonds are converted to C-X bonds (X = C, N, O, S, halogen), rather than removed in desaturation reactions, are included due to space limitations, and examples are chosen to illustrate mechanistic and reactivity principles rather than to serve as an exhaustive list. Readers are directed to many excellent reviews on specific enzyme classes for more in-depth treatments throughout this article. Also covered are cases in which this novel reactivity has translated to practical applications, cases in which protein engineering has facilitated such applications, and a number of emerging technologies involving enzymatic C-H bond fuctionalization.
II. Enzymatic C-H Bond Functionalization in Nature
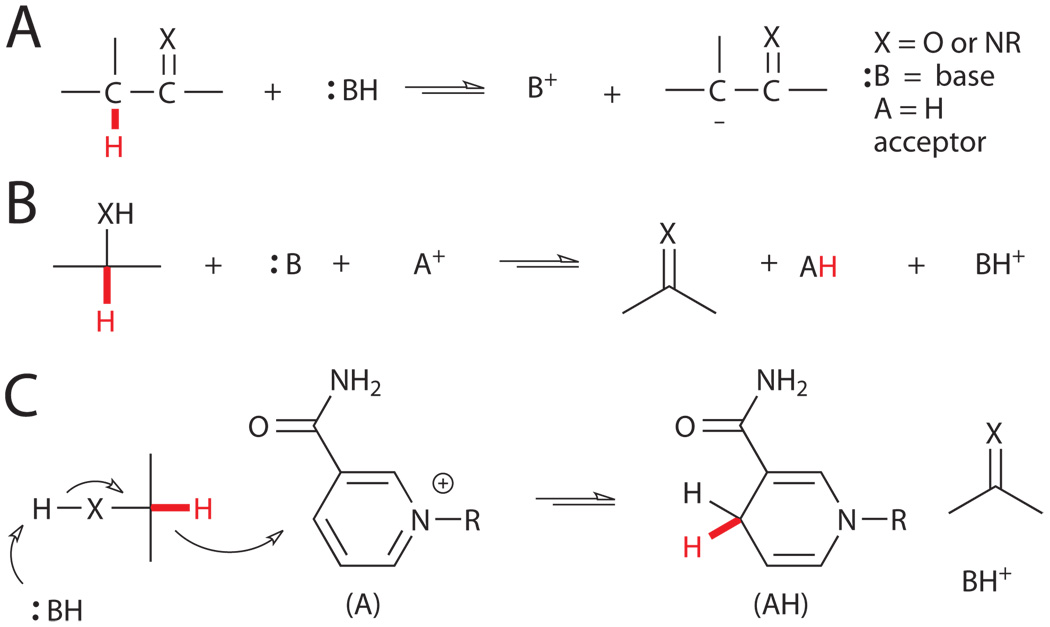
Heterotrophic organisms generate chemical energy (ATP, NAD(P)H, FADH2, etc.) from the conversion of organic molecules into carbon dioxide and water.vii C-H bonds must necessarily be cleaved during this process, but metabolic pathways accomplish this task primarily via deprotonation alpha to carbonyls or imines or via dehydrogenation of alcohols or amines (Fig. 1).viii While these mechanisms can completely account for the metabolism of many compounds (e.g. sugars and lipids), they are powerless against more inert substrates with aryl or unsubstituted-alkyl C-H bonds. However, the abundance of such substrates in certain environments has provided selective pressure for the evolution of enzymes that can activate a range of C-H bonds with pKa’s above 50 and bond dissociation energies (BDEs) above 110 kcal/mol.
Figure 1.
Typical biological routes to C-H bond cleavage include A) deprotonation and B) dehydrogenation. C) Mechanism of alcohol dehydrogenation using NAD(P)H as a hydride acceptor.viii
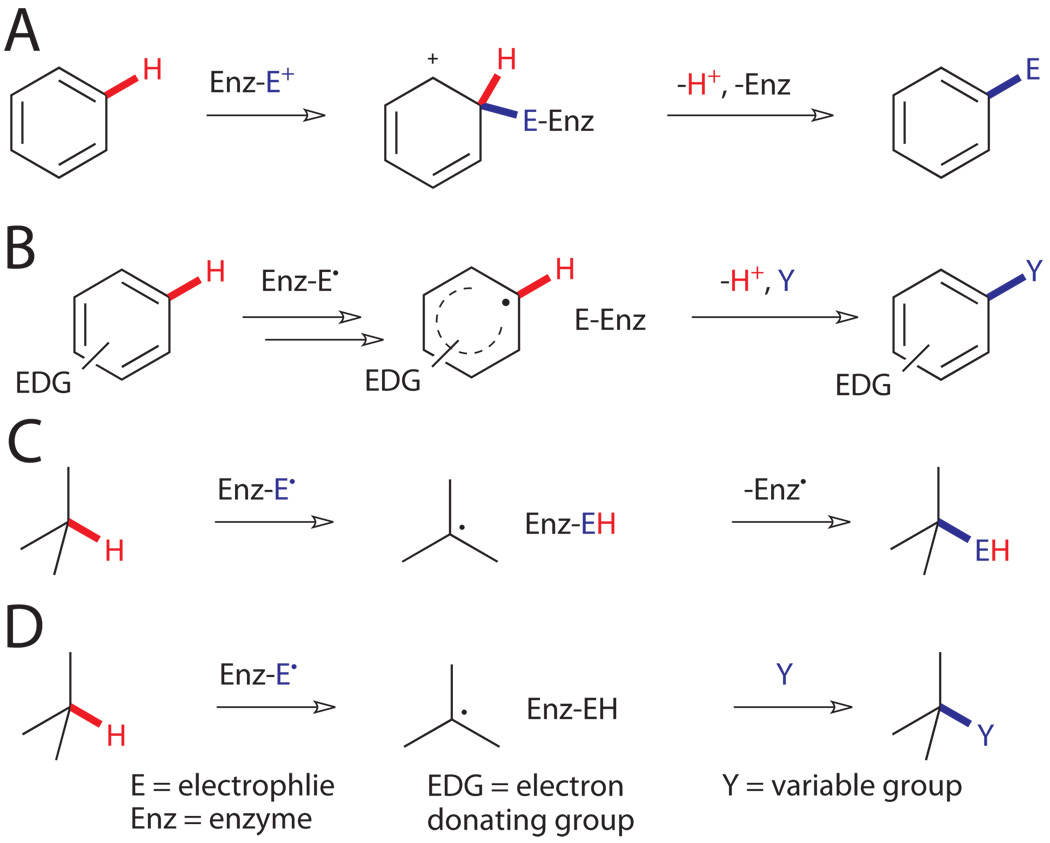
While catalytic activation of such bonds remains largely elusive to synthetic chemists, several different enzymatic mechanisms have emerged in nature to accomplish this task at ambient temperature and pressure in aqueous solution.ix Essentially all of these involve the action of cofactors, frequently metals, positioned within the enzyme active site in a manner that controls the substrate scope and selectivity of the reaction. Two broad classes of reactivity are observed depending on whether functionalization of aromatic or aliphatic C-H bonds is involved. Aromatic substrates are unique in that their C-H bonds can be activated via reaction of their π-electrons.x For example, attack of aromatic π-electrons on an electrophile leads to the formation of a cationic (Wheland) intermediate with significantly enhanced C-H acidity (Fig. 2A). Deprotonation of this intermediate leads to re-aromatization and formal substitution of an aromatic C-H bond by the electrophile to complete the familiar electrophilic aromatic substitution process. Long before chemists began exploiting this reactivity for the synthesis of aniline dyestuffs, enzymes evolved novel means for catalytically generating electrophiles and enforcing high levels of regioselectivity to enable efficient functionalization of aromatic C-H bonds.
Figure 2.
Representative mechanisms for enzymatic functionalization of non-acidic C-H bonds.viii,x,xi A) Electrophilic aromatic substitution. B) Electron abstraction followed by radical coupling. C) H abstraction followed by radical recombination. D) H abstraction followed by radical reaction.
Heterocycles and heteroatom-substituted arenes can also be activated by direct abstraction of their lone-pair electrons to generate radical intermediates (Fig. 2B).xi These can undergo a range of addition or rearrangement reactions, ultimately followed by re-aromatization, to enable functionalization of C-H bonds. Analogous chemical processes for substitution of alkyl C-H bonds are far more energy intensive.x On the other hand, the BDEs of alkyl C-H bonds are lower than those of most arenes, and enzymes have exploited this through the use of radical intermediates to directly abstract H atoms from unactivated alkyl compounds (Fig. 2C, D). The resulting substrate radical can undergo subsequent radical reactions to functionalize the activated position. Again, unlike synthetic radical reactions, high selectivity in both inter- and intramolecular reactions can be enforced by the enzyme active site.
Using such mechanisms, enzymes are able to catalyze the formation of a wide range of C-X bonds (X = C, O, N, S, halogen, etc.). The fate of the reactive intermediates (i.e. cation, radical cation, or radical) varies greatly depending on the enzyme and can include subsequent H-abstractions, radical recombinations, radical or cationic rearrangements, and many others. These reactions are used in both catabolic and anabolic processes, the latter of which enable the synthesis of complex natural products. A variety of examples will be presented in order to provide a sense for their potential synthetic utility. The reactivity of these enzymes is largely dependent on the type of cofactor they employ, so this will serve as a convenient way to group the enzymes.
III. A Survey of Relevant Cofactors and Their Host Enzymes
A. Flavins and Flavoproteins
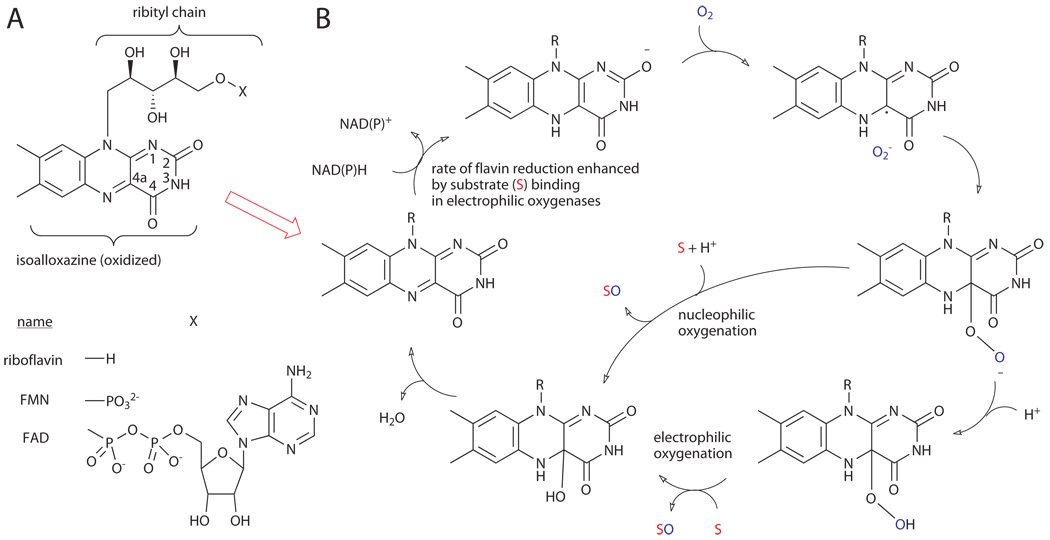
The flavin nucleotides, flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), are redox-active cofactors derived from riboflavin via phosphorylation (FMN) and subsequent adenylation (FAD) (Fig. 3A).xii The chemistry of these compounds derives from their isoalloxazine heterocycle, which can undergo two-electron reduction by NAD(P)H followed by either one-electron-transfer events or two-electron chemical reactions. Because of this ability, flavins are involved in a host of biological processes and serve as an important bridge between polar and radical reactions. Most relevant to the current discussion, however, is their ability to hydroxylate C-H bonds of electron-rich (hetero)arenes.xiii
Figure 3.
A) Structures of riboflavin, FAD, and FMN. B) Mechanism of nucleophilic and electrophilic oxygenation.xiii
Hydroxylation of C-H bonds via insertion of a single oxygen atom derived from dioxygen (monooxygenation) with concomitant reduction of the second oxygen atom to water is a key reaction in the metabolism of a huge number of compounds.xiv This requires reduction of dioxygen to a more reactive state (e.g. superoxide, peroxide, etc.), which in turn requires overcoming the kinetic barrier imposed by the spin mismatch between organic substrates (singlet) and ground state dioxygen (triplet). Flavoproteins accomplish this by promoting single electron transfer from reduced flavin (FADH2) to dioxygen, spin inversion, and radical recombination to generate a nucleophilic flavin peroxide at C4a of the isoalloxazine ring (Fig. 3B).xv This C4a peroxide intermediate can be protonated to generate the corresponding electrophilic hydroperoxyflavin, a mild oxidant (vide infra) that can serve formally as a source of “OH+”. Within the active site of flavoproteins, the regioselectivity of this oxidant toward electron-rich substrates can be controlled. Two broad classes of flavoproteins, the flavin monooxygenases and the FAD-dependent halogenases, utilize this oxidant to functionalize the C-H bonds of electron-rich heterocycles via hydroxylation or halogenation, respectively.xiii
Flavin monooxygenases comprise a highly diverse family of enzymes and are capable of catalyzing many different oxygenation reactions in addition to C-H bond hydroxylation, including epoxidation, Baeyer-Villiger oxidation, and amine oxidation.xiii These variations in reactivity can be roughly correlated with discrete monooxygenase subclasses defined by sequence and structural differences, including number of domains and cofactor requirements (FAD/FMN and NADH/NADPH). Two subclasses defined in this manner have aromatic hydroxylase activity. These enzymes typically exhibit a narrow substrate scope and utilize selective substrate binding to enhance the rate of NAD(P)H-mediated FAD reduction by up to five orders of magnitude (Fig. 3).xii This ensures that FADH2 is generated only in the presence of substrate and minimizes consumption of valuable NAD(P)H and formation of free reactive oxygen species. Such control mechanisms are common in enzyme-catalyzed C-H functionalization reactions and will be highlighted throughout this review.
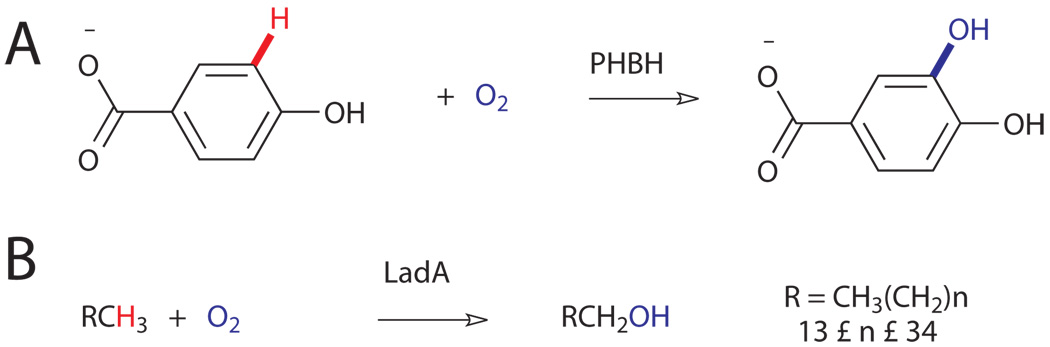
One of the most studied examples of these enzymes is p-hydroxybenzoate 3-hydroxylase (Fig. 4A).xvi As previously mentioned, enzyme-catalyzed reaction of FADH2 with O2 followed by protonation leads to the generation of a flavin C4a hydroperoxide intermediate and places the reactive “OH+” equivalent proximal to a specific site on the aromatic substrate. Electrophilic aromatic substitution of the arene C-H bond with this OH group then provides regioselective hydroxylation of the substrate. Several enzymes with similar activity toward other aromatic substrates are also known.xiii One recent report describes the isolation and characterization of an FMN-dependent long-chain alkane hydroxylase, LadA, most similar to the luciferase subclass of oxygenases (Fig. 4B).xvii Unfortunately, the authors were unable to determine the mechanism through which active site residues might enable use of an FMN-derived oxidant to activate alkyl C-H bonds.
Figure 4.
A) Primary reactions of p-hydroxybenzoate 3-hydroxylase (PHBH)xvi and B) a newly-characterized long-chain alkane hydroxylase, LadA.xvii
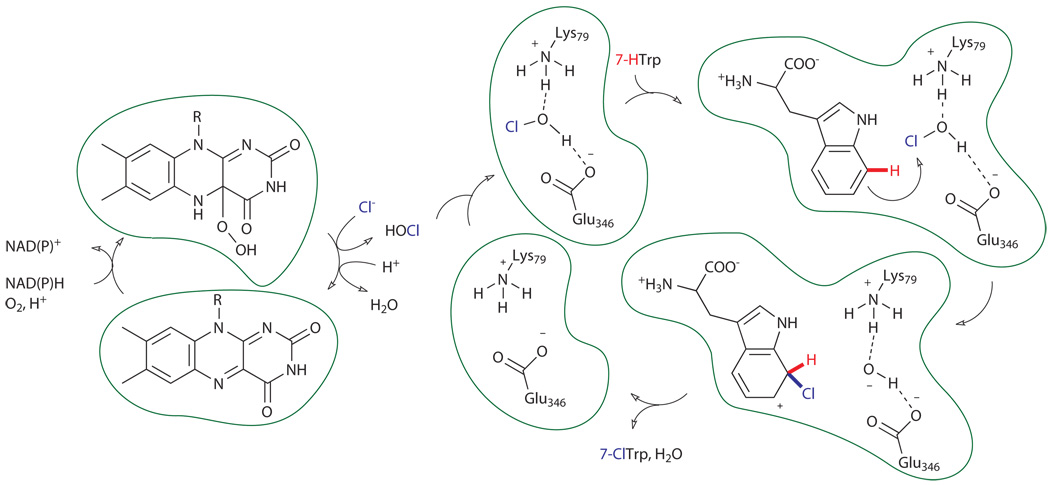
A variation of the monooxygenase mechanism is used by FAD-dependent halogenases to effect halogenation of electron-rich arenes found in a variety of antimicrobial natural products.xviii Several tryptophan halogenases have been characterized, and these are among the most well-studied of the FAD-dependent halogenases (Fig. 5).xix As their name suggests, these enzymes catalyze the halogenation of tryptophan, but they do so at the 5-, 6-, or 7-position of the benzene ring, rather than the more electron-rich 2-position. In analogy to the monooxygenase mechanism described above, these enzymes generate a flavin C4a hydroperoxide intermediate. However, instead of directly hydroxylating aromatic substrates, this species reacts with a specifically bound halide anion (X, X = Br− or Cl−) to generate HOX. In an amazing product of evolution uncovered by several impressive biochemical studies on tryptophan 7-halogenases RebH and prnA, this species is proposed to diffuse through the center of the enzyme where it is intercepted by a conserved active site lysine residue to generate either a haloamine or hydrogen-bonded HOX intermediate adjacent to a conserved glutamate residue.xx Specific substrate binding proximal to this intermediate enables regioselective halogenation of the arene via electrophilic aromatic substitution, with different binding modes giving rise to different regioselective outcomes.
Figure 5.
Proposed mechanism of FAD-dependent halogenases (PrnA/RebH) catalyzing halogenation of tryptophan. A hydrogen-bonded HOX oxidant is depicted.xx
B. The 5’-Deoxyadenosyl Radical (Ado•) and Ado• Enzymes
Flavin-promoted dioxygen activation confers on flavoproteins the ability to hydroxylate or halogenate C-H bonds of aromatic compounds. However, excepting the recent example of alkane hydroxylase activity by LadA, the substrate scope of these enzymes is generally limited to electron-rich aromatic compounds by their use of electrophilic aromatic substitution-based mechanisms. In order to functionalize substrates outside of this range, a more powerful oxidant is required. Several solutions to this problem have evolved in nature, and all of these appear to rely on paramagnetic intermediates to promote single-electron or H-atom abstraction from substrates followed by manifold possibilities for subsequent C-H functionalization.viii
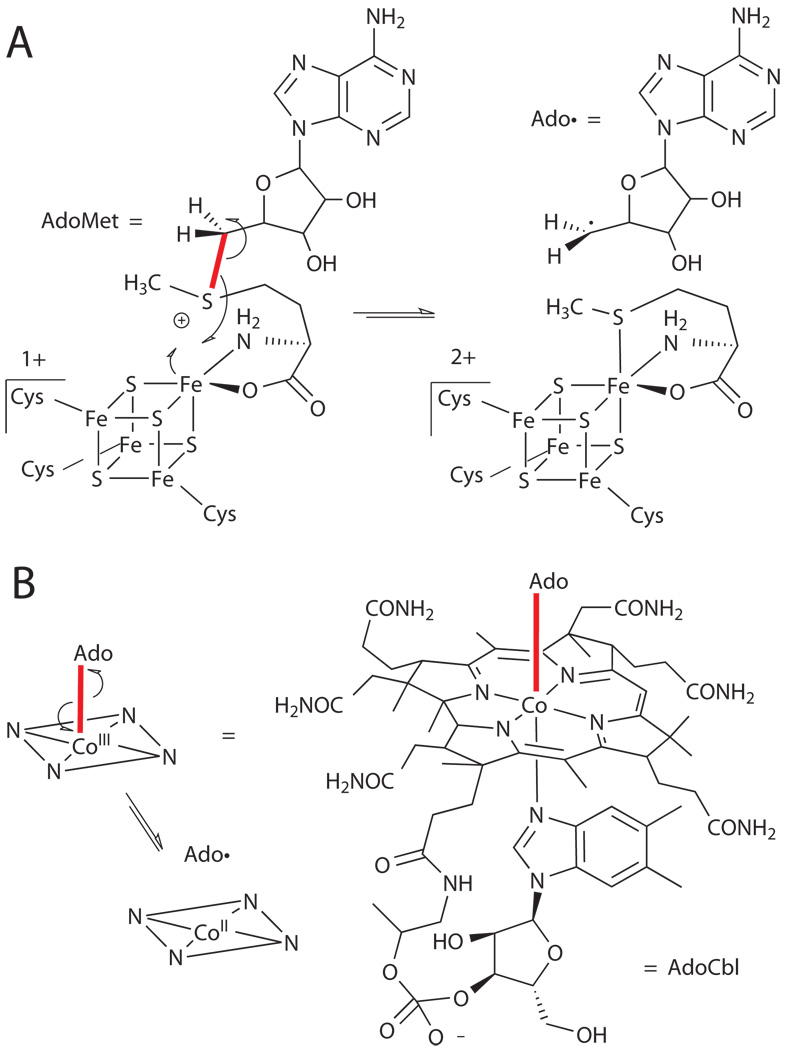
While many of these enzymes utilize oxygen-derived oxidants, at least one class of enzymes appears to have evolved before the oxygenation of earth and are in fact often highly oxygen-sensitive.xxi These utilize a ubiquitous biomolecule, adenosine, albeit in a unique form, the 5’-deoxyadenosyl radical (Ado•), in order to abstract hydrogen atoms from a wide range of substrates, including unsubstituted alkanes (Fig. 6). This cofactor can function either as a stoichiometric reagent that must be regenerated after each catalytic cycle or as a catalyst by reversibly abstracting hydrogen from a substrate. The enzymes containing this cofactor can be broadly classified based on the means by which the adenosyl radical is generated, namely via homolytic cleavage of adenosylcobalamin (vitamin B12, AdoCbl)xxii or one-electron reduction of a [4Fe-4S] cluster-complexed S-adenosylmethionine (AdoMet)xxiii (Fig. 6). Notably, both of these reactions are highly unfavorable, and the free energy for forming Ado• in each case is approximately 30 kcal mol−1.xxia Because the enzyme readily accomplishes this task, it has been hypothesized that this barrier is offset by protein-induced weakening of the Co-C bond (for AdoCbl) or decrease of the reduction potential of the iron-sulfur cluster (for AdoMet) in response to substrate binding, again providing a switch to preserve valuable cofactor in the absence of substrate.
Figure 6.
Structures of AdoMet (A) and AdoCbl (B) and mechanism for generation of Ado• in each case.xxi
The AdoMet enzymes constitute the largest and most primitive class of Ado• enzymes, and sequence analysis has identified approximately 3,000 putative members of this family based on the CxxxCxxC consensus motif.xxib These enzymes are highly oxygen-sensitive due to oxidative decomposition of their [4Fe-4S] cluster and are most commonly found in anaerobes. However, their reactivity enables organisms to activate inert C-H bonds without the need for oxygen-derived oxidants (vide infra). Reactions promoted by the Ado• intermediate generally fall into one of three categories: reversible H abstraction, irreversible H abstraction, and irreversible H abstraction from a glycine residue of a cognate protein which then catalyzes subsequent reversible H abstractions.xxiv
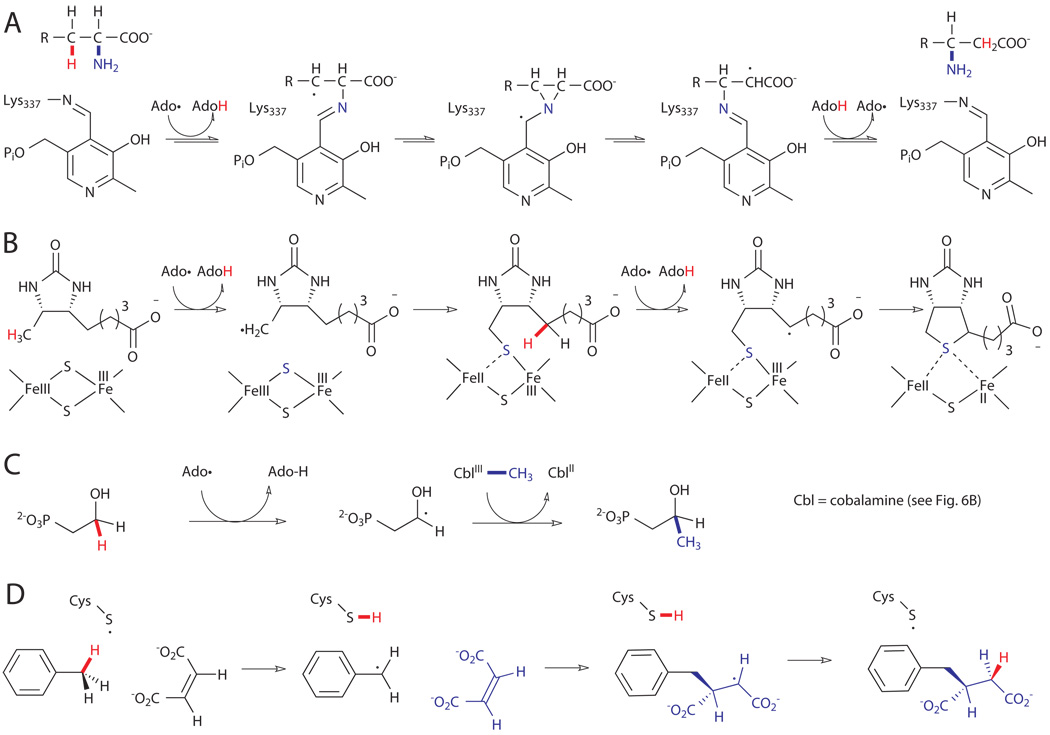
Only a few examples of enzymes in the first category are known, and all of these catalyze the 1,2-interconversion of H and a functional group, X (X = OH, NH3+, etc.).xxia In general, these reactions proceed via initial H abstraction by Ado• to generate AdoH and a substrate radical alpha to X. Migration of X to the substrate radical then generates a second substrate radical, which abstracts H from AdoH to form the rearranged product and regenerate Ado•. Variations on this scheme are known, and additional cofactors are often required to promote the observed rearrangement. This class of enzymes is perhaps best illustrated by lysine-2,3-aminomutase, which involves imine formation between lysine and the coenzyme pyridoxyl phosphate (PLP) followed by radical rearrangement via a three-membered aziridine intermediate (Fig. 7A).xxv
Figure 7.
Examples of Ado• reactions. A) 1,2-rearrangement catalyzed by lysine-2,3-aminomutase.xxv B) Double C-H abstraction and sulfur insertion catalyzed by biotin synthase (BioB).xxvi C) Radical methylation catalyzed by Fom3 in the biosynthesis of fosfomycin.xxvii D) Hydroalkylation of fumarate catalyzed by benzylsuccinate synthase.xxviii
In most cases, AdoMet is utilized as a reagent (i.e. irreversible H abstraction is observed).xxi Enzymes in this class can catalyze reactions analogous to atom transfer processes in synthetic chemistry and enable installation of either sulfur or methyl groups at unactivated C-H bonds. The best example of the former involves the installation of the thiophane ring of biotin (Fig. 7B).xxvi To accomplish this, the AdoMet enzyme, biotin synthase (BioB), utilizes Ado• to abstract a hydrogen atom from the C9 methyl group of dethiobiotin. Interestingly, the resulting substrate radical then reacts with a [2Fe-2S] cluster to form a C9-S bond with concomitant reduction of the proximal Fe center. A second Ado•-mediated H abstraction at C6 generates a substrate radical that reacts with the remaining Fe-S bond to generate the C6-S bond. Several additional enzymes are known to utilize similar mechanisms to install thiols or thioethers at unactivated C-H bonds.xxi Methylation of C-H bonds is also catalyzed by AdoMet enzymes. As before, H abstraction by Ado• generates a substrate radical, but in these cases, methylcobalamine serves as a source of methyl radical to install a methyl group at the activated carbon. One example of this activity, catalyzed by the enzyme Fom3, is observed in the biosynthesis of the natural product fosfomycin (Fig. 7C).xxvii Although the methylene C-H bond is modestly activated by the ipso hydroxyl group, direct methylation of this site is nonetheless an impressive feat.
In the final class of AdoMet enzymes covered here, the Ado• intermediate abstracts a hydrogen atom from a specific glycine residue of a cognate glycyl radical enzyme.xxia The activated glycyl radical enzymes are capable of catalyzing a number C-H functionalization reactions. These typically involve abstraction of a cysteine S-H bond by the glycyl radical and subsequent H abstraction from a substrate by the cysteine radical. A very interesting example of this type of enzyme is the benzylsuccinate synthase used for anaerobic metabolism of toluene (Fig. 7D).xxviii This enzyme catalyzes abstraction of a benzyl H from toluene and subsequent reaction of the tolyl radical with fumarate to generate benzyl succinate. Several additional enzymes are known for anaerobic metabolism of other substituted aromatics and even aliphatic substrates.xxi,xxvii Formally, this reaction is a hydroalkylation of alkenes, a valuable synthetic transformation.xxix Several additional glycyl radical enzymes are known and catalyze dehydration, deoxygenation, and decarboxylation reactions.xxi
While the AdoMet enzymes are a large and functionally diverse class of enzymes, the discovery and characterization of AdoCbl enzymes predated work on the former and has provided a great deal of mechanistic information regarding the activity of Ado• intermediates.xxia A beautiful and highly complex molecule with several discrete chemical domains, AdoCbl is a relatively rare example of an organotransition metal complex in nature. Perhaps not surprisingly, the AdoCbl enzymes are hypothesized to have appeared more recently than the simpler AdoMet enzymes. The AdoCbl cofactor is always used catalytically, probably due to the metabolic expense of its biosynthesis, which makes it a more atom-economical solution to Ado•-mediated C-H functionalization, and it is far less oxygen-sensitive, consistent with its evolution during more oxygen-rich geological past. However, only a handful of these enzymes are known, and all but one are limited to catalysis of 1,2-H/X interconversions similar to those discussed for Class I AdoMet enzymes (e.g. Fig. 7A). The mechanism of these transformations is conceptually identical following generation of the Ado• intermediate. Specific examples of AdoCbl enzymes include lysine-5,6-amino mutase,xxx glutamate mutase,xxxi notable for its ability to promote a carbon skeleton rearrangement, and diol dehydratase,xxxii which promotes a rearrangement followed by spontaneous dehydration to generate a carbonyl compound.
C. Mononuclear Non-Heme Metalloenzymes
As previously mentioned, C-H bond hydroxylation is a key reaction in cellular metabolism. However, the mechanistically-limited substrate scope of flavin monooxygenases and the oxygen sensitivity of Ado• enzymes precludes hydroxylation of many substrates by either of these enzymes. This apparent gap in reactivity is filled by an expansive list of metalloenzymes that activate dioxygen to generate oxidants capable of hydroxylating even the strongest of alkyl C-H bonds and promoting a number of less common oxidative transformations.ix The active sites of these enzymes range from residue-coordinated metal ions, most notably iron and copper,xxxiii to enzyme-bound coordination complexes, such as heme.xxxiv
Among the most prevalent of the former are the mononuclear non-heme iron enzymes,xxxv which utilize as few as two active site residues to coordinate an Fe(II) ion and thus have as many as four sites available to coordinate substrates (Fig. 8A). These enzymes promote a number of oxidative transformations by tuning the ligand environment around the metal. The most common coordination sphere consists of three protein ligands, (His)2-(Asp/Glu), arranged on one face of an octahedron, leaving the three remaining sites of the other face occupied by water in the resting state Fe(II) form of the enzyme. Substrate binding triggers the release of water ligands, opening a vacant coordination site to which dioxygen can be bound and subsequently reduced to generate an active oxidant. Typically, two of the four electrons required for reduction of dioxygen come from a reducing co-substrate (e.g. α-ketoglutarate, tetrahydropterin, ascorbate or NAD(P)H) and the organic substrate being oxidized provides the other two. In most cases, the active oxidant is a ferryl complex, but in some special cases (vide infra) iron-superoxo complexes have been invoked as intermediates for hydrogen abstraction.xxxvi High-valent iron-oxo intermediates are capable of promoting a range of C-H functionalization transformations, namely hydroxylation, halogenation, desaturation/cyclization, and electrophilic aromatic substitution.xxxv
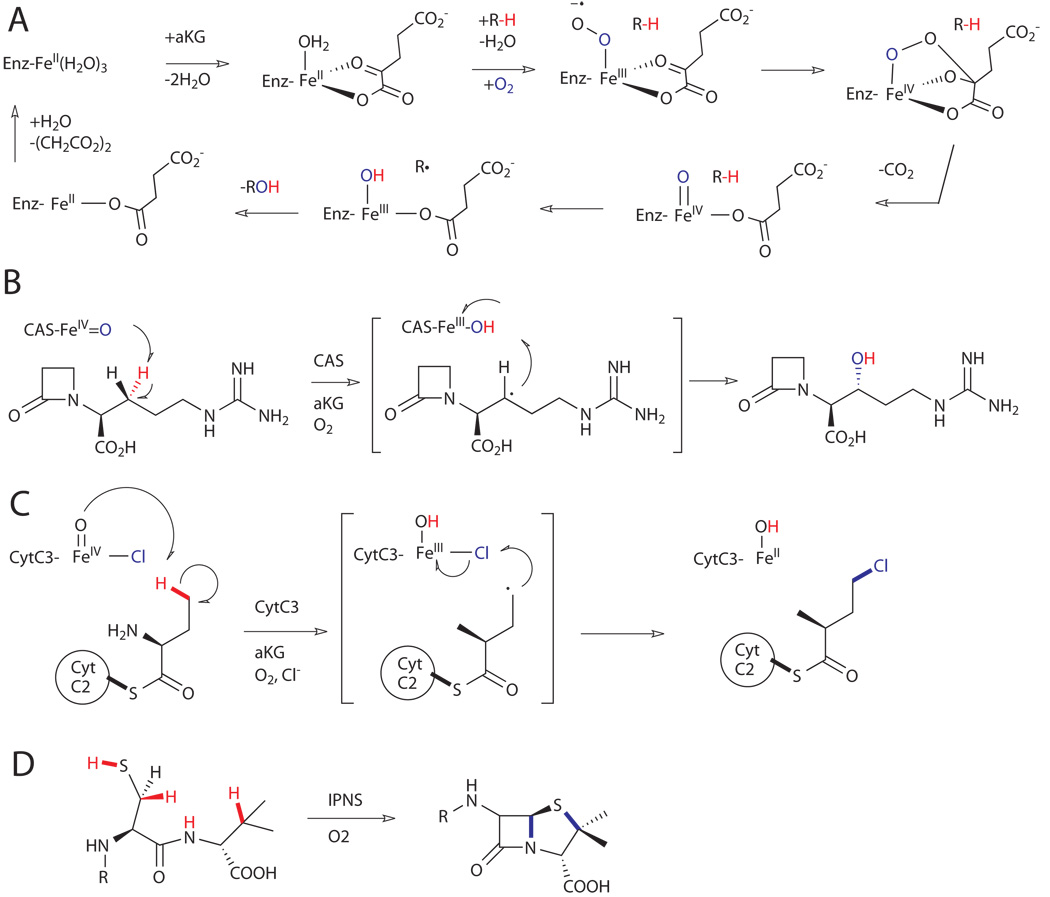
Figure 8.
A) General mechanism for hydroxylation of C-H bonds on substrates (R-H) by αKG-dependent dioxygenases.xxxv B) Hydroxylation reaction catalyzed by clavaminate synthase (CAS).xxxviii C) Chlorination reaction catalyzed by halogenase CytC3.xxxix D) Oxidative cyclization reaction catalyzed by an isopenicillin-N synthase (IPNS).xl
The ubiquitous α-ketoglutarate (αKG)-dependent dioxygenases are the largest and functionally most diverse subgroup of mononuclear non-heme iron enzymes.xxxv These enzymes are involved in the synthesis of a range of primary and secondary metabolites in plants, animals, and microorganisms. αKG-dependent oxidases catalyze the two-electron oxidation (e.g. hydroxylation) of their primary substrate with the concomitant conversion of the αKG co-substrate into succinate and carbon dioxide to provide the other two electrons required for reduction of dioxygen. An important feature for the control of the reactivity of these enzymes is that dioxygen activation can only occur following binding of the primary substrate.xxxvii The mechanism is thought to involve binding of the αKG co-substrate, followed by binding of the primary substrate, which displaces the last iron(II)-bound water to enable oxygen binding and activation (Fig 8A). A well-characterized member of this family is clavaminate synthase (CAS) from Streptomyces clavuligerus, which catalyzes three different steps (hydroxylation, cyclization and desaturation) in the biosynthetic pathway of clavulanic acid (Fig. 8B).xxxviii
Some Fe(II)/αKG-dependent oxygenases also display aliphatic halogenase activity in the chlorination of terminal methyl groups of amino acids tethered via a thioester linkage to the phosphopantetheine arm of peptidyl carrier proteins (PCPs).xviii For example, the non-heme iron halogenase CytC3 from soil Streptomyces, which requires iron, αKG, oxygen and chloride for its activity, is capable of chlorinating the γ-methyl group of L-2-aminobutyric acid (L-Aba) tethered to the PCP domain CytC2 (Fig. 8C).xxxix Exclusive halogenation over hydroxylation presumably ensues due to the lower reduction potential of the chlorine radical (Cl• + e− --> Cl−, 1.36 V) relative to the hydroxyl radical (HO• + e− --> HO−, 2.02 V).
Isopenicillin-N synthases (IPNS) are evolutionarily related to αKG-dependent oxidases, and both families have similar Fe-coordination spheres: (H2O)3Fe(II)(His)2-(Asp).xl IPNS does not require αKG as a co-substrate, but instead catalyzes the formation of penicillin N (IPN) through the four-electron oxidation of the tripeptide, L-δ-aminoadipoyl-L-cysteinyl-D-valine. The 1:1 IPN:O2 stoichiometry requires that the C-H cleavage steps be affected by different intermediates and, in fact, IPNS was the enzyme for which C-H cleavage by an iron-superoxo intermediate was first recognized.xxxvi The first oxidation is promoted by a formally Fe(III)-superoxo intermediate that cleaves the L-Cys C-H, leading to formation of the β-lactam ring (Fig. 8D). The second oxidation event is thought to involve cleavage of the D-Val C-H by an Fe(IV)-oxo intermediate which subsequently completes the β-lactam-thiazolidine core of all penicillins.
D. Carboxylate-Bridged Diiron Centers and Metalloenzymes
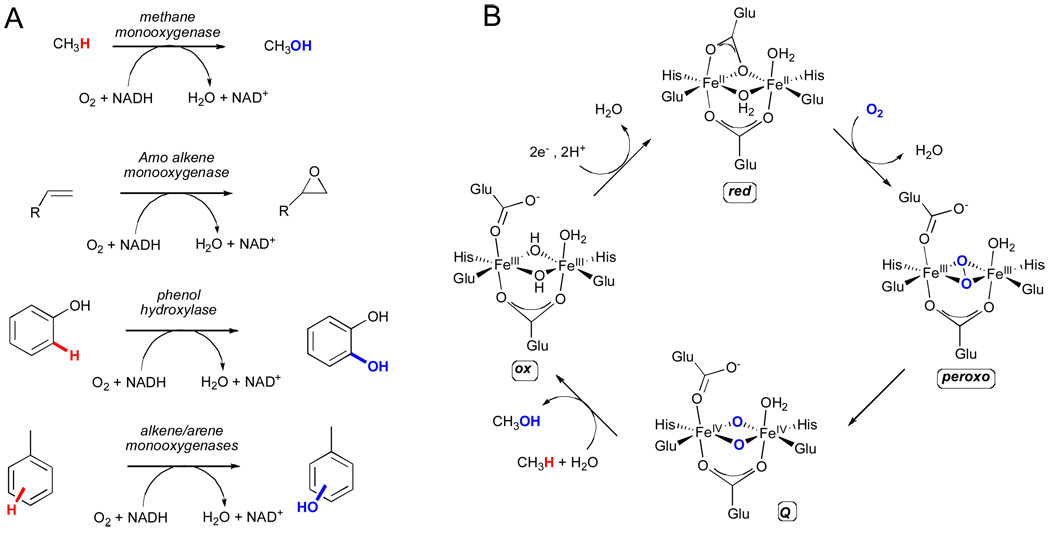
Bacterial multicomponent monooxygenases (BMMs) constitute a second class of enzymes that utilize residue-ligated iron centers, specifically carboxylate-bridged diiron centers, for hydroxylation of unactivated C-H bonds.xli These enzymes are typically found in bacteria living at the aerobic/anaerobic interface where they consume unactivated hydrocarbons as their sole source of carbon and energy. Several classes of evolutionarily-related BMMs have thus far been identified, including methane monooxygenase (MMO), alkene monooxygenase (AMO), phenol hydroxylase, and alkene/arene monooxygenase (Fig. 9A).xlii BMM systems are composed of three or four soluble components, namely: a dimeric hydroxylase protein consisting of two or three subunits in a (αβγ)2 or (αβ)2 quaternary structure with the α subunit harboring the carboxylate-bridged diiron site, an NADH reductase, a small effector protein (necessary for coupling electron consumption with hydrocarbon oxidation), and in some cases, a Rieske-type ferredoxin protein (e.g. arene monooxygenases). All components are required for optimal catalysis and for modulating regiospecificity.xliii
Figure 9.
A) Representative reactions catalyzed by each of the four classes of bacterial multicomponent monooxygenases (BMMs).xli B) The catalytic cycle for the soluble diiron methane monooxygenase.xlv
The mechanism by which BMMs activate dioxygen and subsequently catalyze the hydroxylation of C-H bonds has been studied extensively for the soluble methane monooxygenase from the methanotroph Methylococcus capsulatus.xliv Unlike the α-KG dependent enzymes discussed above, in which iron remained in the +2 oxidation state in both the resting state and O2-activating forms, the resting state for methane monooxygenase consists of an oxidized diiron(III) species (ox) where the positive charge of the two iron atoms is precisely balanced by four glutamate and two bridging hydroxide ligands (Fig. 9B). The octahedral coordination spheres of the two iron atoms are completed by two additional histidine residues. Reduction of the iron(III) centers in the resting state to the diiron (II) form (red) is required prior to dioxygen activation, which is followed by the generation of a peroxo intermediate, subsequently leading to the fully oxidized diiron(IV) species, Q.xlv Species Q is the active hydroxylating agent in BMMs and is capable of inserting an oxygen atom into alkyl C-H bonds, including the 104.5 kcal mol−1 C-H bond of methane.
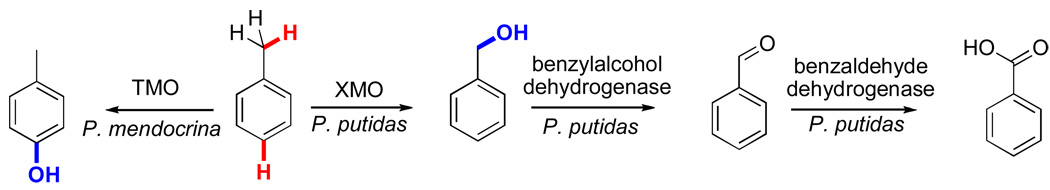
While less mechanistic detail is available for the remaining BMM family members, these enzymes catalyze the hydroxylation of a number of synthetically relevant compounds. For instance, the toluene monooxygenases (TMO of the alkene/arene monooxygenase class) from Pseudomonas mendocrina and Ralstonia picketti have been well characterized and are known to display high regiospecificity for the hydroxylation of toluene to p-cresol (>90% selectivity with only negligible amounts of benzylic oxidation).xliii This preference for aromatic ring hydroxylation provides an interesting contrast with the xylene monooxygenases (XMO) for which benzyl oxidations are predominantly observed.xlvi The XMO from Pseudomonas putida is the first enzyme system involved in the degradation of toluene and xylenes when these hydrocarbons are supplied as a sole source of carbon and energy.xlvii For example, toluene is initially oxidized by XMO to benzylalcohol, which can then be further oxidized to benzoic acid by the benzylalcohol and benzylaldehyde dehydrogenases. These three enzyme systems have been utilized in conjunction to demonstrate the synthetic potential of converting methyl-substituted aromatics into carboxylic acids in one pot (Fig. 10).xlviii
Figure 10.
Toluene monooxygenase (aromatic oxidation) vs xylene monooxygenase (benzylic oxidation) and subsequent dehydrogenation for the synthesis of phenols, alcohols, aldehydes, and acids.xlviii
E. Binuclear Copper Monooxygenases
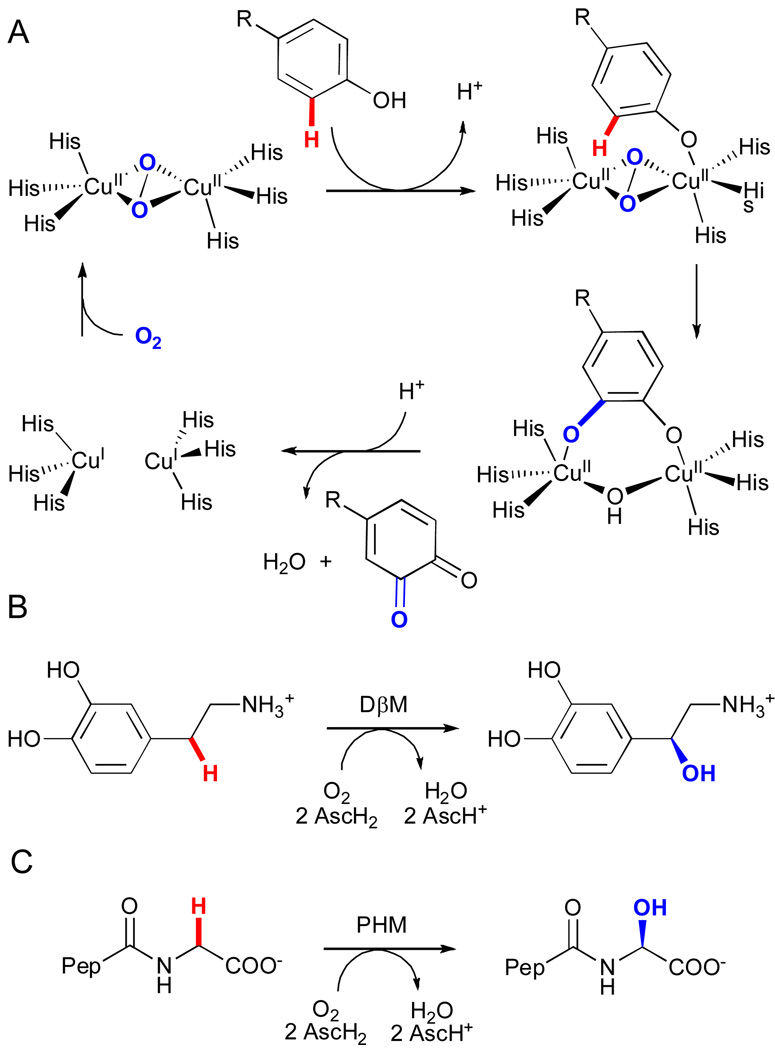
Certain bimetallic copper enzymes also possess the ability to hydroxylate carbon-hydrogen bonds.xiv These monooxygenases can be divided in two classes depending on the extent of magnetic coupling between the two copper ions. In coupled binuclear copper monooxygenases (i.e. tyrosinases), the two copper centers are each ligated by three histidine residues in close proximity (~ 3.6 Å) to one another. Activation of molecular oxygen by the copper centers leads to a μ-η2:η2-peroxo-bridged dicopper(II) species that exhibits strong antiferromagnetic coupling.xlix In contrast, non-coupled binuclear copper enzymes (e.g. dopamine β-monooxygenase (DβM) and peptidylglycine α-hydroxylating monooxygenase (PHM)), utilize two nonequivalent copper centers, (His)2(Met)CuM and (His)3CuH, distant in space (~ 11 Å in PHM) that exhibit no observable magnetic interactions.l, li
Tyrosinases catalyze ortho-hydroxylation of monophenols and subsequent two-electron oxidation of the intermediate catechols to produce quinones, and the mechanism of these transformations has been studied extensively.lii In the resting state of the enzyme, a caddie protein shields the entrance to the active site to regulate substrate access.liii Activation proceeds via cleavage of this caddie protein, which exposes the active site to incoming substrate. η1-Coordination of the phenolic hydroxyl group to the bridging μ-η2:η2-peroxo dicopper(II) intermediate induces electrophilic attack of the phenol (Fig 11A). This generates an o-diphenolate that binds in a bidentate fashion and is finally oxidized to the quinone with concomitant reduction of the remaining oxygen atom from dioxygen to water and formation of the deoxy dicopper (I) state.
Figure 11.
A) The catalytic cycle for tyrosinase.lii Reactions catalyzed by B) dopamine β-monooxygenase (DβM) and C) peptidylglycine α-hydroxylating monooxygenase (PHM) (AscH2 = ascorbic acid, Pep = D-Tyr-Val).l,li
For the non-coupled dicopper monooxygenases (e.g. DβM and PHM), the binding and activation of dioxygen takes place at a single copper site (CuM). Oxygen activation is thought to proceed via one-electron reduction of dioxygen to generate a η2-superoxo-CuIIM intermediate. This species can abstract H atoms from aliphatic C-H bonds to form a CuIIM-hydroperoxo complex and carbon radical.liv Hydroxylation then occurs via recombination of hydroxyl radical from the hydroperoxo complex and the substrate radical and leads to the formation of a highly oxidizing CuIIM-oxyl (O•−) species that provides the driving force for the required long-range intramolecular electron transfer from the second copper (CuIH). Thus, the role of the CuH site is simply to provide the additional electron that is necessary to complete the catalytic cycle.
Recently, the active site of the particulate form of methane monooxygenase (pMMO) was also shown to consist of a dicopper center.lv This membrane-associated enzyme is the most prevalent form of methane monooxygenase and is found in every methanotroph, while sMMO (vide supra) is only expressed by some strains of methanotrophs under copper-limited conditions. The two copper ions in the active site of pMMO are ligated by three histidine residues and are held in close proximity (2.5–2.7 Å) to one another, but little more is known about the mechanism of this enzyme. Computational studies of methane activation by mixed valent bis-(μ-oxo) dicopper (II/III) specieslvi and reports of direct methane oxidation by mono-(μ-oxo) dicopper (II/II) cores in zeoliteslvii illustrate the potential of dicopper centers to promote the oxidation of methane to methanol, but further investigations will be required to better understand the mechanism of methane activation by pMMO.lviii
F. Heme and Heme Enzymes
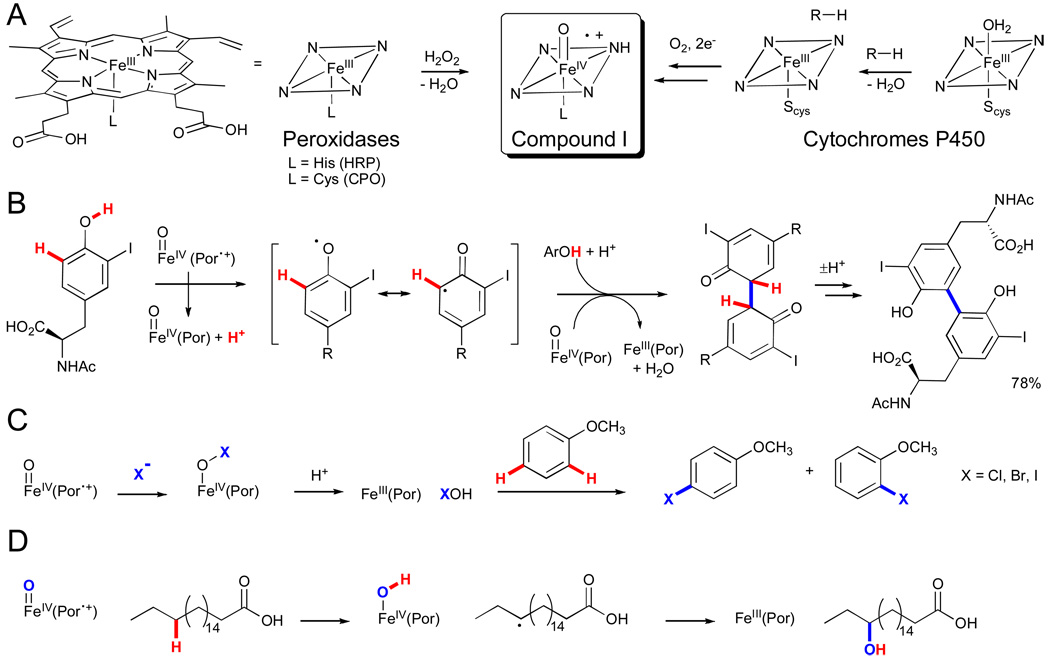
Heme enzymes are perhaps the most familiar of those capable of functionalizing C-H bonds.xxxiv The heme cofactor in these enzymes is comprised of a protoporphyrin IX-bound iron atom that binds to a (proximal) active site residue leaving a single (distal) coordination site available for ligand binding and catalysis. Depending on the nature of the proximal ligand and active site residues, the heme cofactor can react with either peroxides or dioxygen to generate complexes capable of functionalizing C-H bonds.lix Extensive research has been devoted to studying the formation and reactivity of these intermediates in different heme enzymes; however, the transformations relevant to the current discussion can at least formally be described as proceeding via an Fe(IV)-oxo π-radical cation complex known as compound I.lx This intermediate can react with substrates to functionalize C-H bonds via several distinct manifolds characteristic of the different heme enzyme subfamilies. For example, peroxidases catalyze oxidative coupling reactions via sequential one-electron abstractions,lxi haloperoxidases catalyze halogenation of sp2 C-H bonds via halide oxidation,xviii and cytochromes P450 (CYPs) can catalyze hydroxylation of sp2 and sp3 C-H bonds via H atom abstraction, electrophilic aromatic substitution, or direct oxo transfer.lxii
In the case of peroxidases, compound I is readily generated and can be observed by the reaction of hydrogen peroxide with the penta-coordinate ferric resting state (Fig. 11A).lxiii However, even within the peroxidase family, structural variations lead to pronounced differences in reactivity. For example, in horseradish peroxidase (HRP), the axial ligand to the heme is a histidine nitrogen that acts as a hydrogen bond donor to a nearby aspartate.lxiv This is thought to impart a greater imidazolate character on the axial histidine that is considered important in the enzyme’s ability to activate hydrogen peroxide and favor sequential one-electron oxidations over two-electron oxo transfer from compound I. This reactivity, in turn, enables HRP to catalyze oxidative coupling reactions between electron-rich (hetero)arenes or between a (hetero)arene and a heteroatom (e.g. Fig. 11B).lxv
The haloperoxidases are unique among the peroxidases in having a thiolate-ligated heme. Such a strongly donating axial ligand is thought to facilitate heterolytic cleavage of the O-O bond and to lower the reduction potential of the heme iron.lix The chloroperoxidase (CPO) isolated from mold Caldariomyces fumago is the most extensively studied haloperoxidase; its gene has been sequenced and cloned, functional heterologous expression has been achieved, and it is even commercially available as a fine chemical.lxvi Seminal studies on this enzyme revealed that it can catalyze the halogenation of monochlorodimedone, a mimic of the C. fumago natural product caldariomycin.lxvii However, CPO is also capable of promoting oxidative transformations that are characteristic of other heme enzymes, such as dehydrogenation (i.e. peroxidases), H2O2 decomposition (i.e. catalases), and oxo transfer reactions (i.e. CYPs). These reactions, including C-H bond hydroxylation (vide infra), are catalyzed by CPO in the absence of halide at neutral pH.
In the presence of halide (X−, X = Cl, Br, I) and low pH (< 3), however, CPO reacts with halide to generate the corresponding hypohalous acid (HOX).xviii,lxvii This species can in turn halogenate a range of electron-rich substrates, including olefins, aromatics, and β-diketones, via electrophilic substitution pathways (e.g. Fig. 11C).lxvi Most data are consistent with HOX being free to diffuse out of the active site and react with the substrate in the surrounding medium.lxvi This is confirmed by the similar patterns of halogenation observed for various substrates with CPO versus simply using hypohalous acid free in solution. Consequently, regio- or stereoselective halogenation is not possible for CPO, and this feature constitutes an important difference between the haloperoxidases and the FAD-dependent halogenases discussed above.xix
CYPs also possess cysteine as the axial heme ligand but have a much more hydrophobic active site than the haloperoxidases.lxviii And while CYPs can utilize hydrogen peroxide as an oxidant via a so-called “peroxide shunt” pathway, they are distinguished from haloperoxidases by their ability to access compound I via dioxygen activation.lxix The first step in this process involves displacement of a water ligand from the low-spin Fe(III) heme resting state by substrate to generate a predominately high-spin substrate-bound complex (Fig. 11A). In many systems, this spin shift is required to increase the heme reduction potential so that it can be more easily reduced to the corresponding Fe(II) complex, which minimizes the expenditure of metabolically valuable reduced cofactors in the absence of substrate. Oxygen binding to the Fe(II) intermediate leads to the formation of an Fe(III)-oxy complex that undergoes one-electron reduction, two sequential distal oxygen protonation events, and elimination of water to generate compound I. This species can then react with sp3 C-H bonds of the bound substrate via formal H atom abstraction to generate an Fe(IV)-hydroxyl complex and a substrate radical followed by radical rebound to provide the hydroxylated product and the Fe(III) heme complex (e.g. Fig. 11D). Compound I has been proposed to react with substrates via a variety of pathways, including electrophilic substitution and concerted oxo transfer,lxii and similar reactivity is observed for this intermediate in CPO.lxvi
The more than 11,500 distinct CYPs identified thus far are capable of hydroxylating a wide range of organic substrates and play important roles in both biosynthetic and degradative pathways.lxx Despite their broad substrate and reaction scope and low amino acid sequence identities, the CYP super-family is united in having a conserved fold/structural topology. Evolution has demonstrated the versatility of this platform by utilizing CYPs to promote a range of additional transformations including epoxidation, oxidative deformylation, dehydrogenation, rearrangements, Bayer-Villiger oxygenation, and oxidative decarboxylation.lxxi
IV. Enzymatic C-H Functionalization as a Synthetic Tool
A. Synthetic Utility of Enzymes
The enzymes highlighted in the previous section functionalize a range of C-H bond types via a variety of chemical transformations without directing or protecting groups. While the development of such catalysts constitutes a highly sought-after goal of synthetic chemistry,iii practical application of enzymes requires overcoming a variety of challenges regarding enzyme expression/purification, cofactor supply, organic solvent and oxygen tolerance, stability, and substrate scope.lxxii Nonetheless, enzymes, including many capable of functionalizing C-H bonds, are increasingly utilized on an industrial scale for the production of fine chemicals.lxxiii
Here, a number of examples from the literature in which enzymes are used to functionalize C-H bonds of non-natural substrates are presented. Importantly, these examples highlight the fact that an enzyme often exhibits activity on a range of substrates. Although this activity may not be sufficient for practical applications, it provides an important first step toward the development of such catalysts.lxxiv It is also worth noting that many of the enzyme classes discussed above have not yet been successfully employed as biocatalysts due to one or more of the aforementioned technical challenges.xiv The successes outlined herein will, we hope, provide impetus for further research into overcoming these difficulties so that additional enzyme-catalyzed reactions will be available to chemists.
1. FAD-Dependent Hydroxylases and Halogenases
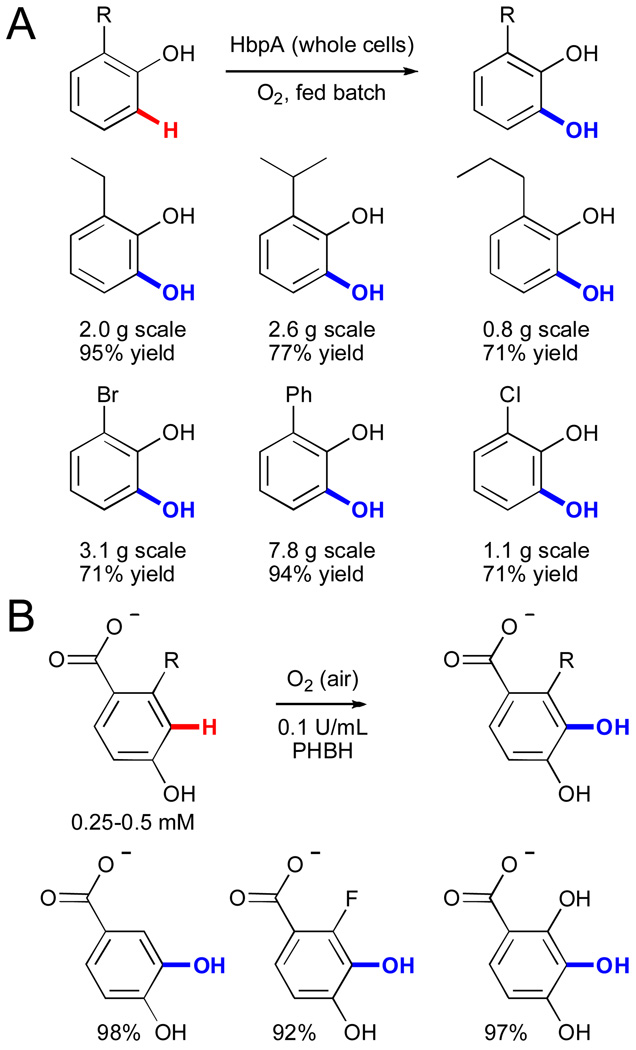
Flavin monooxygenases have been extensively investigated as biocatalysts due to the importance of aromatic hydroxylation and the difficulties associated with chemical methods for this reaction.lxxv Because most of these enzymes are bacterial in origin, they are readily expressed in convenient recombinant hosts such as E. coli, which greatly facilitates their investigation. Several FAD-dependent hydroxylases have been utilized for hydroxylation of electron-rich aromatic compounds structurally similar to their native substrates. In one example, purified 2-hydroxybiphenyl 3-monooxygenase (HbpA) from Pseudomonas azelaica HBP1 was used to selectively hydroxylate a range of 2-alkyl-, aryl-, or halo-substituted phenols to provide the corresponding 3-substituted catechols (Fig. 12A).lxxvi This enzyme was subsequently expressed in E. coli and used as a whole-cell biocatalyst for the production of kilogram quantities of catechols.
Figure 12.
A) Structure of a heme cofactor and general schemes for its conversion to compound I in peroxidases (HRP and CPO) and cytochromes P450 (CYPs).lx B) Oxidative coupling catalyzed by HRP.lxv C) Halogenation catalyzed by CPO.lxvi D) Hydroxylation catalyzed by CYP 102A1.
Synthesis of substituted phenols using A) 2-hydroxybiphenyl 3-monooxygenase (HbpA)lxxvi and B) p-hydroxybenzoate 3-hydroxylase (PHBH).lxxvii
In a second example, the substrate scope of p-hydroxybenzoate 3-hydroxylase from Rhodococcus rhodnii 135 and opacus 557 was explored (Fig. 12B).lxxvii This work demonstrated that a variety of 2- and 3-substituted 4-hydroxybenzoate substrates could be hydroxylated and that the regioselectivity of hydroxylation varied greatly between the two enzymes investigated. Such variation clearly indicates the potential for modification of selectivity for C-H functionalization in enzymes (vide infra). While many additional examples of arene hydroxylation by FAD-dependent enzymes are known,lxxv these two examples represent the types of reactions and substrate scope that have been explored.
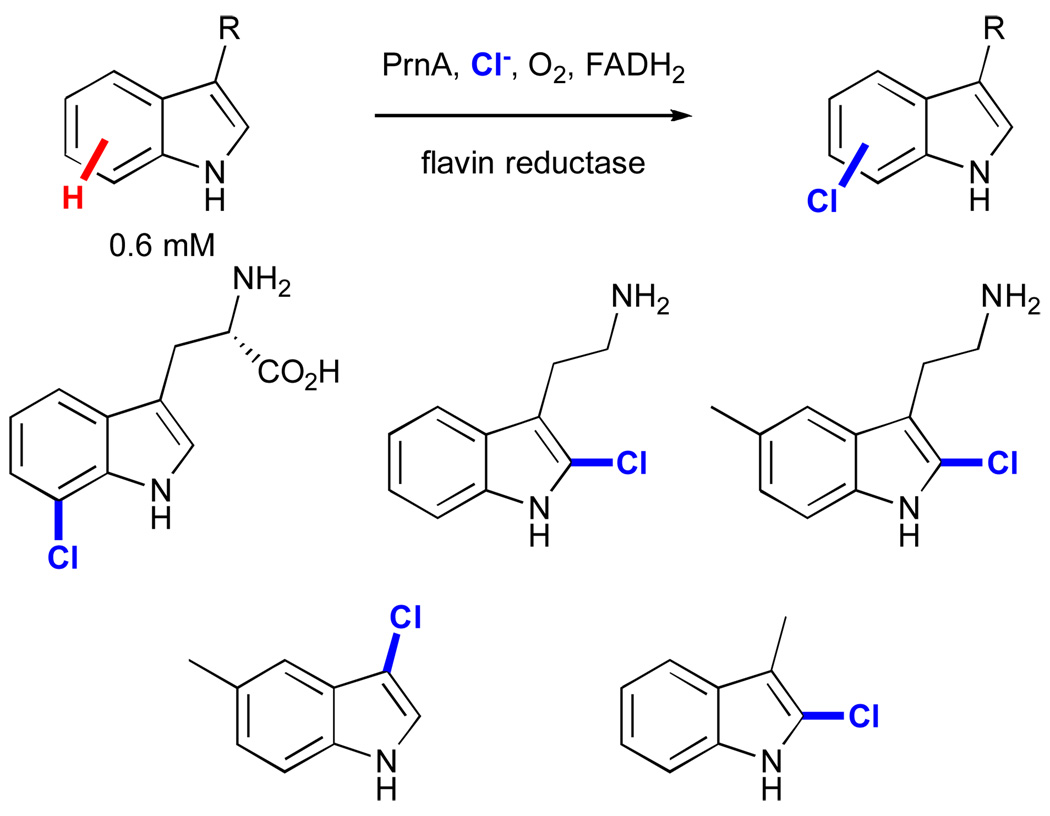
On the other hand, relatively little work has been published regarding synthetic applications of FAD-dependent halogenases. These enzymes were only detected in the late 1990’s, and the first example in which the activity of a member of this family was reconstituted in vitro appeared in 2000.xix Since that time, several such enzymes have been characterized, and extensive mechanistic investigation, particularly in the case of the tryptophan halogenases, has clarified their mode of action as described in the previous section.xx Building on these advances, van Peé and co-workers have explored the substrate scope of one tryptophan-7-halogenase, PrnA (Fig. 13).lxxviii A variety of substituted indoles were accepted by the enzyme, although halogenation invariably occurred at the electronically most activated indole C2-H bond for all but tryptophan.
Figure 13.
Substrate scope and regioselectivity of indole halogenation using the halogenase PrnA.lxxviii
2. Ado• Dependent Dehydratases
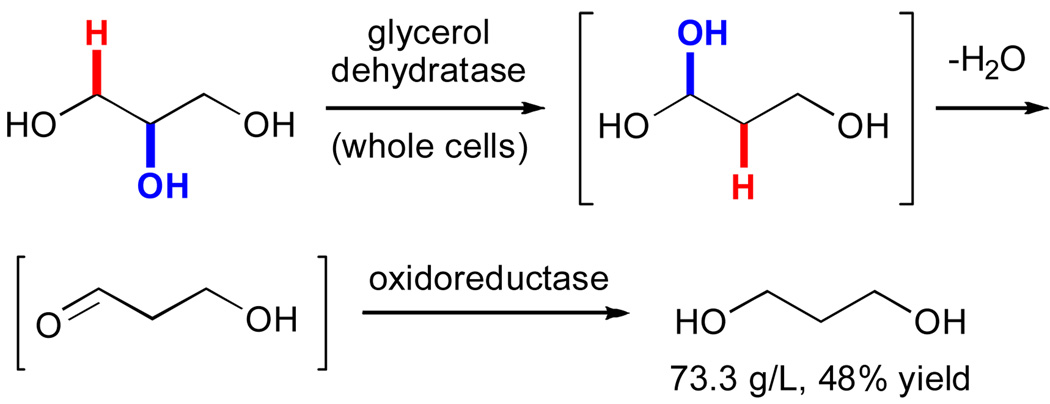
While Ado• enzymes catalyze an enticing array of reactions, their oxygen sensitivity and cofactor (Cbl or Met) requirements certainly pose challenges to their practical application.xxi A notable exception to this trend involves the use recombinantly expressed dehydratases to convert glycerol to 3-hydroxypropionaldehyde.lxxix This reaction has been explored by several researchers and recently utilized in a commercial process for production of 1,3-propoanediol, a valuable commodity chemical (vide infra). Cameron and co-workers provided a thorough description of a similar process employing an AdoCbl-dependent glycerol dehydratase and an oxidoreductase from Klebsiella pneumoniae.lxxx This sequence involves dehydratase-catalyzed conversion of glycerol to 1,1,3-propanetriol via an H-abstraction/rearrangement mechanism similar to those shown in Figure 7.xxxii Spontaneous dehydration and subsequent reduction of the intermediate 3-hydroxypropionaldehyde generates 1,3-propanediol. In addition to its commercial utility, this example provides valuable precedent for the development of other processes employing Ado• enzymes.
3. Non-heme Iron Enzymes
Of the non-heme iron enzymes capable of functionalizing C-H bonds, BMM, and in particular the toluene and xylene monooxygenases (TMO and XMO), have enjoyed the most attention in biocatalysis applications.xliii As previously described, the native activity of these enzymes involves aromatic and benzylic hydroxylation, respectively. Examination of the substrate scope of these enzymes has revealed that both possess activity on a broad range of aromatic and aliphatic compounds.lxxxi
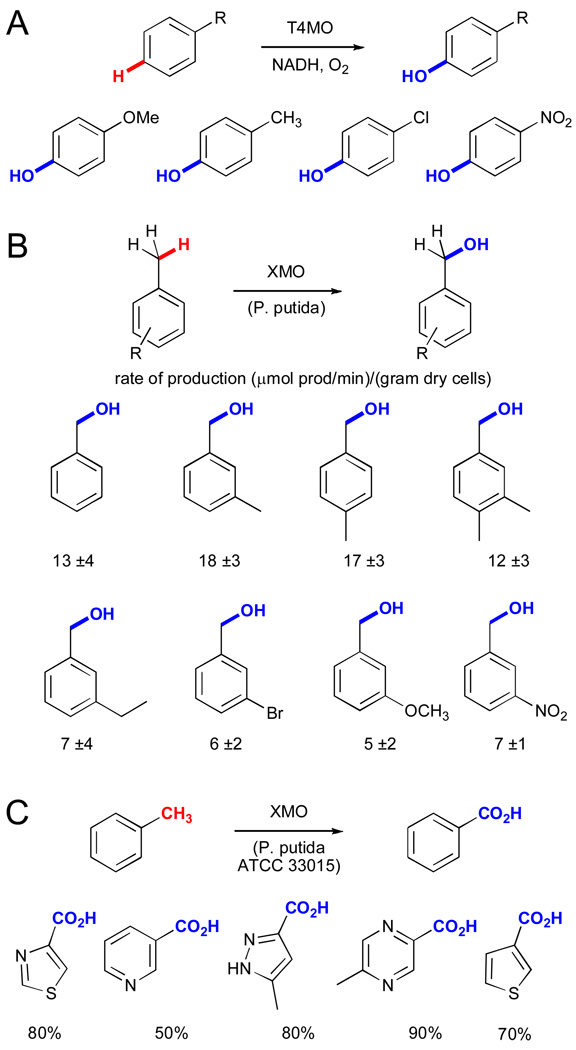
The reactivity of TMO has been exploited by a number of researchers for the hydroxylation of various toluene derivatives. For example, recombinant toluene-4-monooxygenase (T4MO) was used to hydroxylate m-, o-, and p-xylene, benzene, chlorobenzene, anisole, and nitrobenzene with at least 90% selectivity (Fig. 14A).lxxxii Only trace amounts of benzylic hydroxylation were observed, and even the slowest reactions proceeded at a rate within a factor of 3 of the native reaction. This activity was exploited in later syntheses of 4-fluorocatechollxxxiii and hydroxytyrosollxxxi. XMO also accepts a range of toluene derivatives, and whole-cell biocatalysts expressing this enzyme (Pseudomonas putida mt-2) have been used for the benzylic oxidation of these compounds (Fig. 14B).lxxxiv
Figure 14.
Production of 1,3-propane diol by E. coli expressing an AdoCbl-dependent dehydratase and an oxidoreductase.lxxx
A) Synthesis of phenol derivatives using recombinant toluene-4-monooxygenase (T4MO).lxxxii B) Synthesis of substituted benzyl alcohols using cells expressing a xylene monooxygenase (XMO) with reaction rates shown.lxxxiv C). Lonza synthesis of (hetero)arylcarboxylic acids using XMO and additional dehydrogenases.xlviii
These enzymes have also been used for commercial processes. For example, Lonza used Pseudomonas putida ATCC 33015, which harbors an XMO and additional dehydrogenases for benzylic hydroxylation and subsequent alcohol and aldehyde dehydrogenation to convert a range of methyl-substituted heterocycles to the corresponding carboxylic acids (Fig. 14C).xlviii The bioconversions provided high yields (40–90%) for a range of heterocycles in large-scale fermentations (up to 24 g/L, 95% conversion, 1000 L scale). While selective oxidation of C-H bonds is a challenging task in its own right, this example is even more impressive in that it uses heterocycles, which are frequently challenging substrates for synthetic metal-catalyzed reactions.
4. Heme Enzymes
By far the most work on enzymatic functionalization of C-H bonds has focused on heme-containing enzymes and on cytochromes P450 in particular. Several excellent reviews have appeared specifically detailing the use of P450 enzymes as biocatalysts, and readers are directed to those sources for ample discussion of this subject.lxxii,lxxiii It is worth emphasizing that these truly remarkable enzymes can exhibit broad substrate scope while maintaining high reaction rates and selectivities. While aliphatic C-H hydroxylation certainly dominates the reactivity of these enzymes, they are known to catalyze myriad additional transformations, including C-C bond formation via oxidative coupling.lxxi There will undoubtedly be significant practical advances to improve the utility of these reactions, but several examples already highlight their synthetic potential.
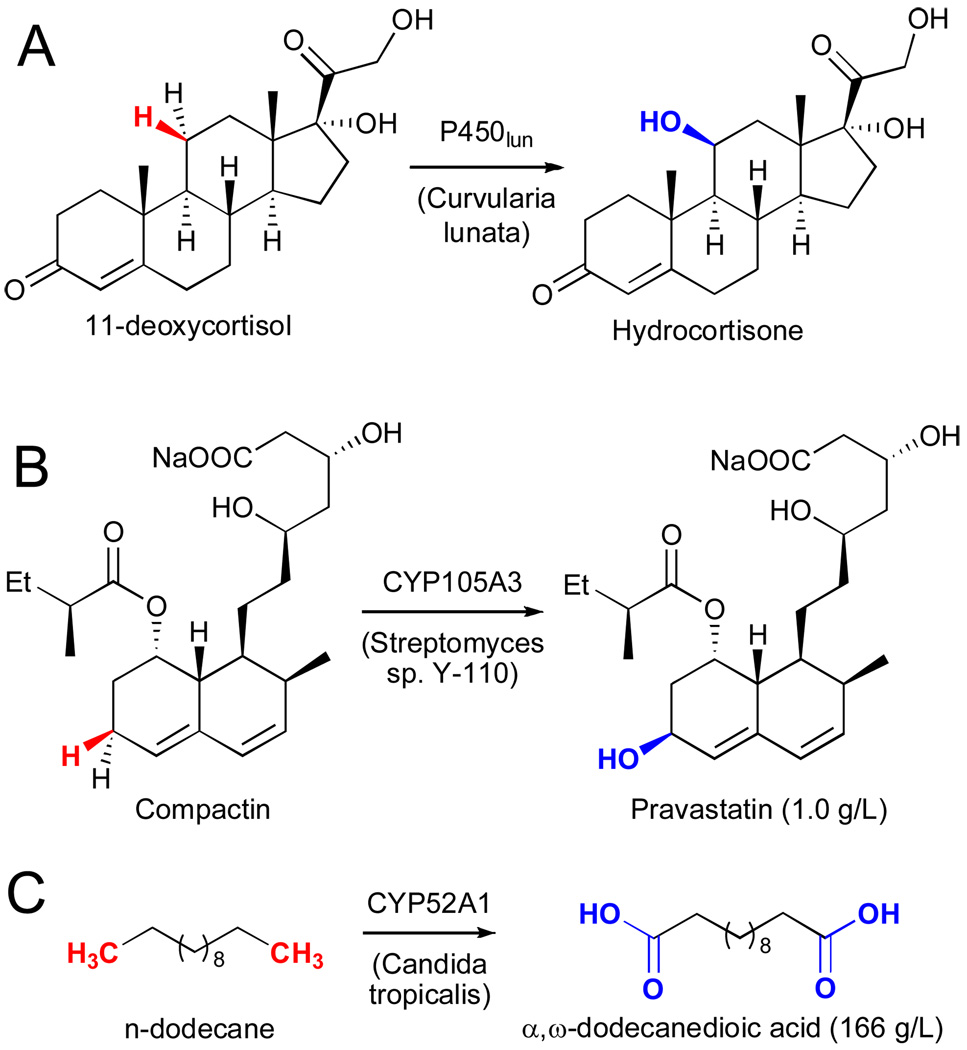
A number of early studies focused on the use of whole-cell catalysts harboring various P450 enzymes for the hydroxylation of steroids. Several of these are used commercially, including for the conversion of 11-deoxycortisol to hydrocortisone using P450lun in Curvularia lunata (Fig. 15A).lxxxv This example clearly shows how remote, unactivated C-H bonds can be efficiently hydroxylated even in the presence of an olefin and acidic, allylic, and tertiary C-H bonds. Bristol Myers Squibb utilized CYP105A3 in Streptomyces sp. Y-110 to convert compactin to Pravastatin (Fig. 15B).lxxxvi And finally, a rare example in which a P450 was used for the synthesis of a bulk chemical is the conversion of n-dodecane to α,ω-dodecanedioic acid by Cognis Inc. (Fig. 15C).lxxxvii
Figure 15.
A) Hydroxylation of 11-deoxycortisol using cytochrome P450lun in Curvularia lunata.lxxxv B) Hydroxylation of compactin using cytochrome 105A3 in Streptomyces sp. Y-110.lxxxvi C) Six sequential oxidations on n-dodecane catalyzed by cytochrome 52A1 in Candida tropicalis.lxxxvii
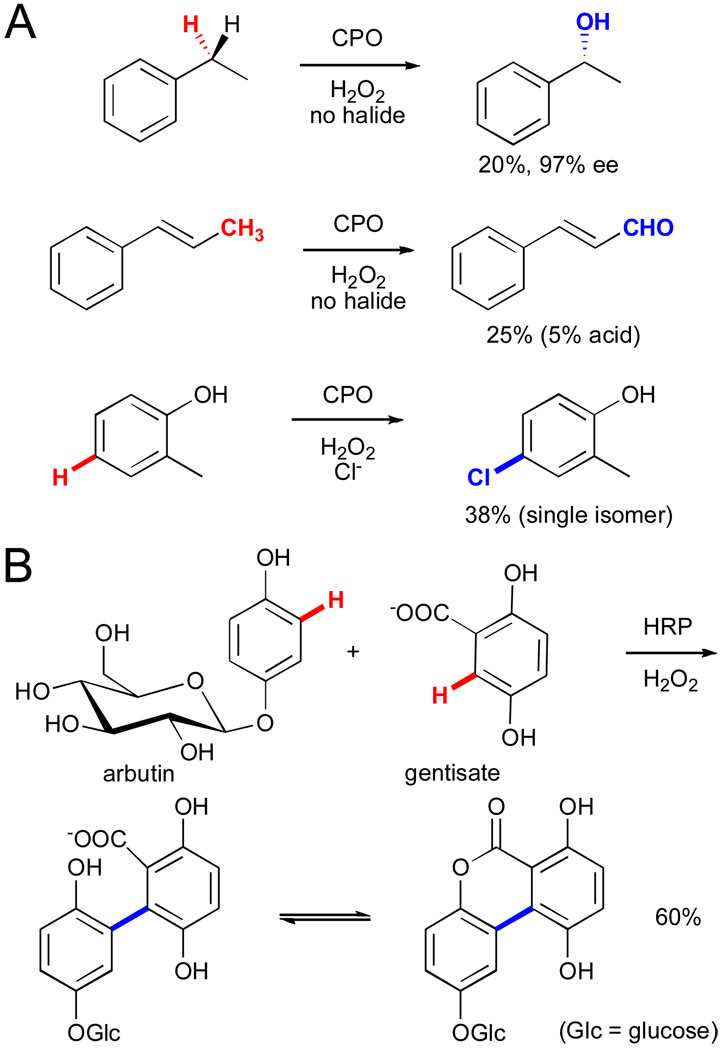
Other heme-containing enzymes, including chloroperoxidase (CPO) and horse radish peroxidase (HRP), have also been explored extensively for a variety of C-H functionalization reactions.lxvi,lxi CPO has been utilized for hydroxylationlxxxviii and halogenationlxxxix (Fig. 16A). On the other hand, HRP can catalyze oxidative coupling of electron-rich arenes, a reaction similar to the impressive arylation chemistry developed by Fagnou and co-workers.xc In one synthetic example, arbutin and gentisate are regioselectively coupled to generate a tricyclic compound (Fig. 16B).xci This example highlights the power of enzymes to provide selectivity to otherwise unselective radical coupling processes and in this case enable selective C-C bond formation between two C-H bonds!
Figure 16.
A) Various hydroxylation and halogenation reactions catalyzed by chloroperoxidase (CPO).lxxxviii,lxxxix B) Oxidative coupling between arbutin and gentisate catalyzed by HRP followed by spontaneous lactonization.xci
B. Optimization of Enzymes via Directed Evolution
Enzymes are genetically encoded polymers of amino acids assembled by the ribosomes of all living organisms. As discussed above, the ubiquity of this biosynthetic machinery often allows the gene that encodes an enzyme to be introduced into heterologous microbial hosts that facilitate its production at high levels and biocatalytic applications of that enzyme. Genes can also be readily amplified from genomic DNA using standard polymerase chain reaction techniques and even synthesized de novo, which provides access to any enzyme for which sequence information has been obtained (although expression of functional enzymes is not always possible). Furthermore, it is possible to alter the amino acid composition of recombinant enzymes in a process conceptually analogous to the synthesis of various small molecule catalysts. Whereas a particular modification to the latter may require tedious alteration of synthetic routes, however, such changes, mutations, are trivial to introduce into enzymes, and a microbe will do the “synthesis” for you (and a little sugar).
The three-dimensional (tertiary) structure of enzymes enables substrate binding, ideal placement of all manner of functional groups to promote reaction of these compounds, and dynamic fluctuations whose implications for catalysis have only recently begun to be unraveled.xcii Rational manipulation of these catalysts is therefore a highly challenging and vigorously pursued goal. On the other hand, the genetic encoding of these catalysts provides a straightforward means to generate libraries of structurally diverse mutants from a selected parent. With a means to assay these mutants for a desired property, improved variants can be identified from a population.vi
Mutations can be made randomly or they can be focused toward particular parts of the protein, based on some hypothesis regarding how those residues might modify enzyme function.xciii Individual library members (i.e. mutant enzymes) can be arrayed in microtiter plates using a variety of automated systems. The biggest challenge is usually assaying these library members to identify those with improved properties relative to the parent. Most commonly, this is accomplished using a screen, in which library members are individually assayed. This is often a highly challenging task and frequently limits the size, although libraries need not be exhaustively screened in order to identify improved variants, and improvements in ultra high pressure liquid chromatography and fast gradient gas chromatography instrumentation are facilitating this process.lxxiv
Most importantly, the genes encoding variants with improved properties can be subjected to subsequent rounds of mutagenesis and screening in order to systematically improve their properties. This process of ‘directed evolution’ has been successful in improving the organic solvent tolerance and stability of enzymes.vi It has also been used to alter or broaden substrate scope and to improve activity or selectivity on a particular substrate. Thus families of enzymes can be engineered to provide starting points for rapid optimization of activity toward a given target.xciv This is a major difference between enzymes and small molecule catalysts, but a potentially advantageous one given that a general small molecule catalyst is often not optimal for a particular application or particularly amenable to optimization for that application.
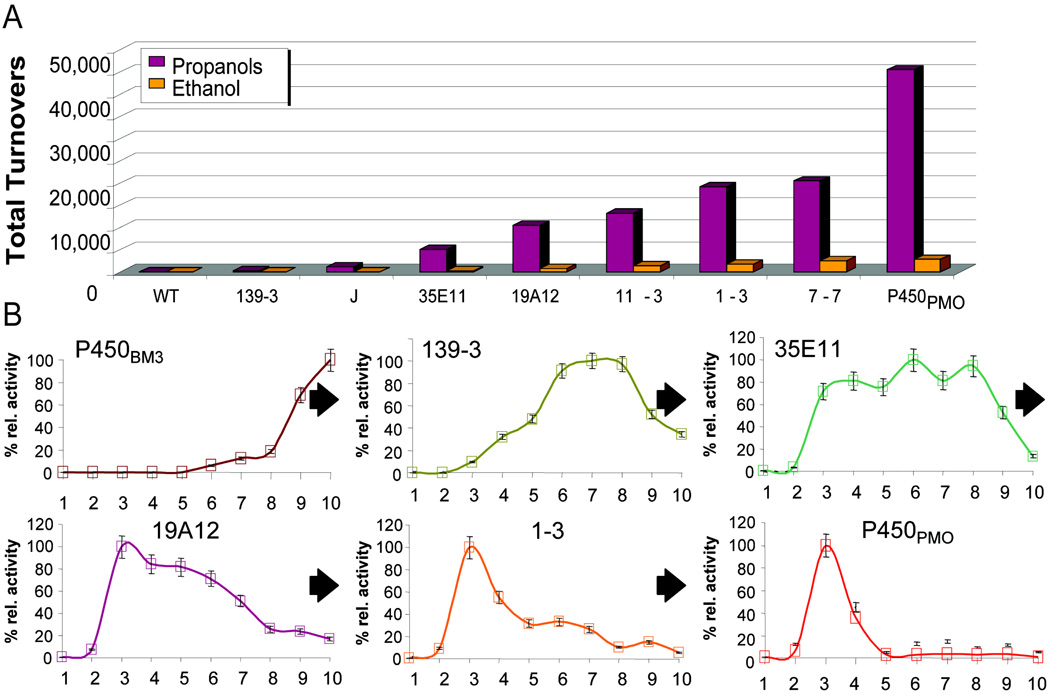
Generalizability is an especially valuable trait of catalysts for C-H functionalization, since such catalysts could potentially be tuned to recognize any desired substrate and provide high selectivity for the functionalization of any C-H bond on that substrate rather than relying on substrate-specific properties for activity or selectivity. An instructive example of this process from our own laboratory involves the directed evolution of cytochrome P450BM3 (CYP 102A1), an enzyme that naturally catalyzes sub-terminal hydroxylation of long-chain fatty acids, to hydroxylate propane (Fig. 17A).xcv A combination of random mutagenesis of just the P450BM3 heme domain, screening, and recombination of beneficial mutations led to mutants with improved activity on medium-chain alkanes. However, the stability of these enzymes and their coupling between NADPH consumption and product formation decreased. These issues were addressed by evolving the heme, FMN, and FAD domains individually in the context of the holoenzyme and then fusing them in a final step to generate P450PMO. P450PMO provided more than 45,000 turnovers on propane and produced 2- and 1-propanol in a 9:1 ratio. This enzyme displayed activity on propane comparable to that of P450BM3 on its natural substrates and 98.2% coupling of NADPH consumption to product hydroxylation. This example shows that enzymes can be evolved to generate near-native activity on completely foreign substrates.
Figure 17.
A) Total turnovers for variants along the P450PMO lineage on propane and ethane.xcv B) Activity of variants along the P450PMO lineage on Cn (n = 1–10) alkanes.xcvi
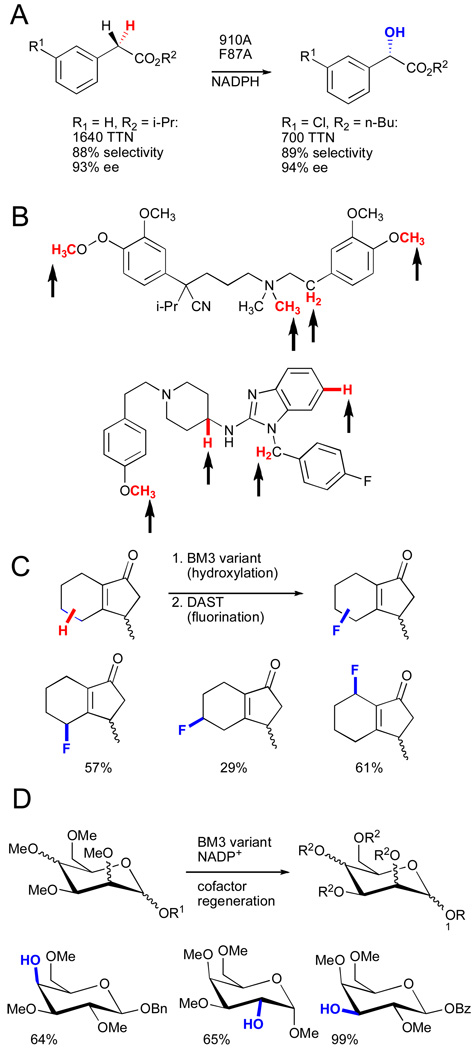
Investigation of the substrate profiles for variants along the evolutionary lineage of P450PMO revealed that the specificity of P450BM3 for hydroxylation of long-chain alkanes was relaxed in intermediate variants (Fig. 17B).xcvi The broad substrate scope of these variants suggested that hydroxylation of substrates not included in the original activity screens might be possible. This proved to be the case, and a panel of enzymes along the P450PMO lineage has enabled rapid hydroxylation and derivatization of common organic scaffolds that would be difficult to accomplish using standard chemical means.xcvii For example, enantioselective hydroxylation of phenyl acetic esters (Fig. 18A),xcviii regioselective hydroxylation of various pharmaceuticals (Fig. 18B),xcix chemoenzymatic fluorination (Fig. 18C),c and chemoenzymatic elaboration of monosaccharides (Fig. 18D)ci have all been accomplished. Specific transformations were easily improved by further directed evolution of variants possessing broad substrate scope. These examples demonstrate the remarkable ability of cytochrome P450 to adapt to new challenges.
Figure 18.
A) Hydroxylation of phenylacetic esters.xcviii B) Regioselectivity of hydroxylation reactions catalyzed by BM3 chimeras.xcix C) Tandem P450BM3-catalyzed hydroxylation/DAST deoxyfluorination.c D) Heteroatom demethylation for regioselective deprotection of monosaccharides.ci
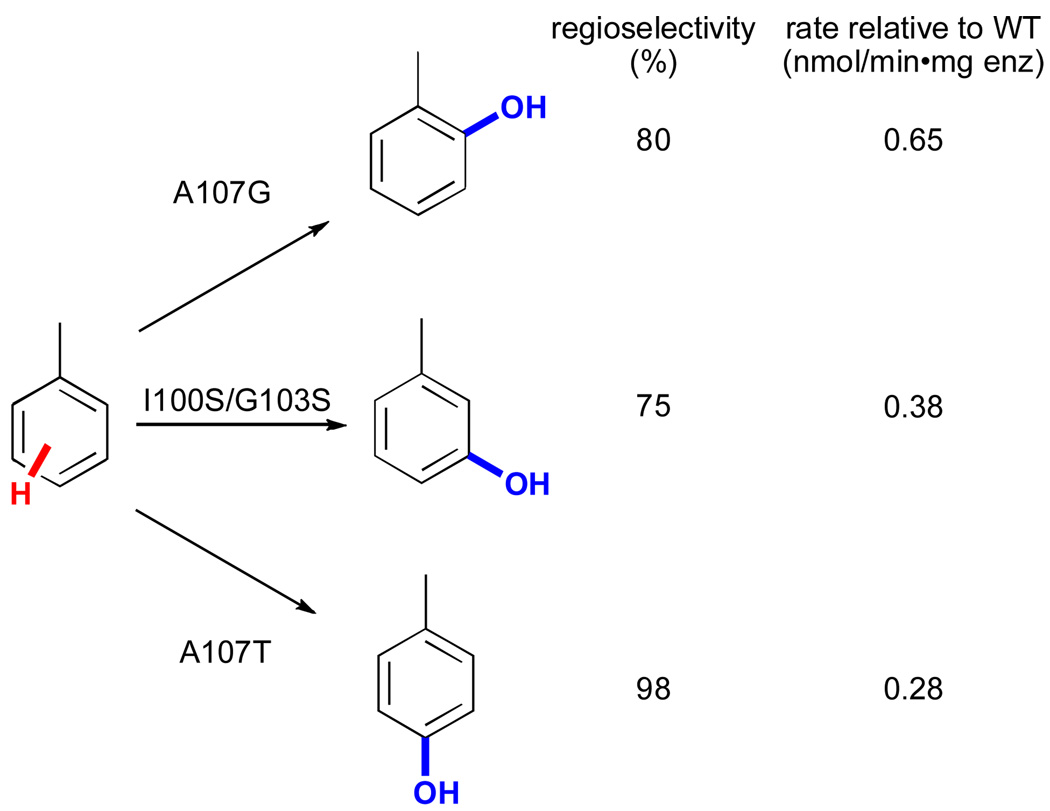
Directed evolution has also been used to improve the activity and alter the regioselectivity of non-heme iron monooxygenases. In one example, Wood and co-workers utilized saturation mutagenesis of sites identified via random mutagenesis and various biochemical studies to identify variants of toluene para-monooxygenase (TpMO) from Ralstonia pickettii PKO1 capable of selectively hydroxylating toluene at the ortho, meta, and para positions (Fig. 19).cii These impressive changes in selectivity were achieved by varying only three active site residues and highlight the ease with which the selectivity of enzymes for activation of a particular C-H bond may be tuned.
Figure 19.
Regioselective hydroxylation of toluene using mutants of toluene para-monooxygenase (TpMO).cii
FAD-dependent monooxygenases have also been improved by directed evolution. As previously discussed, 2-hydroxybiphenyl 3-monooxygenase (HbpA) catalyzes the ortho hydroxylation of various 2-substituted phenols to provide the corresponding catechols (Fig. 12A).ciii Meyer and co-workers utilized 1–2 rounds of random mutagenesis and screening to identify HbpA variants with improved activity on several substrates, including 2-sec-butylphenol, 2-methoxyphenol, and 2-tert-butylphenol. The authors also observed a significant increase in NADH coupling in one case. Such improvements could enable efficient enzymatic hydroxylation of substrates that differ significantly from 2-hydroxybiphenyl.
V. Future Prospects
As discussed above, enzymes enable a range of C-H functionalization reactions that are not possible using small molecule catalysts. While a wide array of such enzymes are known, only a fraction of these have been utilized for biocatalysis, and even fewer have been adopted for commercial applications or have undergone engineering to alter their properties. Additional research on enzyme, process, and metabolic engineering is therefore required in order to understand, optimize, and further exploit this reactivity.lxxii The development and optimization of the enzymes themselves also remains a central challenge. Identifying novel enzymes, developing robust expression protocols, and improving these enzymes for specific applications are all required. Furthermore, evaluating the process compatibility of these catalysts must be a part of these efforts as early as possible.lxxiv Thus, issues regarding enzyme solubility, stability, organic solvent tolerance, cofactor regeneration, and substrate supply/product removal must all be addressed.
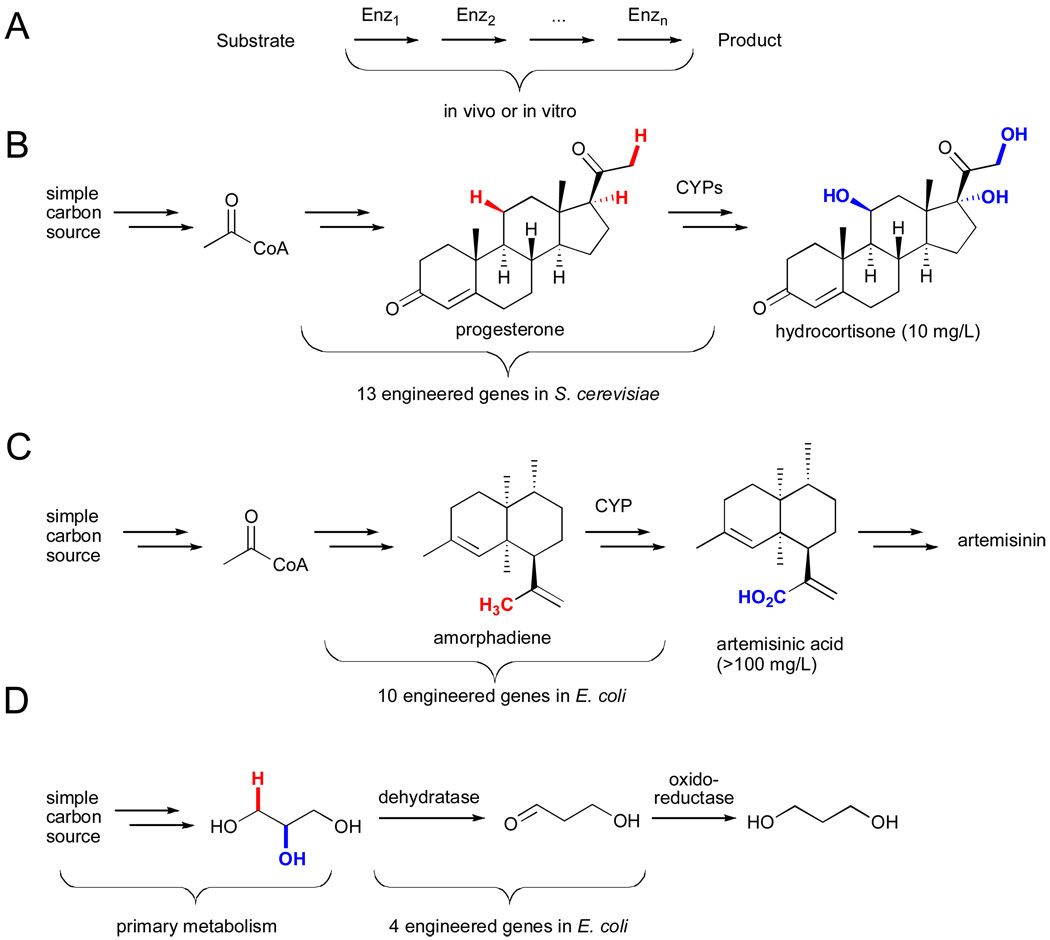
As such issues are resolved, many potential applications will emerge. Among these are the potential to utilize these catalysts in concert with other enzymes, either in vitro or in vivo, for the synthesis of complex molecules (Fig. 20A).civ Enzymatic cascades constructed in vitro enable precise control of all reaction parameters (e.g. pH, concentration, solvents, etc.) while avoiding toxicity and purification issues associated with the use of enzymes in cells. However, this approach also requires expression and isolation of large quantities of the desired enzymes and a means to continually supply appropriate cofactors (e.g. NAD(P)H, FADH2, etc.).
Figure 20.
A) General scheme for in vitro or in vivo biocatalytic cascades.civ Engineered metabolic pathways for production of B) hydrocortisone,cvi C) artemisinic acid,cvii and D) 1,3-propanediolcviii.
Because of these problems, construction of enzymatic cascades in vivo via metabolic engineering is currently more common, and several instances in which C-H bonds on biosynthetic intermediates are functionalized have been reported.cv For example, researchers at the CNRS and Aventis Pharmaceuticals generated an artificial metabolic pathway comprised of 13 engineered genes, including those encoding three cytochromes P450 for C-H hydroxylation, for the total biosynthesis of hydrocortisone from glucose or ethanol (Fig. 20B).cvi Keasling and coworkers reported heterologous expression of a plant cytochrome P450 in yeast to catalyze the allylic hydroxylation and subsequent oxidation of amorphadiene to produce artemisinic acid, an intermediate in the production of the antimalarial compound artemisinin (Fig. 20C).cvii Finally, an AdoCbl-dependent dehydratase-catalyzed reaction similar to that shown in Fig. 14 was utilized in an engineered pathway for production of 1,3-propanediol from glucose in a joint venture between DuPont and Tate & Lyle BioProducts.cviii
In cases where wild-type enzymes are not available or otherwise are incompatible with a given process, engineered enzymes may be of great utility. Furthermore, such enzymes could enable the biosynthesis of natural product derivatives with novel biological activity or other valuable properties without the need for conventional total synthesis. While protein engineering can be used to optimize enzymes and metabolic engineering allows enzymes from different organisms to be combined in robust hosts for the production of desired chemicals, chemists still enjoy a much broader palette of reactions than that found in nature. Many of the enzymes discussed herein utilize the unique reactivity of metal ions or complexes to effect the functionalization of C-H bonds, but many additional C-H functionalization reactions could be enabled through the use of non-biogenic metals, such as rhodium or ruthenium. Efforts to create artificial metalloenzymes by incorporating such catalysts into protein scaffolds have been ongoing for many years.cix Such constructs could provide a means to confer selectivity to otherwise unselective C-H functionalization catalysts. Furthermore, appropriate design of artificial metalloenzymes could allow the optimization of these catalysts using the well-established directed evolution techniques outlined above.cx This would greatly facilitate chemists’ ability to tune the selectivity of a catalyst for functionalization of any desired C-H bond on a given substrate.
Conclusion
The enzymes outlined herein have the potential to significantly improve the efficiency of chemical synthesis by enabling C-H functionalization reactions not possible with small molecule catalysts. Despite their diverse structures and modes of action, many of these enzymes share common themes, including (1) substrate-dependent generation of (2) reactive electrophilic or radical intermediates with (3) selectivity imposed by molecular recognition of the substrate. Perhaps most importantly, these catalysts, like all enzymes, can be evolved to better suit particular applications and can be used to impart microbes with the ability to produce novel compounds. Thus, many of the features that make these catalysts so powerful are now being utilized to develop artificial systems to further expand the range of transformations possible. Continued effort on all of these fronts will lead to improved processes employing enzymes for the functionalization of C-H bonds and for simplifying the synthesis of complex molecules.
Acknowledgments
Prof. Keith Fagnou’s work in the field of C-H bond functionalization and on direct arylation in particular provided many exciting insights into mechanistic aspects of C-H activation chemistry and led to the development of a number of conceptually novel transformations. His constant advances inspired me to work faster and think harder. He will be missed. --JCL
JCL is supported by a U.S. National Institutes of Health Pathways to Independence Award (1K99GM087551-01A1). This work was supported by the U.S. National Institutes of Health (2R01GM068664-05A1) and by the U.S. Department of Energy, Office of Basic Science, grant DE-FG02-06ER15762.
Footnotes
Part of a themed issue on C-H functionalization in organic synthesis
References
- i.(a) Walji AM, MacMillan DWC. Synlett. 2007;10:1477–1489. [Google Scholar]; (b) Newhouse T, Baran PS, Hoffmann RW. Chem. Soc. Rev. 2009;38:3010–3021. doi: 10.1039/b821200g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ii.Hendrickson JB. J. Am. Chem. Soc. 1975;97:5784–5800. [Google Scholar]
- iii.(a) Crabtree RH. J. Chem. Soc., Dalton Trans. 2001;2437 [Google Scholar]; (b) Bergman RG. Nature. 2007;446:391. doi: 10.1038/446391a. [DOI] [PubMed] [Google Scholar]
- iv.Lewis JC, Bergman RG, Ellman JA. Acc. Chem. Res. 2008;41:1013–1025. doi: 10.1021/ar800042p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- v.(a) Yang J, Gabriele B, Belvedere S, Huang Y, Breslow R. J. Org. Chem. 2002;67:5057. doi: 10.1021/jo020174u. [DOI] [PubMed] [Google Scholar]; (b) Das S, Incarvito CD, Crabtree RH, Brudvig GW. Science. 2006;312:1941. doi: 10.1126/science.1127899. [DOI] [PubMed] [Google Scholar]
- vi.Arnold FH. Acc. Chem. Res. 1998;31:125–131. [Google Scholar]
- vii.Michal G, editor. Biochemical Pathways. New York: John Wiley and Sons, Inc.; 1999. [Google Scholar]
- viii.Frey PA. Chem. Rev. 1990;90:1343–1357. [Google Scholar]
- ix.Ragsdale SW. Chem. Rev. 2006;106:3317–3337. doi: 10.1021/cr0503153. [DOI] [PubMed] [Google Scholar]
- x.Shilov AE, Shul’pin GB. Chem. Rev. 1997;97:2879–2932. doi: 10.1021/cr9411886. [DOI] [PubMed] [Google Scholar]
- xi.Sato H, Guengerich FP. J. Am. Chem. Soc. 2000;122:8099–8100. [Google Scholar]
- xii.Massey V. Biochem. Soc. Trans. 2000;28:283–296. [PubMed] [Google Scholar]
- xiii.van Berkel WJH, Kamerbeek NM, Fraaije MW. J. Biotechnol. 2006;124:670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- xiv.Pazmino DET, Winkler M, Glieder A, Fraaije MW. J. Biotechnol. 2010;146:9–24. doi: 10.1016/j.jbiotec.2010.01.021. [DOI] [PubMed] [Google Scholar]
- xv.Massey V. J. Biol. Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- xvi.Entsch B, van Berkel WJH. FASEB J. 1995;9:476–483. doi: 10.1096/fasebj.9.7.7737455. [DOI] [PubMed] [Google Scholar]
- xvii.Li L, Liu X, Yang W, Xu F, Wang W, Feng L, Bartlam M, Wang L, Rao Z. J. Mol. Biol. 2008;376:453–465. doi: 10.1016/j.jmb.2007.11.069. [DOI] [PubMed] [Google Scholar]
- xviii.Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Chem. Rev. 2006;106:3364–3378. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- xix.van Peé KH, Patallo EP. Appl. Microbiol. Biotechnol. 2006;70:631–641. doi: 10.1007/s00253-005-0232-2. [DOI] [PubMed] [Google Scholar]
- xx.(a) Yeh E, Blasiak LC, Koglin A, Drennan CL, Walsh CT. Biochem. 2007;46:1284–1292. doi: 10.1021/bi0621213. [DOI] [PubMed] [Google Scholar]; (b) Flecks S, Patallo EP, Zhu X, Ernyei AJ, Seifert G, Schneider A, Dong C, Naismith JH, van Peé KH. Angew. Chem. Int. Ed. 2008;47:9533–9536. doi: 10.1002/anie.200802466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xxi.(a) Marsh ENG, Patterson DP, Li L. ChemBioChem. 2010;11:604–621. doi: 10.1002/cbic.200900777. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Booker SJ. Curr. Opin. Chem. Biol. 2009;13:58–79. doi: 10.1016/j.cbpa.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xxii.Brown KL. Chem. Rev. 2005;105:2075–2149. doi: 10.1021/cr030720z. [DOI] [PubMed] [Google Scholar]
- xxiii.Frey PA, Magnusson OT. Chem. Rev. 2003;103:2129–2148. doi: 10.1021/cr020422m. [DOI] [PubMed] [Google Scholar]
- xxiv.Frey PA, Booker S. Radical Intermediates in the Reaction of Lysine-2,3-aminomutase. In: Zard SZ, editor. Advances in Free Radical Chemistry. Vol. 2. JAI Press Inc.; 1999. pp. 1–43. [Google Scholar]
- xxv.Song KB, Frey PA. J. Biol. Chem. 1991;266:7651–7655. [PubMed] [Google Scholar]
- xxvi.Jarrett JT. Arch. Biochem. Biophys. 2005;433:312–321. doi: 10.1016/j.abb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- xxvii.Frey PA, Hegeman AD, Ruzicka FJ. Crit. Rev. Biochem. Mol. Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- xxviii.Boll M, Fuchs G, Heider J. Curr. Opin. Chem. Biol. 2002;6:604–611. doi: 10.1016/s1367-5931(02)00375-7. [DOI] [PubMed] [Google Scholar]
- xxix.Chianese AR, Lee SJ, Gagne MR. Angew. Chem. Int. Ed. 2007;46:4042–4059. doi: 10.1002/anie.200603954. [DOI] [PubMed] [Google Scholar]
- xxx.Berkovitch F, Behshad E, Tang KH, Enns EA, Frey PA, Drennan CL. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15870–15875. doi: 10.1073/pnas.0407074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xxxi.Gruber K, Krathy C. Curr. Opin. Chem. Biol. 2002;6:598–603. doi: 10.1016/s1367-5931(02)00368-x. [DOI] [PubMed] [Google Scholar]
- xxxii.Masuda J, Shibata N, Morimoto Y, Toraya T, Yasuoka N. Structure. 2000;8:775–788. doi: 10.1016/s0969-2126(00)00164-7. [DOI] [PubMed] [Google Scholar]
- xxxiii.Decker A, Solomon EI. Curr. Opin. Chem. Biol. 2005;9:152–163. doi: 10.1016/j.cbpa.2005.02.012. [DOI] [PubMed] [Google Scholar]
- xxxiv.Sono M, Roach MP, Coulter ED, Dawson JH. Chem. Rev. 1996;96:2841–2887. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- xxxv.Krebs C, Fujimori DG, Walsh CT, Bollinger JM. Acc. Chem. Res. 2007;40:484–492. doi: 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xxxvi.Bollinger JM, Krebs C. Curr. Opin. Chem. Biol. 2007;11:151–158. doi: 10.1016/j.cbpa.2007.02.037. [DOI] [PubMed] [Google Scholar]
- xxxvii.Holme E. Biochem. 1975;14:4999–5003. doi: 10.1021/bi00693a033. [DOI] [PubMed] [Google Scholar]
- xxxviii.Baggaley KH, Brown AG, Schofield CJ. Nat. Prod. Rep. 1997;14:309–333. doi: 10.1039/np9971400309. [DOI] [PubMed] [Google Scholar]
- xxxix.Ueki M, Galonic DP, Vaillancourt FH, Garneau-Tsodikova S, Yeh E, Vosburg DA, Schroeder FC, Osada H, Walsh CT. Chem. Biol. 2006;13:1183–1191. doi: 10.1016/j.chembiol.2006.09.012. [DOI] [PubMed] [Google Scholar]
- xl.Baldwin JE, Bradley M. Chem. Rev. 1990;90:1079–1088. [Google Scholar]
- xli.Lippard SJ. Philos. Trans. R. Soc. 2005;363:861–877. doi: 10.1098/rsta.2004.1532. [DOI] [PubMed] [Google Scholar]
- xlii.Leahy JG, Batchelor PJ, Morcomb SM. FEMS Microbiol. Rev. 2003;27:449–479. doi: 10.1016/S0168-6445(03)00023-8. [DOI] [PubMed] [Google Scholar]
- xliii.Mitchell KH, Studts JM, Fox BG. Biochemistry. 2002;41:3176–3188. doi: 10.1021/bi012036p. [DOI] [PubMed] [Google Scholar]
- xliv.Baik M-H, Newcomb M, Friesner RA, Lippard SJ. Chem. Rev. 2003;103:2385–2419. doi: 10.1021/cr950244f. [DOI] [PubMed] [Google Scholar]
- xlv.Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Muller J, Lippard SJ. Angew. Chem. Int. Ed. 2001;40:2782–2807. doi: 10.1002/1521-3773(20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- xlvi.Wubbolts MG, Reuvekamp P, Witholt B. Enz. Microb. Technol. 1994;16:608–614. doi: 10.1016/0141-0229(94)90127-9. [DOI] [PubMed] [Google Scholar]
- xlvii.Worsey MJ, Williams PA. J. Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xlviii.Kiener A. Angew. Chem. Int. Ed. 1992;31:774–775. [Google Scholar]
- xlix.Solomon EI, Sundaram UM, Machonkin TE. Chem. Rev. 1996;96:2563–2605. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- l.Klinman JP. Chem. Rev. 1996;96:2541–2561. doi: 10.1021/cr950047g. [DOI] [PubMed] [Google Scholar]
- li.Chen P, Solomon EI. J. Am. Chem. Soc. 2004;126:4991–5000. doi: 10.1021/ja031564g. [DOI] [PubMed] [Google Scholar]
- lii.Itoh S, Fukuzumi S. Acc. Chem. Res. 2007;40:592–600. doi: 10.1021/ar6000395. [DOI] [PubMed] [Google Scholar]
- liii.Decker H, Schweikardt T, Tuczek F. Angew. Chem. Int. Ed. 2006;45:4546–4550. doi: 10.1002/anie.200601255. [DOI] [PubMed] [Google Scholar]
- liv.Decker A, Solomon EI. Curr. Opin. Chem. Biol. 2005;9:152–163. doi: 10.1016/j.cbpa.2005.02.012. [DOI] [PubMed] [Google Scholar]
- lv.Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC. Nature. 2010;465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- lvi.Shiota Y, Yoshizawa K. Inorg Chem. 2009;48:838–845. doi: 10.1021/ic8003933. [DOI] [PubMed] [Google Scholar]
- lvii.Woertink JS, Smeets PJ, Groothaert MH, Vance MA, Sels BF, Schoonheydt RA, Solomon EI. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18908–18913. doi: 10.1073/pnas.0910461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- lviii.Himes RA, Barnese K, Karlin KD. Angew. Chem. Int. Ed. 2010;49:6714–6716. doi: 10.1002/anie.201003403. [DOI] [PubMed] [Google Scholar]
- lix.Green MT. Curr. Opin. Chem. Biol. 2009;13:84–88. doi: 10.1016/j.cbpa.2009.02.028. [DOI] [PubMed] [Google Scholar]
- lx.Harris DL. Curr. Opin. Chem. Biol. 2001;5:724–735. doi: 10.1016/s1367-5931(01)00272-1. [DOI] [PubMed] [Google Scholar]
- lxi.Van Deurzen MPJ, Van Rantwijk F, Sheldon RA. Tetrahedron. 1997;53:13183–13220. [Google Scholar]
- lxii.Groves JT. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3569–3574. doi: 10.1073/pnas.0830019100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- lxiii.Matsunaga I, Shiro Y. Curr. Opin. Chem. Biol. 2004;8:127–132. doi: 10.1016/j.cbpa.2004.01.001. [DOI] [PubMed] [Google Scholar]
- lxiv.Dawson JH. Science. 1988;240:433–439. doi: 10.1126/science.3358128. [DOI] [PubMed] [Google Scholar]
- lxv.Ma Y-A, Guo Z-W, Sih CJ. Tetrahedron Lett. 1998;51:9357–9360. [Google Scholar]
- lxvi.Hofrichter M, Ullrich R. Appl. Microbiol. Biotechnol. 2006;71:276–288. doi: 10.1007/s00253-006-0417-3. [DOI] [PubMed] [Google Scholar]
- lxvii.Wagner C, El Omari M, Konig GM. J. Nat. Prod. 2009;72:540–553. doi: 10.1021/np800651m. [DOI] [PubMed] [Google Scholar]
- lxviii.Loew GH, Harris DL. Chem. Rev. 2000;100:407–419. doi: 10.1021/cr980389x. [DOI] [PubMed] [Google Scholar]
- lxix.Denisov IG, Makris TM, Sligar SG, Schlichting I. Chem. Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- lxx.Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. New York: Kluver Academic/Plenum Publishers; 2005. [Google Scholar]
- lxxi.Guengerich FP. Chem. Res. Toxicol. 2001;14:612–640. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- lxxii.(a) Chefson A, Auclair K. Mol. BioSyst. 2006;2:462–469. doi: 10.1039/b607001a. [DOI] [PubMed] [Google Scholar]; (b) Eiben S, Kaysser L, Maurer S, Kuhnel K, Urlacher VB, Schmid RD. J. Biotechnol. 2006;124:662–669. doi: 10.1016/j.jbiotec.2006.02.013. [DOI] [PubMed] [Google Scholar]; (c) Urlacher VB, Lutz-Wahl S, Schmid RD. Appl. Microbiol. Biotechnol. 2004;64:317–325. doi: 10.1007/s00253-003-1514-1. [DOI] [PubMed] [Google Scholar]
- lxxiii.(a) Julsing MK, Cornelissen S, Buhler B, Schmid A. Curr. Opin. Chem. Biol. 2008;12:177–186. doi: 10.1016/j.cbpa.2008.01.029. [DOI] [PubMed] [Google Scholar]; (b) Guengerich FP. Nat. Rev. Drug. Disc. 2002;1:359–366. doi: 10.1038/nrd792. [DOI] [PubMed] [Google Scholar]; (c) van Beilen JB, Duetz WA, Schmid A, Witholt B. Trends Biotechnol. 2003;21:170–177. doi: 10.1016/S0167-7799(03)00032-5. [DOI] [PubMed] [Google Scholar]
- lxxiv.Savile CK, Janey JM, Mundorff EC, Moore JC, Tam S, Jarvis WR, Colbeck JC, Krebber A, Fleitz FJ, Brands J, Devine PN, Huisman GW, Hughes GJ. Science. 2010;329:305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- lxxv.Alonso DA, Najera C, Pastor IM, Yus M. Chem. Eur. J. 2010;16:5274–5284. doi: 10.1002/chem.201000470. [DOI] [PubMed] [Google Scholar]
- lxxvi.Held M, Suske W, Schmid A, Engesser KH, Kohler HPE, Witholt B, Wubbolts MG. J. Mol. Cat. B: Enz. 1998;5:87–93. [Google Scholar]
- lxxvii.Jadan AP, Moonen MJH, Boeren S, Golovleva LA, Rietjens IMCM, van Berkel WJH. Adv. Synth. Catal. 2004;346:367–375. [Google Scholar]
- lxxviii.Holzer M, Burd W, Rebig HU, van Peé KH. Adv. Synth. Catal. 2001;343:591–595. [Google Scholar]
- lxxix.Kraus GA. Clean. 2008;36:648–651. [Google Scholar]
- lxxx.Cameron DC, Altaras NE, Hoffman ML, Shaw AJ. Biotechnol. Prog. 1998;14:116–125. doi: 10.1021/bp9701325. [DOI] [PubMed] [Google Scholar]
- lxxxi.Brouk M, Fishman A. Food Chem. 2009;116:114–121. and references therein. [Google Scholar]
- lxxxii.Mitchell KH, Studts JM, Fox BG. Biochem. 2002;41:3176–3188. doi: 10.1021/bi012036p. [DOI] [PubMed] [Google Scholar]
- lxxxiii.Nolan LC, O’Connor KE. Biotechnol. Lett. 2007;29:1045–1050. doi: 10.1007/s10529-007-9365-y. [DOI] [PubMed] [Google Scholar]
- lxxxiv.Wubbolts MG, Reuvekamp P, Witholt B. Enz. Microb. Technol. 1994;16:608–614. doi: 10.1016/0141-0229(94)90127-9. [DOI] [PubMed] [Google Scholar]
- lxxxv.Bureik M, Bernhardt R. Steroid Hydroxylation: Microbial Steroid Biotransformations Using Cytochrome P450 Enzymes. In: Schmid RD, Urlacher VB, editors. Modern Biooxidation - Enzymes, Reactions, and Applications. Wiley-VCH; 2007. pp. 155–176. [Google Scholar]
- lxxxvi.Park JW, Lee JK, Kwon TJ, Yi DH, Kim YJ, Moon SH, Suh HH, Kang SM, Park YI. Biotechnol. Lett. 2003;25:1827–1831. doi: 10.1023/a:1026281914301. [DOI] [PubMed] [Google Scholar]
- lxxxvii.Liu SC, Li C, Fang XC, Cao ZH. Enzyme Microb. Technol. 2004;34:73–77. [Google Scholar]
- lxxxviii.Zaks A, Dodds DR. J. Am. Chem. Soc. 1995;117:10419–10424. [Google Scholar]
- lxxxix.Wannstedt C, Rotella D, Siuda JF. Bull. Environ. Contam. Toxicol. 1990;44:282–287. doi: 10.1007/BF01700148. [DOI] [PubMed] [Google Scholar]
- xc.Stuart DR, Fagnou K. Science. 2007;316:1172–1175. doi: 10.1126/science.1141956. [DOI] [PubMed] [Google Scholar]
- xci.Kiso T, Shizuma M, Murakami H, Kiryu T, Hozono K, Terai T, Nakano H. J. Mol. Cat. B Enz. 2007;45:50–56. [Google Scholar]
- xcii.Ringe D, Petsko GA. Science. 2008;320:1428–1429. doi: 10.1126/science.1159747. [DOI] [PubMed] [Google Scholar]
- xciii.Reetz MT. In: Protein Engineering Handbook. Lutz S, Bornscheuer UT, editors. Vol. 2. Weinheim, Germany: Publisher: Wiley-VCH Verlag GmbH & Co. KGaA; 2009. p. 409. [Google Scholar]
- xciv.Khersonsky O, Roodveldt C, Tawfik DS. Curr. Opin. Chem. Biol. 2006;10:498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- xcv.Fasan R, Chen MM, Crook NC, Arnold FH. Angew. Chem. Int. Ed. 2007;46:8414–8418. doi: 10.1002/anie.200702616. [DOI] [PubMed] [Google Scholar]
- xcvi.Fasan R, Meharenna YT, Snow CD, Poulos TL, Arnold FH. J. Mol. Biol. 2008;383:1069. doi: 10.1016/j.jmb.2008.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xcvii.Lewis JC, Arnold FH. Catalysts on Demand: Selective Oxidations by Laboratory-Evolved Cytochrome P450 BM-3. Chimia. 2009;63:309–312. [Google Scholar]
- xcviii.Landwehr M, Hochrein L, Otey CR, Kasrayan A, Bäckvall J-E, Arnold FH. J. Am. Chem. Soc. 2006;128:6058–6059. doi: 10.1021/ja061261x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xcix.Sawayama AM, Chen MM, Kulanthaivel P, Kuo M-S, Hemmerle H, Arnold FH. Chem. Eur. J. 2009;15:11723–11729. doi: 10.1002/chem.200900643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- c.Rentmeister A, Arnold FH, Fasan R. Nat. Chem. Biol. 2008;5:26–28. doi: 10.1038/nchembio.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ci.Lewis JC, Bastian S, Bennett CS, Fu Y, Mitsuda Y, Chen MM, Greenberg WA, Wong C-H, Arnold FH. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16550–16555. doi: 10.1073/pnas.0908954106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- cii.Fishman A, Tao Y, Rui L, Wood TK. J. Biol. Chem. 2005;280:506–514. doi: 10.1074/jbc.M410320200. [DOI] [PubMed] [Google Scholar]
- ciii.Meyer A, Schmid A, Held M, Westphal AH, Rothlisberger M, Kohler HPE, van Berkel WJH, Witholt B. J. Biol. Chem. 2002;277:5575–5582. doi: 10.1074/jbc.M110018200. [DOI] [PubMed] [Google Scholar]
- civ.Lopez-Gallego F, Schmidt-Danner C. Curr. Opin. Chem. Biol. 2010;14:174–183. doi: 10.1016/j.cbpa.2009.11.023. [DOI] [PubMed] [Google Scholar]
- cv.Keasling JD. ACS Chem. Biol. 2008;3:64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- cvi.Szczebara FM, Chandelier C, Villeret C, Masurel A, Bourot S, Duport C, Blanchard S, Groisillier A, Testet E, Costaglioli P, Cauet G, Degryse E, Balbuena D, Winter J, Achstetter T, Spagnoli R, Pompon D, Dumas B. Nat. Biotechnol. 2003;21:143–149. doi: 10.1038/nbt775. [DOI] [PubMed] [Google Scholar]
- cvii.Chang MCY, Eachus RA, Trieu W, Ro DK, Keasling JD. Nat. Chem. Biol. 2007;3:274–277. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- cviii.(a) http://www.duponttateandlyle.com/ [Google Scholar]; (b) Emptage M, Haynie S, Laffend L, Pucci J, Whited G. PCT Int. Appl. 2001:109. WO 2001012833 A2 20010222 CAN 134:188971. [Google Scholar]
- cix.Thomas CM, Ward TR. Chem. Soc. Rev. 2005;34:337–346. doi: 10.1039/b314695m. [DOI] [PubMed] [Google Scholar]
- cx.Reetz MT. Tetrahedron. 2002;58:6595–6602. [Google Scholar]