Abstract
The transforming growth factor β (TGF-β) signaling pathway plays myriad roles in development and disease. TGF-β isoforms initiate signaling by organizing their cell surface receptors TβRI and TβRII. Exploration and exploitation of the versatility of TGF-β signaling requires enhanced understanding of structure–function relationships in this pathway. To this end, small molecule, peptide, and antibody effectors that bind key signaling components would serve as valuable probes. We focused on TβRI-ED as a target for effector screening. The observation that the extracellular domain of TβRI (TβRI-ED) can bind to a TGF-β coreceptor (endoglin), suggests that the TβRI-ED may have multiple interaction sites. Using phage display, we identified two peptides LTGKNFPMFHRN (Pep1) and MHRMPSFLPTTL (Pep2) that bind the TβRI-ED (Kd ~10-5 M). Although our screen focused on TβRI-ED, the hit peptides interact with the TβRII-ED with similar affinities. The peptide ligands occupy the same binding sites on TβRI and TβRII, as demonstrated by their ability to compete with each other for receptor binding. Moreover, neither interferes with TGF-β binding. These results indicate that TβRI and TβRII both possess hot spots for protein–protein interactions that are distinct from those used by their known ligand TGF-β. To convert these compounds into high affinity probes, we exploited the observation that TβRI and TβRII exist as dimers on the cell surface; therefore, we assembled a multivalent ligand. Specifically, we displayed one of our receptor-binding peptides on a dendrimer scaffold. We anticipate that the potent multivalent ligand that resulted can be used to probe the role of receptor assembly in TGF-β function.
Introduction
TGF-β isoforms, TGF-β1, 2, and 3, are disulfide-linked homodimers with molecular weights of approximately 25 kDa (Fig. 1A).1, 2, 3 TGF-β signaling occurs upon formation of a quinary complex that consists of TGF-β and two copies each of the transmembrane Ser/Thr kinase receptors, TβRI and TβRII.2, 4 Signaling complex formation occurs when TGF-β1 or TGF-β3 binds with high affinity (Kd ~5-30 pM) to two copies of TβRII. The resulting TGF-β:TβRII complex then recruits two copies of TβRI to form a hetero-oligomeric complex. The TGF-β2 homolog is lacking two key arginine residues present in TGF-β1 and TGF-β3 that facilitate high affinity interactions with TβRII;5 therefore, TGF-β2 requires a coreceptor (β-glycan or TβRIII) to assemble a signaling complex. Once TβRI and TβRII are proximal, the cytoplasmic domain of TβRII catalyzes the phosphorylation of multiple TβRI threonine and serine residues within a conserved juxtamembrane GS domain (a 30-amino acid region that contains a characteristic SGSGSG sequence). GS domain phosphorylation promotes activation of the adjacent TβRI kinase domain. The activated enzyme then catalyzes the phosphorylation of the receptor-regulated Smad proteins (R-Smad),6 Smad2 and Smad3, with the help of an adaptor protein SARA (Smad anchor for receptor activation).7 R-Smads are critical regulators of TGF-β signaling that shuttle between the cytoplasm and nucleus. Upon growth factor stimulation, the phosphorylated Smad2 or Smad3 dissociates from SARA and binds to the common Smad (co-Smad), Smad4; this complex undergoes nuclear translocation. Once in the nucleus, the Smad complex interacts with various DNA binding partners to activate or repress the expression of hundreds of genes.
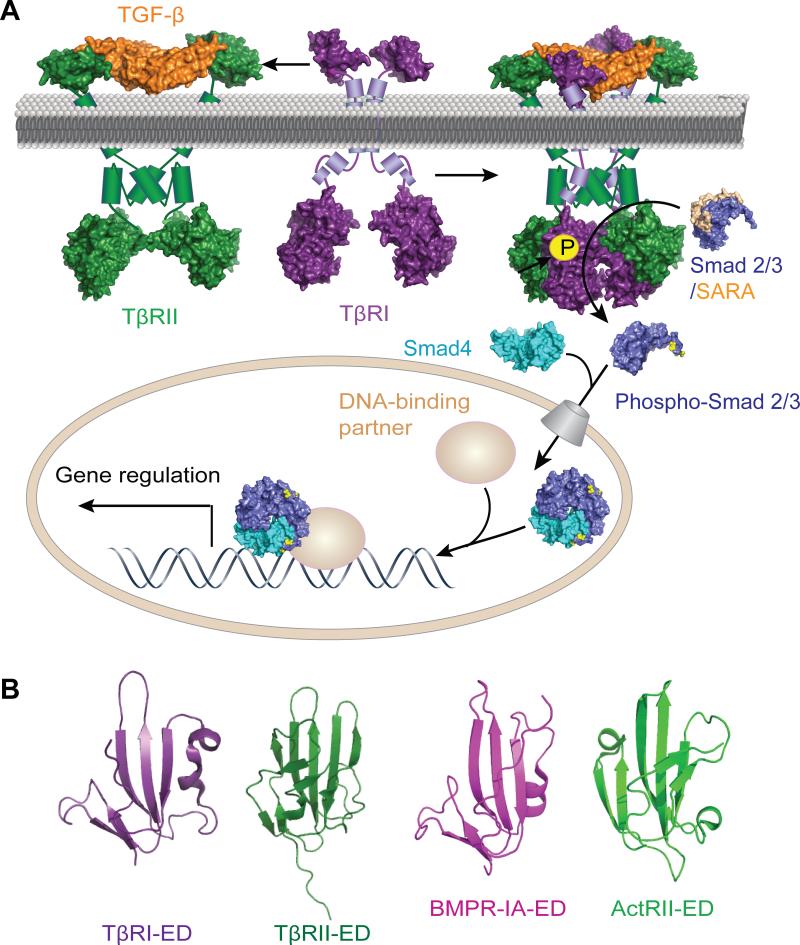
FIG. 1.
(A) Schematic depiction of the TGF-β signaling pathway. The covalently linked TGF-β homodimer (orange) binds to two copies of TβRII (green) which forms non-covalent homodimers as well as higher order oligomers.18 The TGF-β/TβRII complex then recruits two copies of TβRI (purple). This quinary complex enables the constitutively active TβRII to catalyze the phosphorylation of the serine residues in the juxtamembrane GS domain of TβRI. Upon GS domain phosphorylation, the adjacent kinase domain catalyzes the phosphorylation and activation of receptor-regulated Smad proteins (R-Smad), Smad2 and Smad3 (blue) with the help of an adaptor protein SARA (Smad anchor for receptor activation, light brown). Phosphorylated Smad2 and Smad3 dissociate from SARA and bind to common Smad (co-Smad), Smad4 (teal), which facilitates the translocation of this complex into the nucleus. Once in the nucleus, the Smads bind to different DNA binding partners to control gene expression. Structures used in the creation of this Fig. were determined by X-ray crystallographic analysis and rendered using PyMOL molecular graphics. PDB files used to construct this scheme follow: PDB ID 3KFD19 (for TGF-β1:TβRI-ED:TβRII-ED ternary structure), 2QLU20 (for activin receptor type IIB cytoplasmic domain residues 188-483, which is homolous to TβRII residues 267-592), 1IAS21 (for TβRI cytoplasmic domain residue 171-503), 1DEV22 and 1U7F23 (for unphosphorylated Smad3 bound to the Smad binding-domain of SARA), 1U7F (for phosphorylated Smad3 and Smad3:Smad3:Smad4 trimeric complex). For extracellular and intracellular segments whose structures have not been determined by X-ray crystallography, online software NetSurfP (http://www.cbs.dtu.dk/services/NetSurfP/) was used to predict secondary structures, and α-helices and transmembrane domains are represented by cylinders. (B) Structures of the extracellular domains of TβRI (purple, PDB ID 2PJY: C), TβRII (dark green, PDB ID 2PJY: B),24 BMPR-IA (magenta, PDB ID 2GOO: B) and ActRII (light green, PDB ID 2GOO: C)25, indicate these proteins share a common three-finger toxin fold stabilized by four disulfide bonds.
TGF-β-induced changes in gene expression elicit a wide range of cellular responses, including cell adhesion, migration, extracellular matrix deposition, proliferation, apoptosis, and differentiation.3, 8 Depending on the cellular context, TGF-β can play essential or deleterious roles in development, immunity, wound healing, or cancer. For example, the growth factor controls embryonic stem cell self-renewal as well as important developmental processes such as the epithelial to mesenchymal transition.9 Loss of TGF-β signaling is associated with autoimmunity, which highlights its role in immune suppression.10 TGF-β is crucial for wound healing, but its prolonged presence causes inflammation and scar formation.11 Another role for the growth factor is as a strong tumor suppressor, yet it is also implicated in the late stage metastasis of many cancer types.12
Because of the important and myriad roles of TGF-β, its ligands would be valuable tools. They could be used to probe its diverse cellular functions and facilitate the identification of potential therapeutics. Hence, TGF-β isoforms and their receptors are popular targets for small molecule screens and antibody-based therapeutics.13 Compounds that inhibit the TβRI kinase domain and the highly related kinase domains of another two type I receptors, Activin A and Nodal, have been sought.14 One such compound, the kinase inhibitor SB-431542, has become a powerful tool for assessing the involvement of TGF-β signaling in specific biological processes. TGF-β2 antisense oligonucleotides,15 neutralizing antibodies16 and peptide ligands for the growth factor17 have been developed to dissect the roles of each individual TGF-β isoform. These investigations highlight the utility of compounds that act on targets within the TGF-β pathway for dissecting the function of TGF-β signaling components in development and disease.
These valuable tools, combined with structures of the TGF-β:TβRI-ED:TβRII-ED complex determined by X-ray crystallography,19, 24 have led to insight into the function of this canonical signaling complex. The growth factor and its receptors, however, have additional binding partners, and the roles of these interactions in TGF-β signaling are less explored. For example, TGF-β isoforms bind to β-glycan and endoglin. Endoglin is another type III receptor (sharing 71% amino acid identity in the transmembrane and cytoplasmic domain with β-glycan) that is highly expressed in proliferating endothelial cells. The receptors, TβRI and TβRII, also interact with endoglin through both their extracellular domains and cytoplasmic domains.26 Thus, although TβRI and TβRII have small extracellular domains (~150 residues) and large surface areas are buried in the TGF-β:TβRI-ED:TβRII-ED ternary complex,24 they possess other hot spots27 for protein-protein interactions. We found this feature of the receptors intriguing. Because phage-display has been used to identify preferred binding sites for protein–protein interactions, 28, 29 we envisioned applying this method to the TβR extracellular domains to indentify novel TβR ligands.
Phage-displayed peptide library screening is a technique used to identify ligands for protein targets.29, 30 Compounds that disrupt31 or promote protein–protein interactions,32 function as hormone or growth factor mimetics,33 or serve as ligands for whole cells34 have been discovered using this technology. We screened a phage-displayed peptide library using TβRI-ED as bait to identify TβR ligands. Intriguingly, this screen yielded peptide ligands that recognize both TβRI-ED and TβRII-ED, yet do not compete with TGF-β. Thus, our data indicate that TβRI and TβRII share a novel binding site that may serve as a target for probing and modulating TGF-β function.
Experimental
Materials
All reagents for phage panning experiments and solution phase synthesis were purchased from Sigma Aldrich (Milwaukee, WI) and used without further purification unless specified otherwise. BSA (albumin, bovine pH 7.0, biotechnology grade) was purchased from Research Organics (Cleveland, OH). M13 Ph.D 12 phage display library kits were purchased from New England Biolabs (Ipswich, MA). The 96-well plates used for immobilization of targets in phage panning experiments and ELISA assays were purchased from Nunc Thermo Fisher Scientific (Rochester, NY). Anti-M13 antibody conjugated with HRP (horseradish peroxidase) was purchased from GE healthcare (Piscataway, NJ). The substrate for HRP, 2,2'-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) or ABTS was purchased from Invitrogen (Carlsbad, CA). Peptides were either purchased from Biomatik (Wilmington, DE) or synthesized at the Peptide Synthesis Facility at the University of Wisconsin-Madison and purified by HPLC to > 80% purity. The mink lung epithelial (Mv1Lu) cell line stably transfected with SBE (CAGA)12-Luc reporter gene was a generous gift from professor F. M. Hoffmann (University of Wisconsin-Madison). Fetal bovine serum (FBS) and DMEM were purchased from Invitrogen. The Bright-Glo™ Luciferase Assay System and CellTiter-Glo® Luminescent Cell Viability Assay were purchased from Promega (Madison, WI). LumiNunc™ Plates were purchased from Nunc Thermo Fisher Scientific. Recombinant human BMPR-IA (Gln24-Arg152) Fc chimera, recombinant human ActRII (Ser25-Pro134) Fc chimera, recombinant human endoglin-ED (residues 26-586) and recombinant human β-glycan-ED (residues 21-781) were purchased from R&D Systems (Minneapolis, MN).
Protein preparation
Recombinant human TGF-β1 was a generous gift from Professor F. M. Hoffmann (University of Wisconsin-Madison). Recombinant human TβRI-ED (residues 1-101) and recombinant human TβRII-ED (residues 1-137) were expressed in E. coli, refolded and purified as described.35
Phage display and phage ELISA
TβRI-ED (residues 1-101) was immobilized in microtiter wells by incubating 100 μL of a solution of TβRI-ED (residues 1-101, 15 μg, 10 kDa) 4 °C for 12 h. The wells were exposed to 200 μL of blocking buffer, which consists of 2% BSA in Tris-buffered saline with detergent (TBST: 50 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 7.4). The M13 Ph.D. phage library (100 μL at 1012 pfu/mL) was allowed to bind to TβRI-ED coated wells for 2 h. Unbound and weakly bound phage particles were removed by washing 6 × 5 min with 200 μL of washing buffer (0.1% Tween 20 in TBST for the first round and 0.5% Tween 20 in TBST for rounds 2–4). Phage particles that bound to TβRI-ED were eluted after 10 min treatment with 100 μL of 0.1 M glycine buffer (pH 2.2); the samples were neutralized immediately with 10 μL of 2 M Tris-HCl buffer (pH 8.2). The resulting phage were amplified after the first and second rounds of panning. The output phage from the third round was used as the input for the fourth round without amplification. After four rounds of panning, 24 phage plaques were sequenced and 7 unique clones were identified.
Phage clones were evaluated for binding to TβRI and TβRII using a phage-based ELISA. Equimolar concentration of TβR solutions were used in immobilization experiments, in which 100 μL of TβRI-ED (1.0 μg) or TβRII-ED (residues 1-137, 1.5 μg, 15 kDa) were immobilized in microtiter wells at 4 °C for 12 h. The wells were subsequently blocked with a solution of 6% BSA for 2 h. Phage clones at various concentrations in a solution of 2% BSA in phosphate buffer saline (PBS) with 0.5% Tween 20 were incubated in TβRI-ED, TβRII-ED or BSA-coated wells for 1 h at room temperature. In the ELISA based competition assay, phage clones (305 pM for clone 1 and 350 pM for clone 2) and varying concentrations of Pep1 or Pep2 (in a solution of 2% BSA in PBS with 0.5% Tween 20) were mixed together and added to immobilized TβRI-ED and TβRII-ED. After 4 × 5 min washes with 200 μL of 0.5% Tween 20 in PBS, phage that bound to these wells were detected by exposure to with anti-M13 antibody conjugated with HRP for 1 h. Followed by incubation with the substrate ABTS in the presence of H2O2 for 30 min, the absorbance at 405 nm of each well was measured on an ELx800 absorbance microplate reader (BioTek).
Reporter gene assay
Mink lung epithelial (Mv1Lu) cells stably transfected with a TGF-β responsive reporter gene SBE (CAGA)12-Luc was used. The gene construct consists of twelve repeats of a Smad binding element with a sequence of CAGA (SBE(CAGA)12) engineered immediately upstream of a gene encoding luciferase.36 The transfected cells were cultured in 10% FBS in DMEM.37 About 4000 cells were plated into 24-well plates and allowed to attach overnight in the normal cell culture media. The media was switched to a low serum media (0.2% FBS in DMEM) 4 hours before TGF-β treatment to eliminate the effect of TGF-β the serum. Cells were treated with TGF-β1, TGF-β1 with serial dilutions of the peptides and peptides alone. Non-treated cells were used as a control. Three replicates were performed for each condition. After 18-24 h, the media was removed and cells were washed once with PBS. Luciferase production was quantified using a Bright-Glo™ Luciferase Assay System. More specifically, cells were lysed by incubating with 75 μL of Glo Lysis Buffer for 5 min. The cell lysate (25 μL) was transferred to a 96-well white plate (LumiNunc™ Plate). Bright-Glo™ Assay Reagent containing the luciferin substrate (25 μL) was added to the cell lysate, followed by immediate quantification using a luminometer plate reader (Perkin Elmer Victor 3 from MTX lab systems). The luminescence reading from each well was normalized by the cell number, which was determined separately using a CellTiter-Glo® Luminescent Cell Viability Assay.
Dendrimer synthesis
Because the region of sequence variability on phage particles is at the N-terminus of the PIII coat protein, peptides identified from the screen were coupled to dendrimers through a C-terminal modification. A cysteine was installed at the C-terminus and its nucleophilicity was exploited. A 20% methanol solution of PAMAM dendrimer (generation 3)38 with an ethylenediamine core presenting 32 surface amino groups (Sigma Aldrich) was diluted in 1 M HEPES buffer (pH 7). A bifunctional N-hydroxysuccinimidyl ester (NHS)-PEG8-maleimide linker (Thermo Scientific) was dissolved in dimethyl sulfoxide (DMSO) at 200 mg/mL and 64 molar equivalents were added to the dendrimer solution; this mixture was allowed to react for 16 h. The remaining NHS-PEG8-maleimide was then removed using a PD-10 size-exclusion column. The cysteine-extended Pep1 was appended to the reactive dendrimer through conjugate addition of the thiolate to the maleimide in 1 M HEPES buffer (pH 7). Cysteine was then added to block any remaining reactive maleimide groups. The final product was dialyzed in water (Milli-Q) overnight and then subjected to lyophilization. The resulting dendrimer was characterized by SDS-PAGE and MALDI (matrix-assisted laser desorption/ionization) mass spectrometry and has a molecular weight of ~35 kDa.
Surface plasmon resonance (SPR) experiments to evaluate dendrimer binding
HEPES-buffered saline (HBS from Biacore, pH 7.4) at a flow rate of 5 μL/min was used as the running buffer to generate protein surfaces on a CM5 sensor chip (Biacore). The sensor chip was preconditioned by injecting two consecutive 10 s pulses of each of the following solutions in the order listed: 10 mM aqueous HCl, 50 mM aqueous sodium hydroxide, 0.1% SDS, and water. The flow rate was maintained at 100 μL/min. Three separate flow cells were functionalized with TβRI-ED (residues 7-91), TβRII-ED (residues 1-137) and endoglin-ED (residues 26-586). A protein-free flow cell was generated as a negative control. The flow rate was maintained at 5 μL/min for surface generation. The carboxymethyl dextran surfaces were activated through an injection of 25 μL of a 1:1 aqueous mixture of 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (75 mg/mL) and N-hydroxysuccinimide (NHS) (11.5 mg/mL). Protein attachment presumably occurs via coupling of the succinimidyl ester functionalized flow cells to the protein Lys side chains. Injections of 50 μL of 20 μg/mL protein solutions in 10 mM NaOAc (pH 5.0) buffer were used for these reactions. An injection of 50 μL of ethanolamine (1 M in H2O, pH 8.5) was added to block any remaining succinimidyl esters. Approximately 3600 RU of TβRI-ED, 1900 RU of TβRII-ED, 7300 RU of endoglin-ED were immobilized. In a separate flow cell, ethanolamine was coupled directly to the activated surface to evaluate non-specific interactions. In a separate experiment, three distinct flow cells were functionalized with either 1600 RU of β-glycan-ED, 3300 RU of BMPR-IA-ED, or 4300 RU of ActRII-ED. Dendrimer binding was tested at concentrations ranging from 2.9 nM–6 μM. Serial dilutions of dendrimers in HBS buffer were injected (KINJECT) over all four flow cells for 5 min and allowed to dissociate for 5 min at a flow rate of 10 μL/min. Signals from the negative control surface were subtracted from signals from protein-immobilized surfaces using BIAevaluation version 4.1 software. The surfaces were regenerated after each injection to remove the bound dendrimer. To optimize the regeneration conditions, solutions of high salt, high pH, or low pH were tested. The optimal regeneration condition, which sufficiently removes bound dendrimer without compromising activities of immobilized proteins, was determined to be a 30 s pulse of 100 mM HCl at a flow rate of 100 μL/min.
Results and discussion
Identification of peptide ligands for TβRI using phage display
The features of protein–protein interfaces render them challenging targets for ligand identification. In the case of TGF-β for example, the structure of the TGF-β:TβRI-ED:TβRII-ED ternary complex indicates that more than 2000 Å2 of solvent accessible area on each receptor is buried.24, 39 Given the large sizes of protein interaction interfaces, it is not surprising that phage panning experiments tend to yield successful ligands when libraries composed of peptide sequences longer than 10-residues are employed. We therefore screened a library of 1011 random, 12-residue peptides displayed on the N-terminus of the PIII protein of M13 phage against immobilized TβRI-ED. Because the goal of our study was to discover novel ligands for TβRI-ED, we recovered bound phage particles using acid elution rather than competitive ligand-based elution. After four rounds of panning, 24 clones were sequenced, and 7 unique phage-borne peptides were identified. We evaluated these clones using a phage enzyme-linked immunosorbent assay (ELISA).40 In this way, we could identify clones that exhibit specificity for the target over BSA. Two such clones emerged; they display the peptide sequence LTGKNFPMFHRN (clone 1) or MHRMPSFLPTTL (clone 2) (Supplementary Fig. S1). We further characterized these two peptides.
Evaluation of the affinity and specificity of the peptide ligands using ELISA
We assessed the affinity of phage clone 1 (Fig. 2A) and clone 2 (Fig. 2B) for immobilized TβRI-ED using a phage ELISA. Both clones bind TβRI-ED with high affinity (apparent Kd ~10-10 M) (Supplementary Table S1A). We also tested their ability to interact with TβRII-ED. Although this receptor does not share obvious sequence homology with TβRI-ED, the active clones also interact with the TβRII-ED. Indeed, the measured affinities were similar to those for the target TβRI-ED. We suspected that these phage clones bind avidly to the receptors because each phage particle displays an average of 3-5 copies of the peptide ligand.41 Thus, the Kd values measured represent their “apparent affinity”.42
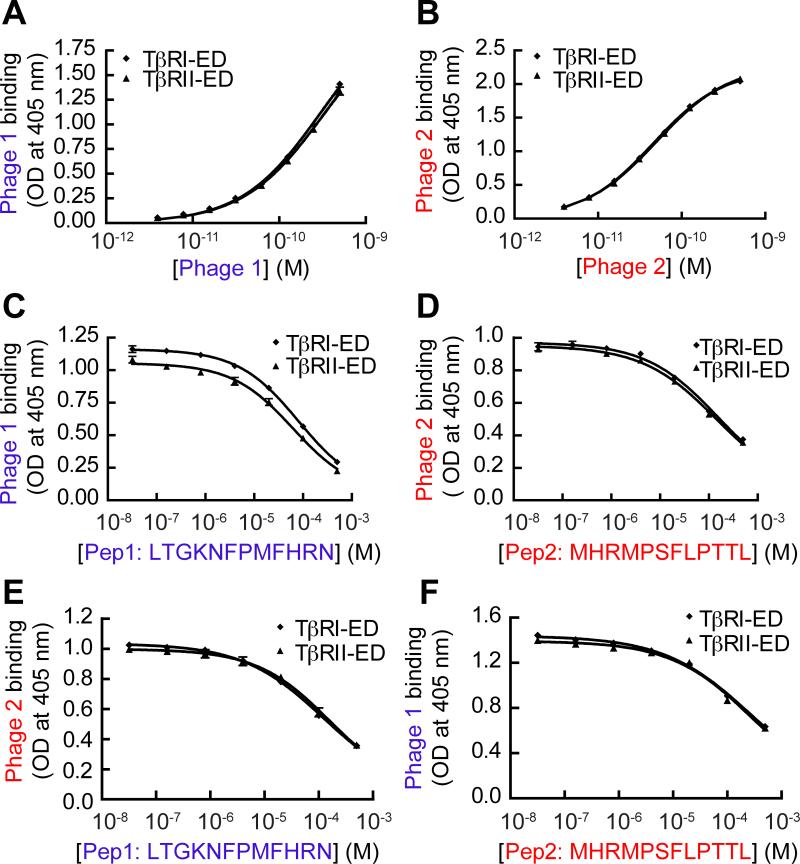
FIG. 2.
Phage display against TβRI yields peptides that bind TβRI-ED and TβRII-ED indistinguishably. The binding of (A) phage clone 1 and (B) phage clone 2 to immobilized TβRI-ED and TβRII-ED was assessed using a phage-based ELISA. (C) ELISA-based competition binding assay. Pep1 derived from phage clone 1 and (D) Pep2 derived from clone 2 were tested for inhibition of phage clone binding to immobilized receptors (550 pM of clone 1 and 39 pM of clone 2 were used). (E) An assay with Pep1competing with phage clone 2 (39 pM) for binding to either immobilized TβRI-ED or TβRII-ED. The IC50 value for Pep1 with phage clone 2 and TβRI-ED is 110 μM; the corresponding value for TβRII-ED is 156 μM. (F) An assay with Pep2 competing with phage clone 1 (550 pM) for binding to either immobilized TβRI-ED or TβRII-ED. The IC50 value for Pep2 inhibiting phage clone 1 binding to TβRI-ED is 256 μM; the corresponding value for TβRII-ED is 274 μM. Error bars represent the mean ± the standard deviation in (A) to (F).
To evaluate the affinity and specificity of the monovalent peptides, we synthesized peptide LTGKNFPMFHRN (Pep1) and MHRMPSFLPTTL (Pep2) as well as the corresponding N-terminal fluorophore-labeled counterparts. The use of these materials in direct binding assays, such as ELISA, SPR, or fluorescence polarization (FP), should require high micromolar to millimolar concentrations. These conditions, however, can result in aggregation, which would interfere with the mass transfer or fluorescence polarization output of SPR or FP assays. Moreover, low affinity ligands tend to have fast rates of dissociation; therefore, their binding is difficult to observe in assays that require washing steps, such as ELISAs. Because the phage clones are multivalent, they will bind avidly rendering their interactions readily monitored. To take advantage of the phage detection system and avoid the problems associated with directly measuring synthetic peptide binding, we carried out competition ELISAs.31 We reasoned that adding a peptide that interacts selectively with a given TβR should cause a decrease in phage binding. A key requirement for competition is that the observed signal cannot arise from phage aggregation or nonspecific binding to the plastic well, but rather from specific interactions with the immobilized receptors. To minimize non-specific phage binding, we tested the influence of two different detergents on binding. The nonionic detergent, Tween 20, is more effective at disrupting non-specific phage binding and aggregation. Utilization of 0.5% Tween 20 allowed for competition of synthetic peptides with the phage-borne peptides (Supplementary Fig. S2A-C).
After optimization of the assay conditions, we tested whether Pep1 or Pep2 could block phage clone binding to either TβRI-ED or TβRII-ED. Phage (at a constant concentration close to their apparent Kd value) were mixed with increasing concentrations of the corresponding synthetic peptide in TβR-coated wells. Both peptides exhibited dose-dependent competition with either phage clone (Fig. 2C, D). These data indicate that both Pep1 and Pep2 bind to TβRI and TβRII. We determined the IC50 values of the peptides by fitting the competition curves and then derived their Kd using the Cheng-Prusoff equation: Kd =IC50/(1+[phage]/ Kd phage)43 (Supplementary Table S1B). This analysis revealed that the monovalent ligands, Pep1 and Pep2 exhibit reasonable affinities (Kd ~10-5 M) for both TβRI and TβRII. These results demonstrate that competition ELISAs of this type can be used to determine the binding affinities of low-affinity phage-derived peptides. Importantly for our goals, phage panning against TβRI-ED yielded peptide ligands that bind to TβRI-ED and TβRII-ED with similar affinities.
Probing the peptide binding sites on TβRI and TβRII
There is no apparent sequence homology between Pep1 and Pep2, yet both bind to TβRI and TβRII. These results prompted us to ask whether they share binding sites on TβRI and TβRII or whether each occupies a unique site on each receptor. To this end, we carried out a cross competition assay. Interestingly, Pep2 inhibited not only phage clone 2 but also clone 1, and Pep1 similarly inhibited phage clone 2. Thus, each peptide occupies the same binding site on a given receptor (Fig. 2E, F).
That seemingly unrelated peptides can bind the same site on each TβR is a finding that has parallels in other systems. Specifically, phage panning experiments focused on the vascular endothelial growth factor (VEGF) yielded three classes of sequences.31 Although no sequence homology is apparent among these classes, they all compete with each other for receptor binding. Structures of two of these peptides in complex with VEGF have been determined by X-ray crystallography;44 one peptide binds VEGF using side chain contacts while the other acts through backbone interactions. These results emphasize two features of phage display screening. First, ligands can be found that use very different binding modes to occupy the same site, and second, these ligands tend to bind at protein–protein interaction sites.
To narrow the pool of potential peptide binding sites on TβRI-ED, we employed a truncated version of TβRI-ED (residues 7-91), which lacks structurally disordered segments on the N- and C-termini. The affinities for both clones for the truncated TβRI-ED were similar (Supplementary Fig. S3A-C), and analogous results were obtained with the synthetic peptides (Supplementary Fig. S3E-F). Given these affinities and observations indicating that TβRI-ED (residues 7-91) is more soluble than TβRI-ED (residues 1-101), we employed the former in all subsequent experiments.
Although TGF-β directly contacts both TβRI and TβRII in the oligomeric complex, the TGF-β binding site on each receptor is quite distinct.24, 39 Given that Pep1 and Pep2 interact with both receptors and compete with each other for binding, it seems unlikely they occupy the same regions as TGF-β. Consistent with this analysis is the observation that phage binding to TβRI and TβRII is unaffected by the addition of TGF-β (data not shown). We used surface plasmon resonance (SPR) to further explore peptide versus growth factor binding. Specifically, TβRI-ED and TβRII-ED were immobilized onto the sensor chip to test for competition of the peptides with TGF-β. It is known that TGF-β alone has weak affinity for the TβRI-ED.24 Consistent with our expectations, TGF-β1 did not bind detectably to the TβRI-ED surface (Fig. 3B). In contrast, the TGF-β interaction with the TβRII-ED surface was readily monitored. From the observed dose dependent SPR responses, an apparent Kd value of approximately 1.4 nM was determined (Fig. 3A). Previous SPR studies have found that TGF-β1 binds to the monomeric TβRII-ED with a Kd value of ~100 nM,45 while it binds to artificially dimerized TβRII-ED with Kd of ~5 pM.46 The intermediate dissociation constant for TGF-β with our TβRII-ED surface indicates that the surface presents the receptor as a mixture of monomeric and dimeric forms. Notably, when Pep1 was added as a potential competitor, no significant changes in TGF-β binding to TβRII-ED were observed (Fig. 3B). These results suggest that Pep1 and Pep2 share a previously unknown binding site on TβRII.
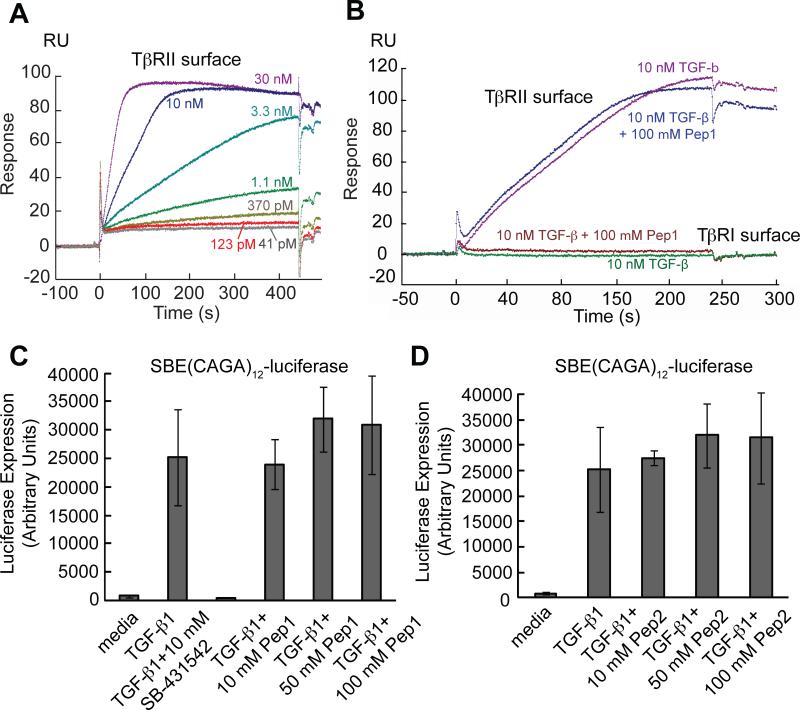
FIG. 3.
Pep1 and Pep2 do not compete with TGF-β in binding to either TβRI-ED or TβRII-ED. (A) Binding of TGF-β1 (41 pM to 30 nM) to TβRII-ED was tested using SPR. TβRI-ED and TβRII-ED were immobilized through their lysine residues. A protein-free flow cell was used as control. TGF-β binds to TβRII-ED with a saturating concentration of 10 nM. At the concentrations tested, TGF-β1 has no observable affinity to TβRI-ED (data not shown). (B) Pep1 does not compete with TGF-β in binding to TβRII-ED. (C) TGF-β1 initiated luciferase gene expression in an Mv1Lu reporter cell line stably transfected with a SBE(CAGA)12-luciferase reporter gene. TβRI kinase inhibitor SB-431542 inhibited TGF-β regulated luciferase gene expression. Pep1 and (D) Pep2 do not alter the cellular response to TGF-β1. In this competition assay, 10 pM TGF-β1 was used.
As stated earlier, a direct binding assay cannot be used to ascertain whether the peptide ligands compete with TGF-β for binding to TβRI-ED. We therefore employed a cell-based functional assay. If the peptide ligands occupy the TGF-β binding site on either receptor, TGF-β1-regulated gene expression should be affected. This possibility was evaluated using a mink lung epithelial cell line (Mv1Lu) stably transfected with a TGF-β responsive reporter gene. The gene construct consists of twelve repeats of a Smad binding element (SBE) with a sequence of CAGA (SBE(CAGA)12) immediately upstream of the luciferase sequence.36 When the transfected cells are treated with TGF-β1, Smad3 translocates into the nucleus and binds to the SBE(CAGA)12 sequence thereby promoting the expression of a gene encoding luciferase. The production of luciferase is readily quantified. TGF-β1 regulated luciferase gene expression with an EC50 value of approximately 10 pM (Supplementary Fig. 4A). As expected, the addition of 10 μM TβRI kinase inhibitor SB-431542 completely blocked the TGF-β-induced luciferase gene expression (Fig. 3C), demonstrating TGF-β indeed functions through TβRI. To test whether Pep1 and Pep2 compete with TGF-β1, a titration with each peptide ligand was conducted with 10 pM of TGF-β1. Neither Pep1 (Fig. 3C) nor Pep2 (Fig. 3D) affects TGF-β1-regulated luciferase gene expression. Additionally, the peptides alone had no effect on the baseline luciferase gene expression (Supplementary Fig. 4B).47 Together, our results demonstrate that Pep1 and Pep2 occupy the same binding sites on both TβRI-ED and TβRII-ED, and these sites are distinct from those used by the natural growth factor.
These findings indicate that our phage panning experiment has identified hot spots for ligand interactions within TβRI and TβRII. Previous studies using phage display have suggested that natural protein-binding sites have intrinsic properties that predispose them to ligand binding.27, 28, 48, 49 It is therefore likely that the sites identified by our phage-derived peptide ligands are used by endogenous proteins.
Dendrimers as platforms to display multiple copies of Pep1
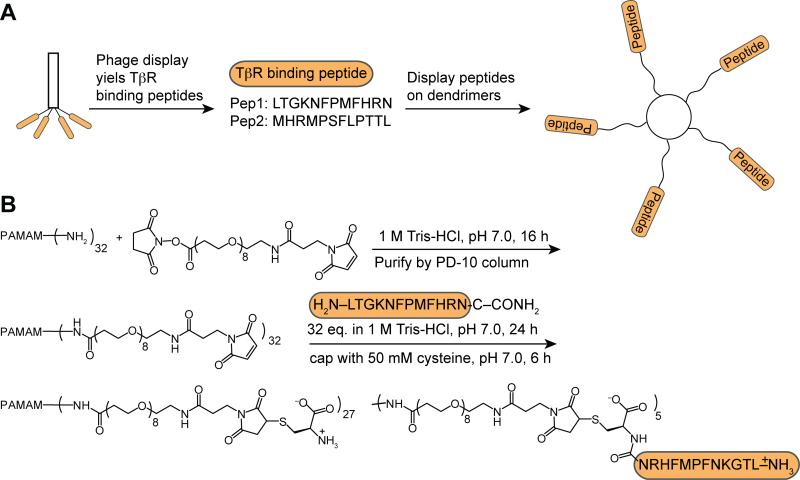
The observed difference (105-fold) in binding affinities between monovalent synthetic peptides and that of the phage particles (bearing 3-5 copies of the peptides) suggests that multivalent ligands for TβRI and TβRII will be more potent. Multivalent binding is an intrinsic feature of TGF-β receptor signaling, as the active complex involves 2 copies of each receptor and both TβRI and TβRII form dimers or oligomers on the plasma membrane of the cell surface.18 Thus, we postulated that the functional affinities of the peptides could be increased by multivalent display50, 51 and that multivalent ligands would serve as valuable probes. To this end, we employed generation 3 PAMAM dendrimer38 as a scaffold for peptide attachment (Fig. 4A). This framework was chosen because the dendrimer is extremely water-soluble and possesses many (a maximum of 32) primary amino groups as potential peptide conjugation sites. Its high molecular weight also is valuable because its binding can be detected readily by using SPR.52, 53 The strategy for functionalizing the dendrimer involved mimicking the presentation of the peptide on phage, which is displayed as a fusion to the N-terminus of the PIII coat protein. Accordingly, we appended the C-terminus of Pep1 to the dendrimer. Conjugation was mediated through a PEG8 crosslinker which contains a succinimidyl ester at one end and a maleimide at the other. While the linker contains two electrophilic groups, the dendrimer amino groups react preferentially with the succinimidyl ester moieties. Pep1 was subsequently coupled to the maleimide-displaying dendrimer through conjugate addition of the C-terminal cysteine residue (Fig. 4B). The cysteine thiolate is an excellent nucleophile that can undergo selective and rapid conjugation to the maleimide.54 The resulting dendrimer has a molecular weight of ~35 kDa based on the SDS-PAGE and MALDI mass spectrometry analysis, indicating that it bears approximately 5 peptide moieties.
FIG. 4.
Multivalent display of Pep1 on G3 dendrimer. (A) Peptides identified from phage display can be displayed on multivalent scaffolds to afford ligands with increased avidity. (B) Synthetic scheme for conjugating Pep1 to G3 PAMAM dendrimer.
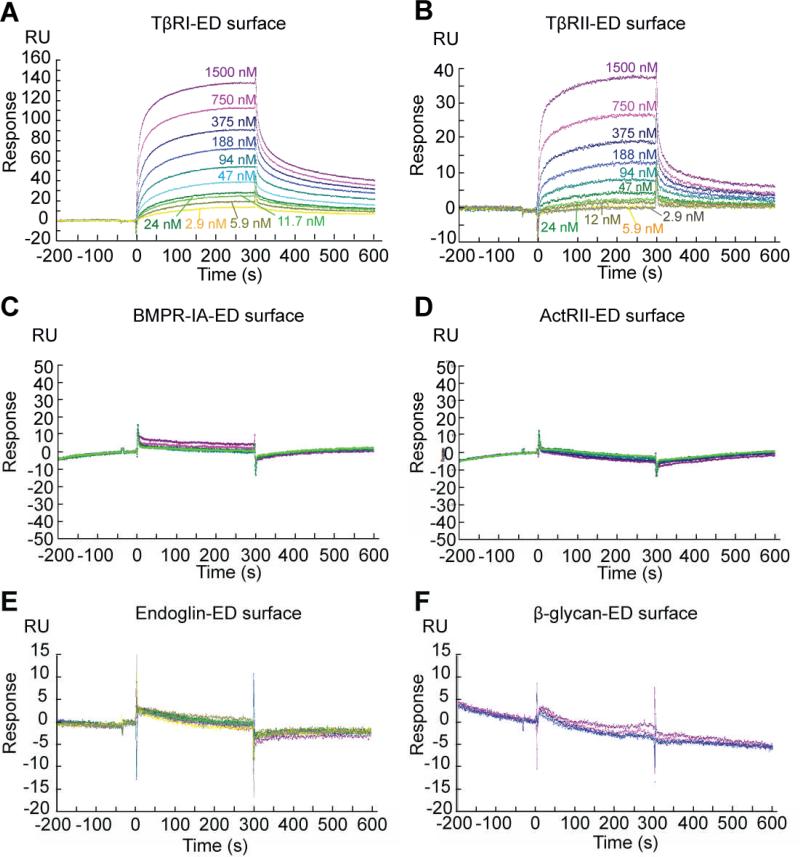
The avidity of Pep1-presenting dendrimers for the TβRI and TβRII was evaluated using SPR. The dendrimer was injected over TβRI-ED and TβRII-ED-functionalized flow cells, as well as an ethanolamine-functionalized control. Binding of the dendrimer was detected when it was used at nanomolar concentrations (Kd ~10-7 M), indicating that it is an excellent ligand (Fig. 5A, B). This dendrimer binds to TβRI-ED and TβRII-ED with similar affinities, consistent with phage ELISA results. This observation is intriguing. Indeed, although type I and type II receptors in the TGF-β superfamily are distinct by sequence comparison, they are structurally related. Specifically, they have a common pattern of four disulfide bonds, stabilizing a structure feature named the “three-finger toxin fold”24, 55 (Fig. 1B). This structural feature also is shared with other TGF-β superfamily members, including the bone morphogenic protein receptor IA (BMPR-IA) and the activin receptor II (ActRII).25 These observations raised the possibility that our peptide ligands recognize other receptors in the TGF-β superfamily.
FIG. 5.
(A) Binding affinities of the dendrimer to TβRI-ED, (B) TβRII-ED, (C) BMPR-IA-ED, (D) ActRII-ED, (E) endoglin-ED and (F) β-glycan-ED were assessed by SPR. All proteins were immobilized through their lysine residues. A protein-free flow cell was used as control. The dendrimer binds to TβRI-ED and TβRII-ED, but not to Endoglin-ED at concentrations ranging from 2.93 nM to 1.5 μM. In a separate experiment, the dendrimer did not interact with BMPR-IA-ED, ActRII-ED or β-glycan-ED at concentrations ranging from 47 nM to 6 μM.
To test whether binding of the dendrimer is specific for TβRI-ED and TβRII-ED, we assessed its affinity for two of the aforementioned TGF-β family members: BMPR-IA and ActRII. The extracellular domain of BMPR-IA (BMPR-IA-ED) and ActRII (ActRII-ED) were immobilized on the SPR sensor chip. The activity of these immobilized receptors was verified by their ability to bind BMP-4, a known ligand56 (Supplementary Fig. 5). Interestingly, even at high dendrimer concentrations (6 μM), no interaction of the dendrimer with BMPR-IA-ED nor ActRII-ED could be detected (Fig. 5C, D). These results demonstrate that Pep1 interact specifically with TβRI-ED and TβRII-ED but not with the closely related BMPR-IA or ActRII.
In addition to receptors closely related to TβRI and II, we also tested if our dendrimer binds to β-glycan and endoglin. These proteins were chosen as controls for two reasons. First, although they are coreceptors for TGF-β signaling, β-glycan and endoglin are not related to TβRI and TβRII; therefore, a peptide ligand for the receptors should not show any affinity to these receptor ligands. Secondly, β-glycan and endoglin are members of a large class of proteoglycans that are modified with heparan sulfate- or condroitin sulfate-containing glycosaminoglycans (GAGs). Anionic GAGs are abundant at the plasma membrane of eukaryotic cells and in the extracellular matrix; a specific ligand for TβRI and TβRII should not interact with these species. The extracellular domain of endoglin (endoglin -ED) and β-glycan-ED were immobilized on the SPR sensor chip. As expected, no interaction of the dendrimer with endoglin-ED or β-glycan-ED could be detected at any concentration tested (Fig. 5E, F). These results indicate that Pep1 binds to TβRI-ED and TβRII-ED specifically. Thus, the multivalent display of Pep1 can increase its functional affinity by approximately 100-fold while retaining high specificity. This finding is consistent with our previous observations indicating that a small change in ligand affinity for different targets can be amplified when a ligand is displayed multivalently.57 As a result, multivalent ligands can show enhanced functional affinity and specificity.57
We anticipate that dendrimer display can serve as a general strategy to facilitate the characterization of low affinity peptide ligands. Peptide hits from a first generation phage library screening can have relatively weak affinities (e.g. 10-4 M), which complicates characterizing their relative affinities and specificities. Indeed, peptide characterization is often the rate-limiting step in ligand optimization. False positives, as well as false negatives can arise that undermine the design of effective second generation libraries. Dendrimer-displaying peptides can overcome this limitation because their increased affinity and molecular weight render them useful probes in SPR assays. The peptide-substituted dendrimers provide other attractive features such as their size and the opportunities they present for introducing multifunctionality. For example, steric effects from dendrimer binding might result in an increase in its potency.50 In addition, because a dendrimer molecule can display many sites for functionalization, a label such as a fluorophore or a nanoparticle can also be appended.53 Such a label could facilitate the characterization of the peptide ligands, as well as their target. For example, such a conjugate could be used to visualize58 or manipulate51 the targeted protein on a cell surface. We note that dendrimeric probes like the ones we describe that do not directly compete with the growth factor ligand might be especially useful for probing signaling and endocytosis.
Conclusions
In summary, we have used phage display to uncover peptide ligands for the TβR-EDs. Although our screen focused on the TβRI-ED, the peptides we found also bind to TβRII-ED with similar affinities. To facilitate the characterization of the peptide ligands, we displayed Pep1 on a dendrimer scaffold to afford a ligand with excellent functional affinity. The resulting dendrimer interacts with TβRI-ED and TβRII-ED, but not with related receptors. This finding suggests that there are intrinsic ligand-binding hot spots on TβRI-ED and TβRII-ED uncovered by phage panning. These sites are distinct from those occupied upon TGF-β binding, suggesting that the peptide ligands target novel binding sites. Based on the hot spot theory in protein–protein interactions,27, 48 it is likely that these newly identified binding sites are exploited by endogenous proteins. Specifically, they may be used by coreceptors that enhance or modulate TGF-β signaling. Given the importance of cell-surface receptor oligomerization in TGF-β signaling, the identification of peptides that bind to both TβRI and TβRII suggest that multivalent ligands might be used to control TGF-β signaling.59
Supplementary Material
Acknowledgements
This research was supported by the University of Wisconsin, Materials Research Science and Engineering Center (DMR0520527), NIAID (AI055258), NIH (GM58670) and the Robert A. Welch Foundation (AQ1431). We thank Dr. Eric S. Underbakke, Adam H. Courtney and Dr. F. Michael Hoffmann for helpful discussions on phage display and TGF-β signaling. We thank Dr. Gary L. Case for help with automated peptide synthesis and Dr. Matthew R. Levengood for help with MALDI analysis. SPR data were obtained at the University of Wisconsin-Madison Biophysics Instrumentation Facility (BIF). We thank Dr. Darrell R. McCaslin for helpful conversations on SPR experiments.
Footnotes
† Electronic Supplementary Information (ESI) available: five suplementary figures and one supplementary table are included. See DOI: 10.1039/b000000x/
Notes and references
- 1.Hinck AP, Archer SJ, Qian SW, Roberts AB, Sporn MB, Weatherbee JA, Tsang MLS, Lucas R, Zhang BL, Wenker J, Torchia DA. Biochemistry. 1996;35:8517–8534. doi: 10.1021/bi9604946. [DOI] [PubMed] [Google Scholar]; Mittl PRE, Priestle JP, Cox DA, McMaster G, Cerletti N, Grutter MG. Protein Sci. 1996;5:1261–1271. doi: 10.1002/pro.5560050705. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shi YG, Massague J. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 2.Hart PJ, Deep S, Taylor AB, Shu ZY, Hinck CS, Hinck AP. Nat. Struct. Biol. 2002;9:203–208. doi: 10.1038/nsb766. [DOI] [PubMed] [Google Scholar]
- 3.Massague J. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 4.Deep S, Walker KP, Shu ZY, Hinck AP. Biochemistry. 2003;42:10126–10139. doi: 10.1021/bi034366a. [DOI] [PubMed] [Google Scholar]; Boesen CC, Radaev S, Motyka SA, Patamawenu A, Sun PD. Structure. 2002;10:913–919. doi: 10.1016/s0969-2126(02)00780-3. [DOI] [PubMed] [Google Scholar]; Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]; Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massague J. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 5.De Crescenzo G, Hinck CS, Shu ZY, Zuniga J, Yang JH, Tang YP, Baardsnes J, Mendoza V, Sun LZ, Lopez-Casillas F, O'Connor-McCourt M, Hinck AP. J. Mol. Biol. 2006;355:47–62. doi: 10.1016/j.jmb.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Derynck R. Trends Cell Biol. 1999;9:274–279. doi: 10.1016/s0962-8924(99)01579-2. [DOI] [PubMed] [Google Scholar]; Derynck R, Zhang YE. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]; Derynck R, Zhang Y, Feng XH. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]; Feng XH, Derynck R. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]; Yingling JM, Datto MB, Wong C, Frederick JP, Liberati NT, Wang XF. Mol. Cell. Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]; Panopoulou E, Gillooly DJ, Wrana JL, Zerial M, Stenmark H, Murphy C, Fotsis T. J. Biol. Chem. 2002;277:18046–18052. doi: 10.1074/jbc.M107983200. [DOI] [PubMed] [Google Scholar]; Runyan CE, Schnaper HW, Poncelet AC. J. Biol. Chem. 2005;280:8300–8308. doi: 10.1074/jbc.M407939200. [DOI] [PubMed] [Google Scholar]
- 8.Massague J. Annu. Rev. Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 9.Waite KA, Eng C. Nat. Rev. Genet. 2003;4:763–773. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]; Pires-daSilva A, Sommer RJ. Nat. Rev. Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- 10.WojtowiczPraga S. J. Immunother. 1997;20:165–177. [Google Scholar]; Levings MK, Bacchetta R, Schulz U, Roncarolo MG. Int. Arch. Allergy Appl. Immunol. 2002;129:263–276. doi: 10.1159/000067596. [DOI] [PubMed] [Google Scholar]; Fontana A, Constam DB, Frei K, Malipiero U, Pfister HW. Int. Arch. Allergy Appl. Immunol. 1992;99:1–7. doi: 10.1159/000236328. [DOI] [PubMed] [Google Scholar]; Weiner HL. Immunol. Rev. 2001;182:207–214. doi: 10.1034/j.1600-065x.2001.1820117.x. [DOI] [PubMed] [Google Scholar]; Li MO, Wan YY, Sanjabi S, Robertson AKL, Flavell RA. Annu. Rev. Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 11.Okane S, Ferguson MWJ. Int. J. Biochem. Cell Biol. 1997;29:63–78. doi: 10.1016/s1357-2725(96)00120-3. [DOI] [PubMed] [Google Scholar]
- 12.Massague J, Blain SW, Lo RS. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]; Derynck R, Akhurst RJ, Balmain A. Nat. Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]; Bierie B, Moses HL. Nat. Rev. Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]; Massague J. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yingling JM, Blanchard KL, Sawyer JS. Nat. Rev. Drug Discovery. 2004;3:1011–1022. doi: 10.1038/nrd1580. [DOI] [PubMed] [Google Scholar]
- 14.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. Mol. Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]; Peng SB, Yan L, Xia XL, Watkins SA, Brooks HB, Beight D, Herron DK, Jones ML, Lampe JW, McMillen WT, Mort N, Sawyer JS, Yingling JM. Biochemistry. 2005;44:2293–2304. doi: 10.1021/bi048851x. [DOI] [PubMed] [Google Scholar]; Uhl M, Aulwurm S, Wischhusen J, Weiler M, Ma JY, Almirez R, Mangadu R, Liu YW, Platten M, Herrlinger U, Murphy A, Wong DH, Wick W, Higgins LS, Weller M. Cancer Res. 2004;64:7954–7961. doi: 10.1158/0008-5472.CAN-04-1013. [DOI] [PubMed] [Google Scholar]; Callahan JF, Burgess JL, Fornwald JA, Gaster LM, Harling JD, Harrington FP, Heer J, Kwon C, Lehr R, Mathur A, Olson BA, Weinstock J, Laping NJ. J. Med. Chem. 2002;45:999–1001. doi: 10.1021/jm010493y. [DOI] [PubMed] [Google Scholar]; Li HY, Wang Y, Yan L, Campbell RM, Anderson BD, Wagner JR, Yingling JM. Biorg. Med. Chem. Lett. 2004;14:3585–3588. doi: 10.1016/j.bmcl.2004.04.065. [DOI] [PubMed] [Google Scholar]; Mendel DB, Laird AD, Xin XH, Louie SG, Christensen JG, Li GM, Schreck RE, Abrams TJ, Ngai TJ, Lee LB, Murray LJ, Carver J, Chan E, Moss KG, Haznedar JO, Sukbuntherng J, Blake RA, Sun L, Tang C, Miller T, Shirazian S, McMahon G, Cherrington JM. Clin. Cancer Res. 2003;9:327–337. [PubMed] [Google Scholar]; Singh J, Chuaqui CE, Boriack-Sjodin PA, Lee WC, Pontz T, Corbley MJ, Cheung HK, Arduini RM, Mead JN, Newman MN, Papadatos JL, Bowes S, Josiah S, Ling LE. Bioorg. Med. Chem. Lett. 2004;14:2991–2991. doi: 10.1016/j.bmcl.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 15.Schlingensiepen KH, Schlingensiepen R, Steinbrecher A, Hau P, Bogdahn U, Fischer-Blass B, Jachimczak P. Cytokine Growth Factor Rev. 2006;17:129–139. doi: 10.1016/j.cytogfr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JE, Vaughan TJ, Williams AJ, Wilton J, Johnson KS, Bacon L, Green JA, Field R, Ruddock S, Martins M, Pope AR, Tempest PR, Jackson RH. J. Immunol. Methods. 1999;227:17–29. doi: 10.1016/s0022-1759(99)00060-5. [DOI] [PubMed] [Google Scholar]; Dasch JR, Pace DR, Waegell W, Inenaga D, Ellingsworth L. J. Immunol. 1989;142:1536–1541. [PubMed] [Google Scholar]
- 17.Dotor J, Lopez-Vazquez AB, Lasarte JJ, Sarobe P, Garcia-Granero M, Riezu-Boj JI, Martinez A, Feijoo E, Lopez-Sagaseta J, Hermida J, Prieto J, Borras-Cuesta F. Cytokine. 2007;39:106–115. doi: 10.1016/j.cyto.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Chen RH, Derynck R. J. Biol. Chem. 1994;269:22868–22874. [PubMed] [Google Scholar]; Luo KX, Lodish HF. EMBO J. 1997;16:1970–1981. doi: 10.1093/emboj/16.8.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gilboa L, Wells RG, Lodish HF, Henis YI. J. Cell Biol. 1998;140:767–777. doi: 10.1083/jcb.140.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radaev S, Zou ZC, Huang T, Lafer EM, Hinck AP, Sun PD. J. Biol. Chem. 2010;285:14806–14814. doi: 10.1074/jbc.M109.079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han S, Loulakis P, Griffor M, Xie Z. Protein Sci. 2007;16:2272–2277. doi: 10.1110/ps.073068407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huse M, Muir TW, Xu L, Chen YG, Kuriyan J, Massague J. Mol. Cell. 2001;8:671–682. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- 22.Wu G, Chen YG, Ozdamar B, Gyuricza CA, Chong PA, Wrana JL, Massague J, Shi YG. Science. 2000;287:92–97. doi: 10.1126/science.287.5450.92. [DOI] [PubMed] [Google Scholar]
- 23.Chacko BM, Qin BY, Tiwari A, Shi GB, Lam S, Hayward LJ, de Caestecker M, Lin K. Mol. Cell. 2004;15:813–823. doi: 10.1016/j.molcel.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP. Mol. Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 25.Allendorph GP, Vale WW, Choe S. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7643–7648. doi: 10.1073/pnas.0602558103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerrero-Esteo M, Sanchez-Elsner T, Letamendia A, Bernabeu C. J. Biol. Chem. 2002;277:29197–29209. doi: 10.1074/jbc.M111991200. [DOI] [PubMed] [Google Scholar]
- 27.Jones S, Thornton JM. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lo Conte L, Chothia C, Janin J. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]; Wells JA. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]; Moreira IS, Fernandes PA, Ramos MJ. Proteins Struct. Funct. Bioinf. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 28.Kay BK, Kurakin AV, Hyde-DeRuyscher R. Drug Discov. Today. 1998;3:370–378. [Google Scholar]; Sidhu SS, Fairbrother WJ, Deshayes K. ChemBioChem. 2003;4:14–25. doi: 10.1002/cbic.200390008. [DOI] [PubMed] [Google Scholar]
- 29.Sidhu SS, Lowman HB, Cunningham BC, Wells JA. Applications of Chimeric Genes and Hybrid Proteins, Pt C. Academic Press Inc; San Diego: 2000. pp. 333–363. [Google Scholar]
- 30.Smith GP, Petrenko VA. Chem. Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]; Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Annu. Rev. Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 31.Fairbrother WJ, Christinger HW, Cochran AG, Fuh C, Keenan CJ, Quan C, Shriver SK, Tom JYK, Wells JA, Cunningham BC. Biochemistry. 1998;37:17754–17764. doi: 10.1021/bi981931e. [DOI] [PubMed] [Google Scholar]
- 32.Orner BP, Liu L, Murphy RM, Kiessling LL. J. Am. Chem. Soc. 2006;128:11882–11889. doi: 10.1021/ja0619861. [DOI] [PubMed] [Google Scholar]
- 33.Wrighton NC, Farrell FX, Chang R, Kashyap AK, Barbone FP, Mulcahy LS, Johnson DL, Barrett RW, Jolliffe LK, Dower WJ. Science. 1996;273:458–463. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]; Ballinger MD, Shyamala V, Forrest LD, Deuter-Reinhard M, Doyle LV, Wang JX, Panganiban-Lustan L, Stratton JR, Apell G, Winter JA, Doyle MV, Rosenberg S, Kavanaugh WM. Nat. Biotechnol. 1999;17:1199–1204. doi: 10.1038/70746. [DOI] [PubMed] [Google Scholar]; Sato A, Sone S. Biochem. J. 2003;371:603–608. doi: 10.1042/BJ20020993. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lowman HB, Chen YM, Skelton NJ, Mortensen DL, Tomlinson EE, Sadick MD, Robinson I, Clark RG. Biochemistry. 1998;37:8870–8878. doi: 10.1021/bi980426e. [DOI] [PubMed] [Google Scholar]
- 34.Brown KC. Curr. Opin. Chem. Biol. 2000;4:16–21. doi: 10.1016/s1367-5931(99)00045-9. [DOI] [PubMed] [Google Scholar]; Giordano RJ, Cardo-Vila M, Lahdenranta J, Pasqualini R, Arap W. Nat. Med. 2001;7:1249–1253. doi: 10.1038/nm1101-1249. [DOI] [PubMed] [Google Scholar]; Derda R, Musah S, Orner BP, Klim JR, Li LY, Kiessling LL. J. Am. Chem. Soc. 2010;132:1289–1295. doi: 10.1021/ja906089g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP. Mol Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 36.Zawel L, Dai JL, Buckhaults P, Zhou SB, Kinzler KW, Vogelstein B, Kern SE. Mol. Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 37.Zhao BM, Hoffmann FM. Mol. Biol. Cell. 2006;17:3819–3831. doi: 10.1091/mbc.E05-10-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esfand R, Tomalia DA. Drug Discov. Today. 2001;6:427–436. doi: 10.1016/s1359-6446(01)01757-3. [DOI] [PubMed] [Google Scholar]
- 39.Radaev S, Zou Z, Huang T, Lafer EM, Hinck AP, Sun PD. J. Biol. Chem. 2010;285:14806–14814. doi: 10.1074/jbc.M109.079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham BC, Lowe DG, Li B, Bennett BD, Wells JA. EMBO J. 1994;13:2508–2515. doi: 10.1002/j.1460-2075.1994.tb06540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowman HB, Bass SH, Simpson N, Wells JA. Biochemistry. 1991;30:10832–10838. doi: 10.1021/bi00109a004. [DOI] [PubMed] [Google Scholar]
- 42.Kiessling LL, Gestwicki JE, Strong LE. Curr. Opin. Chem. Biol. 2000;4:696–703. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 43.Cheng Y, Prusoff WH. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]; Cheng HC. J. Pharmacol. Toxicol. Methods. 2001;46:61–71. doi: 10.1016/s1056-8719(02)00166-1. [DOI] [PubMed] [Google Scholar]; Cheng HC. Pharmacol. Res. 2004;50:21–40. doi: 10.1016/j.phrs.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Wiesmann C, Christinger HW, Cochran AG, Cunningham BC, Fairbrother WJ, Keenan CJ, Meng G, de Vos AM. Biochemistry. 1998;37:17765–17772. doi: 10.1021/bi9819327. [DOI] [PubMed] [Google Scholar]; Pan B, Li B, Russell SJ, Tom JYK, Cochran AG, Fairbrother WJ. J. Mol. Biol. 2002;316:769–787. doi: 10.1006/jmbi.2001.5370. [DOI] [PubMed] [Google Scholar]
- 45.De Crescenzo G, Grothe S, Zwaagstra J, Tsang M, O'Connor-McCourt MD. J. Biol. Chem. 2001;276:29632–29643. doi: 10.1074/jbc.M009765200. [DOI] [PubMed] [Google Scholar]
- 46.De Crescenzo G, Pham PL, Durocher Y, O'Connor-McCourt MD. J. Mol. Biol. 2003;328:1173–1183. doi: 10.1016/s0022-2836(03)00360-7. [DOI] [PubMed] [Google Scholar]
- 47.Fan F, Binkowski BF, Butler BL, Stecha PF, Lewils MK, Wood KV. ACS Chem. Biol. 2008;3:346–351. doi: 10.1021/cb8000414. [DOI] [PubMed] [Google Scholar]
- 48.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 49.Pillutla RC, Hsiao KC, Beasley JR, Brandt J, Ostergaard S, Hansen PH, Spetzler JC, Danielsen GM, Andersen AS, Brissette RE, Lennick M, Fletcher PW, Blume AJ, Schaffer L, Goldstein NI. J. Biol. Chem. 2002;277:22590–22594. doi: 10.1074/jbc.M202119200. [DOI] [PubMed] [Google Scholar]; Cwirla SE, Balasubramanian P, Duffin DJ, Wagstrom CR, Gates CM, Singer SC, Davis AM, Tansik RL, Mattheakis LC, Boytos CM, Schatz PJ, Baccanari DP, Wrighton NC, Barrett RW, Dower WJ. Science. 1997;276:1696–1699. doi: 10.1126/science.276.5319.1696. [DOI] [PubMed] [Google Scholar]
- 50.Mammen M, Choi SK, Whitesides GM. Angew. Chem. Int. Ed. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 51.Kiessling LL, Gestwicki JE, Strong LE. Angew. Chem. Int. Ed. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munoz EM, Correa J, Fernandez-Megia E, Riguera R. J. Am. Chem. Soc. 2009;131:17765–17767. doi: 10.1021/ja9074826. [DOI] [PubMed] [Google Scholar]
- 53.Helms BA, Reulen SWA, Nijhuis S, de Graaf-Heuvelmans P, Merkx M, Meijer EW. J. Am. Chem. Soc. 2009;131:11683–+. doi: 10.1021/ja902285m. [DOI] [PubMed] [Google Scholar]
- 54.Underbakke ES, Zhu YM, Kiessling LL. Angew. Chem. Int. Ed. 2008;47:9677–9680. doi: 10.1002/anie.200803378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenwald J, Fischer WH, Vale WW, Choe S. Nat. Struct. Biol. 1999;6:18–22. doi: 10.1038/4887. [DOI] [PubMed] [Google Scholar]
- 56.Chen D, Zhao M, Mundy GR. Growth Factors. 2004;22:233–80241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]; Lavery K, Swain P, Falb D, Alaoui-Ismaili MH. J. Biol. Chem. 2008;283:20948–20958. doi: 10.1074/jbc.M800850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mortell KH, Weatherman RV, Kiessling LL. J. Am. Chem. Soc. 1996;118:2297–2298. [Google Scholar]
- 58.Shukla R, Thomas TP, Peters J, Kotlyar A, Myc A, Baker JR. Chem. Commun. 2005:5739–5741. doi: 10.1039/b507350b. [DOI] [PubMed] [Google Scholar]
- 59.Gestwicki JE, Kiessling LL. Nature. 2002;415:81–84. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]; Stockwell BR, Schreiber SL. Curr. Biol. 1998;8:761–770. doi: 10.1016/s0960-9822(98)70299-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.