Abstract
Objective
Endothelial Kruppel-like factor 2 (KLF2) mediates endothelium-dependent vascular homeostasis by differentially regulating endothelial genes, leading to an anti-inflammatory and anti-thrombotic endothelial surface with normal vasodilatory function. In contrast, the tumor suppressor p53 leads to inflammatory gene expression and impairs endothelium-dependent vasodilatation, thus promoting endothelial dysfunction. We asked whether p53 decreases KLF2 expression, and determined whether p53-mediated suppression of KLF2 plays a role in p53-induced endothelial dysfunction.
Methods and results
The effect of p53 on KLF2 expression was determined. P53 inhibited KLF2 transcription in a histone deacetylase-dependent and histone acetyltransferase-independent fashion. KLF2 expression was suppressed by p53 via a conserved p53-binding repressor sequence in its promoter. P53 bound to, and stimulated deacetylation of histone-3 at, the KLF2 promoter. The effect of p53 on endothelial KLF2 target genes was examined. Down-regulation of p53 increased expression of endothelial nitric oxide synthase (eNOS) and thrombomodulin(TM), while inhibiting expression of plasminogen activator inhibitor 1 (PAI-1). Conversely, overexpression of p53 suppressed eNOS and TM expression, and stimulated PAI-1 and endothelin-1 (ET-1) expression. Knockdown of KLF2 abolished p53-induced decrease in TM, and increase in ET-1 expression. Both, overexpression of p53 and knockdown of KLF2 in endothelial cells, increased blood coagulation on an endothelial cell monolayer. P53-induced increase in coagulation was rescued by forced expression of KLF2. P53 also impaired endothelium-dependent vasodilatation and decreased bioavailable vascular nitric oxide, both of which were rescued by forced KLF2 expression.
Conclusions
These findings illustrate a novel p53-dependent mechanism for the regulation of endothelial KLF2 expression. In addition, they show that down-regulation of KLF2, in part, mediates a p53-stimulated dysfunctional endothelium.
Keywords: KLF2, Endothelial Dysfunction, p53, Thrombosis
Introduction
The Kruppel-like factor (KLF)/Sp is a subclass of the zinc-finger family of DNA-binding transcriptional factors. To date, there are 17 KLF factors (KLF1–17) and 4 Sp factors (S1–4). These family members are characterized by DNA binding domains containing the conserved sequence CX2CX3FX5LX2HX3H (X is any amino acid; underlined cysteine and histidine residues coordinate zinc binding) 1. Members of this family can bind to the consensus DNA sequence CACCC. One member of this family, Lung Kruppel-like factor (LKLF)/KLF2 is highly expressed in vascular endothelium, and serves as a molecular switch to regulate a range of endothelial functions. KLF2 expression in the endothelium is upregulated by laminar shear stress and inhibited by pro-inflammatory cytokines 2, 3. Forced expression of KLF2 in endothelial cells results in induction of endothelial specific nitric oxide (eNOS) and thrombomodulin (TM) expression while abrogating cytokine mediated expression of vascular cell adhesion molecule (VCAM-1) and tissue factor (TF) 4, 5. In addition, KLF2 inhibits endothelial cell migration and angiogenesis and this is partly attributed to its ability to inhibit VEGF receptor VEGFR2/ KDR expression 5. KLF2 also protects endothelial cells from oxidative stress mediated injury and apoptosis 3, consistent with the capacity of endothelial KLF2 to regulate cellular levels of reactive oxygen species 6.
The p53 tumor suppressor gene is a transcription factor which is induced and activated in response to various cellular stresses. It plays a crucial role in coordinating cell cycle arrest and apoptosis in response to such stresses 7. Whether vascular p53 promotes or retards the atherosclerotic process remains controversial. P53 expression has been reported in vascular smooth muscle cells (VSMCs), endothelial cells, and macrophages of human atherosclerotic lesions, but is undetectable in normal vascular specimens 8. On the one hand, super-p53 transgenic mice carrying an extra allele of p53 have decreased neointimal thickening induced by mechanical injury, but are not protected from diet-induced atherosclerosis 9. Further supporting a role for p53 in retarding atherosclerosis are observations that p53 deletion in apoE −/− mice leads to increased aortic atherosclerosis in response to high fat diet 10. This has been attributed to increased rate of cell proliferation and reduced apoptosis in multiple cell types in the plaques of p53 deficient mice. On the other hand, p53 in the endothelium promotes oxidative stress and impairs endothelium-dependent vasorelaxation, suggesting that in this capacity it may promote atheroma formation 11. Finally, although the role of p53 in vascular inflammation has not been examined, p53 does stimulate inflammatory signaling and inflammatory gene expression in several cell types 12, 13.
Transcriptional regulation of KLF2 expression is complex. The majority of studies have examined a 140 bp of the proximal promoter of KLF2 14, 15, a region which is highly conserved between mouse and human. Within this region, a binding site for the Myocyte Enhancing Factor 2 (MEF2) transcription factor plays a pivotal role in regulation of KLF2 expression in endothelial cells induced by several stimuli 2, 3, 16, 17. In addition to MEF2, additional mechanisms have been implicated in the flow-mediated induction of KLF2. For example, P300/CBP-associated factor (PCAF), heterogeneous nuclear ribonucleoprotein D, and nucleolin have all been identified as factors that bind to the proximal KLF2 promoter as a part of shear stress regulatory complex 14, 15.
Recognizing that endothelial p53 impairs vasorelaxation and KLF2 has salutary effects on endothelial function, we hypothesized that p53 may exert some of its deleterious effects on endothelium-dependent vascular function by suppressing endothelial expression of KLF2. Using overexpression and knockdown approaches we provide evidence that p53 transcriptionally down-regulates KLF2 expression, and demonstrate that this down-regulation is responsible for some of the phenotypic changes induced by p53 in the endothelium.
Methods
Cell culture and reagents
Human umbilical vein endothelial cells (HUVEC) were obtained from Cambrex Company (Baltimore, MD). 293 cells were obtained from ATCC. HCT116 cells were kind gift Dr Bert Vogelstein. Antibodies recognizing p53 (DO-1), beta-actin, Thrombomodulin, PAI-1 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); anti-p53 (Pab- 421) from Calbiochem, USA; anti-acetylated p53 and anti-acetylated Histone H3 from Millipore, USA; anti-KLF2 was kind gift of Dr Huck Hui Ng, (Genome Institute of Singapore, Singapore). Ad-LacZ, Ad-p53 and Ad-KLF2 adenoviruses have been described previously 18 4; Methyl methanesulfonate (MMS) from Sigma-Aldrich; TNFα from R & D System, Minneapolis, MN.
KLF2 promoter firefly luciferase reporter constructs have been described previously 2. Additional 117 and 88 bp fragments of KLF2 proximal promoters were generated by PCR and subsequently cloned in pGL3 firefly luciferase reporter (Promega). Point mutations in the MEF2 site described previously 2 were created in full length KLF2 promoter using site directed mutagenesis kit from Stratagene, USA. The 27 bp putative p53-response element was deleted and replaced by the restriction site sequence TTACGCGTGCTAGCCCGGGCTCGAG in full-length KLF2 promoter by restriction digestion. All deletions and mutations were confirmed by sequencing. Expression plasmid for p53 in pCMV vector backbone was gift from Dr P. Hwang (NIH, Bethesda). Mutant p53 plasmids (p53-H175 and p53H175A392) were from Dr Xin Lu (Ludwick Institute for Cancer Research, UK). SR-IκBα plasmid was gift from Dr Alex Baldwin (The University of North Carolina at Chapel Hill, North Carolina, USA).
Transient transfection reporter assays
Cells were plated at a density of 5×104/well in 24 well plates one day before transfection. Transient transfection was performed using Lipofectamine2000 (InVitrogen, CA USA). A total 1–2 µg of plasmid DNA was used in transfections and total DNA was kept constant. Co-transfected Renilla luciferase was used as an internal control. Cells were harvested 24 h after transfection, and firefly and renilla luciferase activity were measured using the Dual Luciferase reporter kit (Promega), and firefly values were normalized to renilla values in each sample. All transfections were performed in triplicate and results are representative of at least three independent experiments.
Immunoblotting
Protein lysates were are boiled in SDS-PAGE gel loading buffer, subjected to SDS-PAGE, transferred to nitrocellulose filter, and probed with the specified primary antibody and the appropriate peroxidase-conjugated secondary antibody (Santa Cruz Biotech). Chemiluminescent signal is developed using Super Signal West Femto substrate (Pierce), blots imaged with a Gel Doc 2000 Chemi Doc system (BioRad), and bands quantified using Quantity One software (BioRad).
Electrophoretic mobility shift assays (EMSA)
EMSA were performed as previously described 2. Approximately 5 µg of the nuclear extract were used in each incubation. A p53 binding site from the KLF2 promoter was included using the following radio-labeled oligonucleotide: 5’-CCAGGCTTATATACCGCGGCTAAATTTAGGCTGAGC CCGGA-3’. Anti-p53 (Pab-421) or unlabeled oligonucleotide was pre-incubated with nuclear extract for the super shift and competition, respectively.
siRNA transfection
Human p53 and KLF2 siRNAs and a non-specific control siRNA were purchased from InVitrogen, USA and were transfected into HUVECs using Lipofectamine2000 (InVitrogen, CA) according to the manufacturer’s protocol.
Real time RT-PCR
Total RNA from cultured cells is isolated by the TRIZOL (InVitrogen, CA USA) method. Real-time PCR is performed using the Prism 7000 Sequence Detection System (Applied Biosystems) with the SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (InVitrogen). The following primers are used:
-
Human KLF2: forward 5’-tgcggcaagacctacaccaagagt-3’, reverse 5’-agccgcagccgtcccagtt-3’
Human p53: forward 5’-ttgagcactacagacctcacg-3’, reverse 5’-tgcaccagtatttccaatcaa-3’
Human eNOS: forward 5’-ctcatg ggcacggtgatg-3’, reverse 5’-accacg tcatactcatccatacac-3’
Human TM: forward 5’-tgagcgttattggtcggcagcct-3’, reverse 5’-cacaggtagggtgactcagg-3’
Human PAI-I: forward 5’-gcatgacctaccaggacagaact-3’, reverse 5’-tccgagctgcctgtctctct-3’
Human GAPDH: forward 5’-ccacatcgctcagacaccat-3’, reverse 5’-ccaggcgcccaatacg-3’
Human ET-1: forward 5’-tggacatcatttgggtcaaca-3’, reverse 5’-tctcttggacctagggcttcc-3’
Chromatin Immunoprecipitation (ChIP) assays
ChIP assays were performed using a ChIP Assay Kit (Upstate) using indicated antibodies 2. Briefly, DNA associated with specific immunoprecipitates or with negative control mouse non-immune IgG were isolated and used as a template in a PCR reaction amplifying a 169 bp region of the KLF2 promoter with the putative p53-binding element, or the GAPDH gene. Primer sequences were: KLF2: forward 5’-gcagtccgggctcccgcagtag-3’, reverse 5’-cttataggcgcggcaggcac-3’; GAPDH forward 5’-atgacatcaagaaggtggtg-3’, reverse 5’-cataccaggaaatgagcttg-3’.
Ex Vivo Adenoviral Infection of Rat Aortic Rings
Aortic rings from 3–4-month old Wistar Kyoto rats were incubated with the specified adenovirus ex vivo for 24 hours as described previously 19 before measurement of vascular reactivity.
In vitro Clotting Assay
HUVEC were plated in 96-well dishes and infected with indicated adenovirus 48 h. Cells were rinsed twice with warm phosphate-buffered saline and 100 µl of 37 °C human plasma (Sigma-Aldrich) added to each well. Immediately thereafter, 100 µl of 25 mM CaCl2 was added, and plates were placed in a Vmax kinetic plate reader (Molecular Devices) and read at 405 nm every 20 s for 30 min 20. Fibrin clot formation is indicated when maximum absorbance is reached. Clotting times are reported at half-maximal absorbance.
Vascular Reactivity measurement
3–4-month old rats are anesthetized using ketamine-acepromazine (100 and 10 mg/kg ip, respectively). The animals were rapidly euthanized by cardiac excision. The thoracic aorta was carefully dissected, rapidly removed, and placed in ice-cold oxygenated Krebs-Ringer bicarbonate solution. The vessel was carefully cleared of loose connective tissue. In some experiments, the endothelium was removed by moving the arterial ring around two tungsten wires. It is then cut into 5–10 1.5 mm rings. A single ring from each rat was suspended between two wire stirrups (150 µm) in one of the 25-ml organ chambers of the four-chamber myograph system (DMT Instruments) in 5 ml Krebs-Ringer (95% O2-5% CO2, pH 7.4, 37°C). One stirrup was connected to a three-dimensional micromanipulator, and the other to a force transducer. The mechanical force signal was amplified, digitalized, and recorded (PowerLab 8/30). All concentration-effect curves were performed on arterial rings beginning at their optimum resting tone. This was determined by stretching arterial rings at 10 min intervals in increments of 500 mg to reach optimal tone (~3000 mg). One dosage of KCl (60mM) was administered to verify vascular smooth muscle viability. Cumulative dose-response curve for phenylephrine (PE) (10−9-10−5 M) was obtained by administering the drug in one-half log doses. Endothelium-dependent and –independent vasodilatation was determined by generating dose-response curves to acetylcholine (ACh 10−9-10−5 M) and sodium nitroprusside (SNP 10−9-10−5 M), respectively. Vasorelaxation evoked by ACh and SNP is expressed as percent relaxation determined by the percentage of inhibition to the pre-constricted tension. NO bioavailability was measured physiologically by determining the increase in contractile response to NOS inhibition (L-NAME10−4M) in rings pre-constricted with PE (10−6M). Vasorelaxation evoked by ACh and SNP is expressed as percent relaxation determined by the percentage of inhibition to the pre-constricted tension.
Results
P53 inhibits KLF2 expression
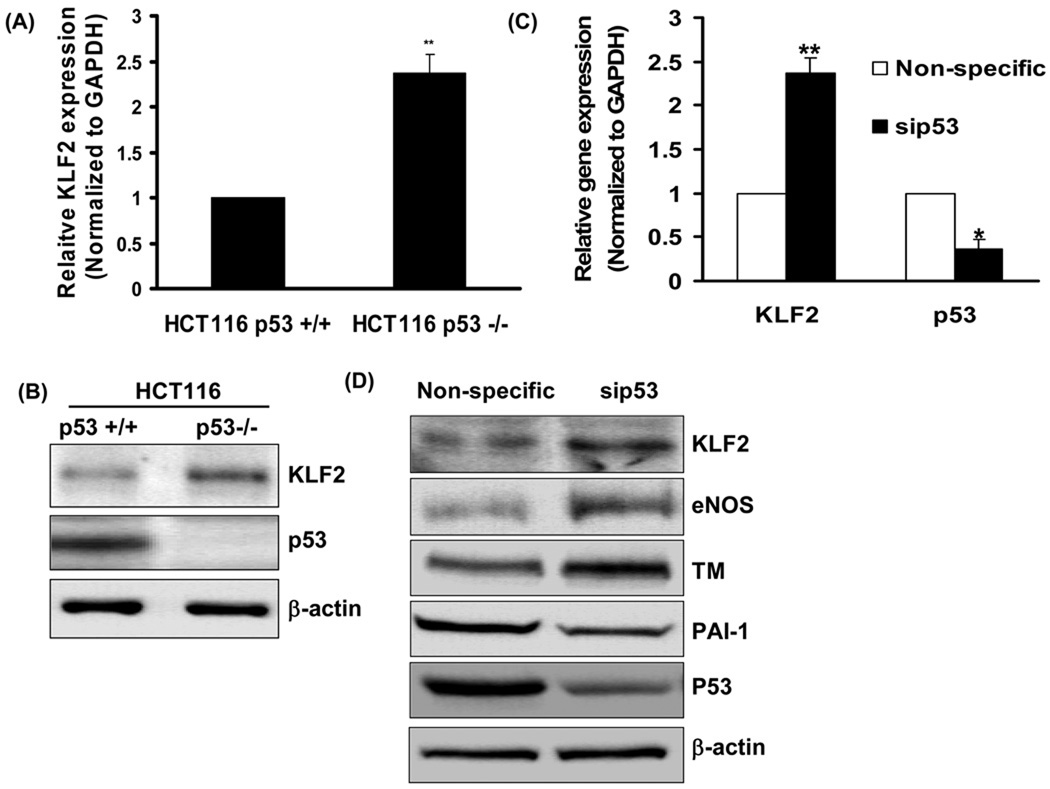
To determine whether p53 regulates KLF2, we first compared KLF2 expression in a HCT116 p53+/+ colon cancer cell line and its isogenic p53 −/− cell line. KLF2 expression, both at the mRNA (Fig 1A) and protein (Fig 1B) level, was significantly higher in the p53−/− HCT116 cells when compared to the p53+/+ HCT116, suggesting transcriptional down-regulation of KLF2 by p53. To determine if a similar role for p53 exists in endothelial cells, KLF2 expression was assessed in human umbilical vein endothelial cells (HUVEC) following siRNA mediated downregulation of p53. Knockdown of p53 in HUVEC was accompanied by upregulation of KLF2 expression at the mRNA and protein levels (Fig 1C and 1D). Thus p53 inhibits KLF2 expression at the transcriptional level.
Figure-1. Endogenous p53 inhibits KLF2 expression.
(A, B) Genetic deficiency of p53 increases KLF2 expression. KLF2 mRNA (A) and protein (B) were compared in HCT116 p53+/+ and HCT116 p53−/− cells. Normalized mRNA expression values are shown relative to p53 +/+ cells (**p<0.01, n=3). (C, D) Knockdown of endogenous p53 in endothelial cells increases KLF2 expression. Expression of KLF2 and p53 mRNA (C) and protein (D) were measured in HUVEC in which endogenous p53 was knocked down with p53 siRNA. Normalized mRNA expression values are shown relative to cells transfected with non-specific siRNA (**p<0.01, *<0.05, n=3). Representative immunoblots from three independent experiments are shown.
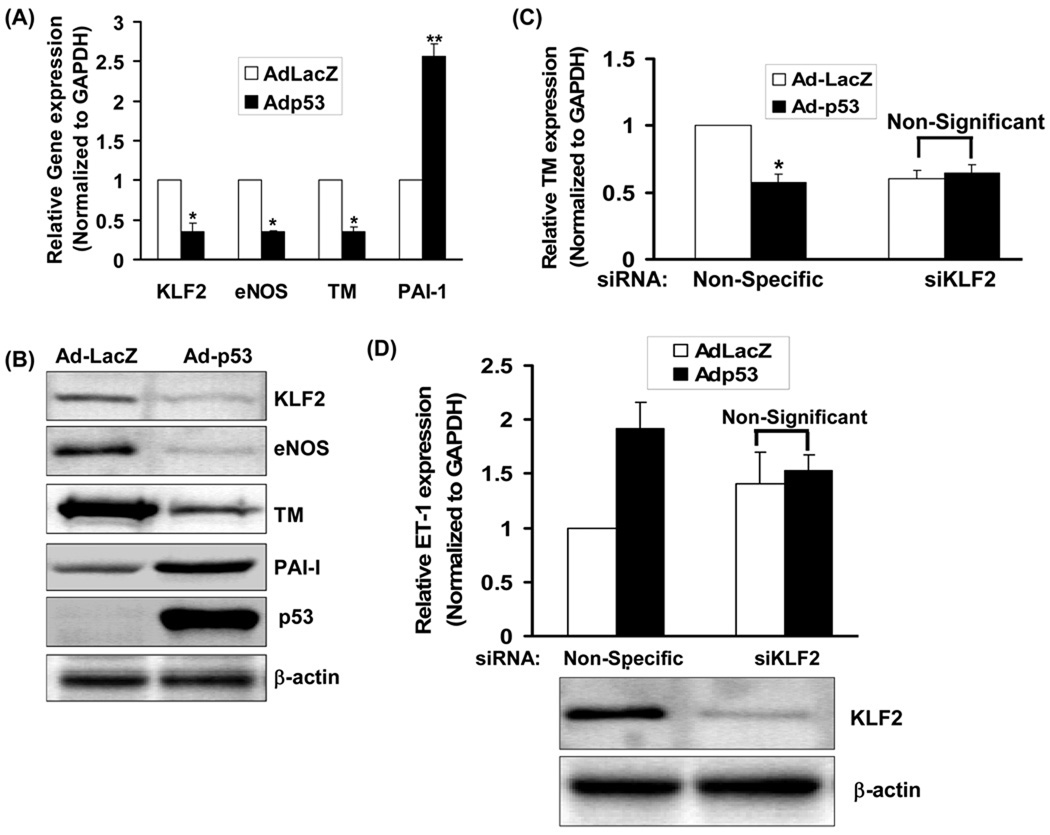
To examine if KLF2 down-regulation by p53 leads to a change in endothelial KLF2 target genes, we measured the expression of thrombomodulin (TM) and endothelial nitric oxide synthase (eNOS), genes that are upregulated by KLF2, and plasminogen activator inhibitor-1 (PAI-1), a gene that is down-regulated by KLF2. Knockdown of p53 in HUVEC led to an increase in TM and eNOS expression, and a decrease in PAI-1 expression (Fig 1D). Conversely, adenoviral overexpression of p53 in HUVEC decreased KLF2, eNOS and TM expression, while increasing PAI-1 and Endothelin-1 (ET-1) expression (Fig 2A, 2B, 2C, 2D). Thus, in keeping with its effect on KLF2 expression, p53 also impacts on KLF2 target genes in endothelial cells.
Figure-2. P53 overexpression suppresses endothelial KLF2 expression and regulates endothelial KLF2 target genes.
(A, B) P53 down-regulates KLF2 in endothelial cells and modulates KLF2 target gene expression. Expression of KLF2, eNOS, TM and PAI-1 mRNA (A) and protein (B) were measured in HUVEC infected with an adenovirus encoding p53 (Adp53) or a control adenovirus encoding LacZ (AdLacZ). Immunoblots representative of three experiments are shown (*p<0.05; **p<0.01; n=3). (C, D) P53-mediated modulation of endothelial TM and ET-1 is KLF2 dependent. KLF2, TM and ET-1 mRNA were measured in HUVEC transfected with KLF2 siRNA or non-specific siRNA, followed by infection with AdLacZ or Adp53. Normalized mRNA expression relative to AdLacZ control is shown (*p<0.05; n=3). Knockdown of KLF2 is shown at bottom.
Next, we determined the mediating role of KLF2 in p53-induced suppression or induction of KLF2 target genes. The effect of p53 overexpression on endothelial TM and ET-1 expression in cells in which KLF2 was knocked down was examined. Compared with control siRNA-transfected cells, adenoviral p53 overexpression in KLF2 siRNA-transfected HUVEC did not lead to a decrease in TM expression (Fig 2C). Similarly, p53 overexpression did not increase ET-1 expression in endothelial cells in which KLF2 was knocked down (Fig 2D). Thus, KLF2 mediates p53-induced regulation of TM and ET-1 expression in endothelial cells.
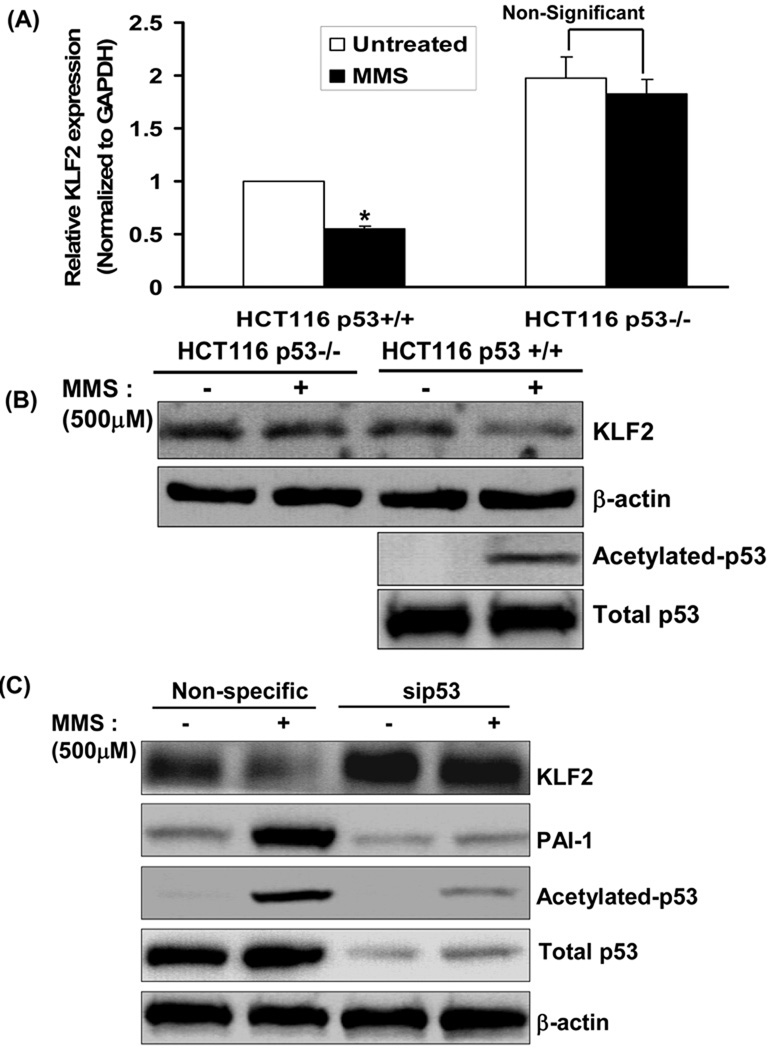
We next asked if an extrinsic stimulus that activates p53 also represses KLF2 expression. Some genotoxic agents lead to cell death or cell cycle arrest by increasing p53 expression or inducing post-translational modifications on p53 that increase its DNA-binding activity 21. We investigated the effect of the genotoxic drug Methyl methanesulfonate (MMS) on KLF2 expression, and asked whether p53 mediates this effect. MMS stimulated acetylation of p53 in the colon cancer cell line HCT116 p53+/+ (Fig 3A). In addition, HCT116 p53+/+ cells challenged with MMS displayed decreased expression of KLF2 at the RNA (Fig 3A) and protein (Fig 3B) levels. However, MMS-induced decrease in KLF2 expression was completely abolished in the p53 deficient (p53−/−) HCT116 cell line (Fig 3A and 3B). Similarly, in endothelial cells, siRNA-mediated knockdown of p53 abrogated the down-regulation of KLF2, and up-regulation of the KLF2 target gene PAI-1, by MMS (Fig 3C). Taken together, this data shows that p53 is indispensable for genotoxic stress-induced down-regulation of KLF2.
Figure-3. Genotoxic stress-mediated inhibition of KLF2 is p53-dependent.
(A, B) P53 mediates MMS-induced down-regulation of KLF2. KLF2 mRNA (A) and protein (B) were measured in HCT116 p53+/+ and p53−/− following treatment with Methyl methanesulfonate (MMS 500 µM) for 4 hours. Data is shown relative to KLF2 mRNA in untreated HCT 116 +/+ cells (*p<0.05, n=3) (C) P53 mediates MMS-induced modulation of KLF2 target genes in endothelial cells. Expression of PAI-1 was measured in whole cell lysates of HUVEC transfected with p53 siRNA or non-specific (NS) siRNA, followed by treatment with MMS (500 µM) for 4 hours. Representative immunoblots from three experiments are shown
P53 regulates KLF2 promoter activity via a novel p53 response element
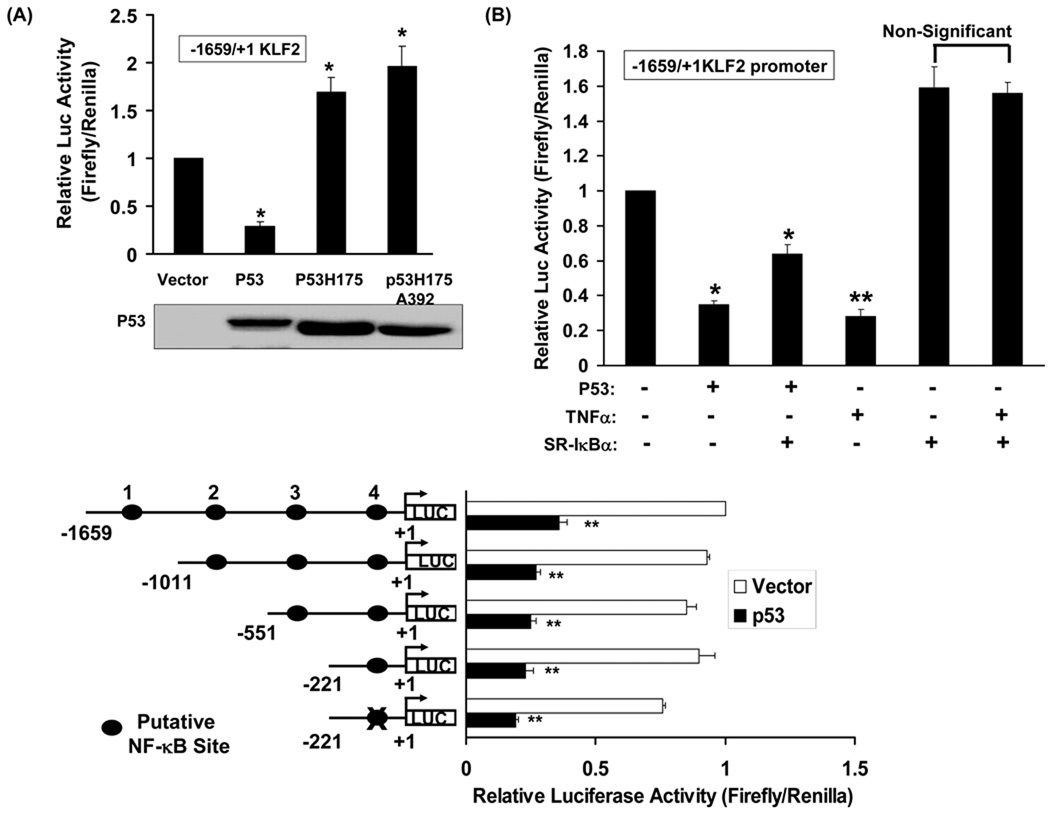
We next assessed the effect of p53 on KLF2 promoter activity. A 1659 bp region upstream of the transcription start site (−1659) in the KLF2 gene was used in promoter-reporter assays. P53 inhibited KLF2 promoter activity (Fig 4A). In contrast, two DNA-binding-deficient mutants of p53 (H175p53 and HA175A392p53) which act in a dominant inhibitory fashion, increased KLF2 promoter activity. These findings indicate a role for p53 in suppressing KLF2 transcription.
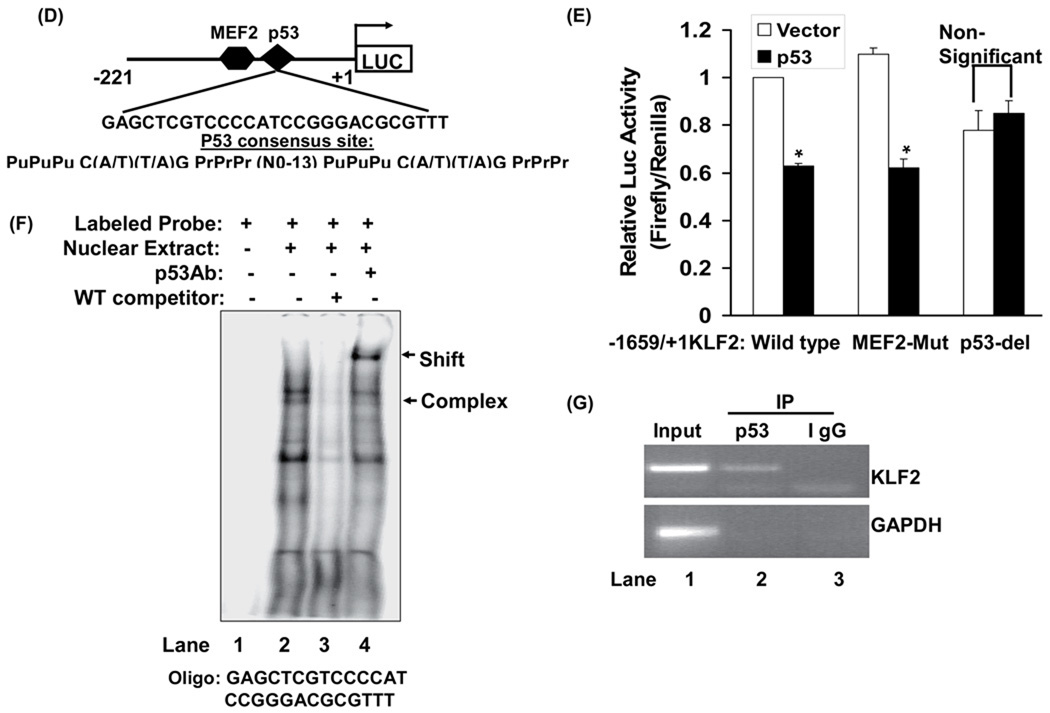
Figure-4. P53 binds to the KLF2 promoter and regulates its activity.
(A) p53 represses KLF2 promoter activity. Activity of the −1659 bp KLF2 promoter-reporter construct was measured in HEK 293 cells with and without expression of wild-type p53 or dominant negative mutants of p53 (H175 and H175A392). Normalized (firefly/renilla) promoter activity is expressed relative to cells not expressing p53 (p*<0.05; n=3). (B, C) p53-induced inhibition of KLF2 promoter activity is NF-kB-independent. (B) Activity of −1659 bp KLF2 promoter was measured in HEK 293 cells in presence of p53, SR-IκBα or TNFα alone or in combination as indicated. Normalized (firefly/renilla) promoter activity is expressed relative to vector transfected control (*p<0.05; **p<0.01; n=3) (C).Activity of the indicated KLF2 promoter-reporter constructs with sequential deletions of putative NF-kB response elements were measured in HEK 293 cells with and without expression of p53. Normalized (firefly/renilla) promoter activity is expressed relative to cells not expressing p53 (*p<0.05; **p<0.01; n=3) (D, E) p53 mediates KLF2 repression through p53 response element. (D) Schematic representation of the 27 bp p53 response element in the KLF2 promoter. The consensus p53 response element is shown. Pu indicates purines; Pr indicates pyrimidine; N is any nucleotide. (E) Activity of wild-type, MEF2-Mut (MEF2 site AATT mutated to TCGG) or p53-del (27 bp of p53 response element deleted) of KLF2 promoters (−1659/+1KLF2) were measured in 293 cells in presence or absence of p53. Luciferase activities are expressed relative to wild-type KLF2 promoter. (F, G) P53 binds to the p53 response element in the KLF2 promoter. (F). P53 binds in vitro to an oligonucleotide with a sequence corresponding to the p53 response element in the KLF2 promoter. Electrophoretic mobility shift assay (EMSA) was performed using HEK 293 nuclear extract (NE) overexpressing p53 and a radiolabeled oligonucleotide with a sequence corresponding to the putative p53 binding site in the KLF2 promoter. P53Ab: p53 antibody. The data is representative of three independent experiments (G) P53 occupies the KLF2 promoter. ChIP assay was performed on sonicated p53-overexpressing HEK 293 cell chromatin with p53 antibody (lane 2) or non-immune IgG (lane 3), and a 169 bp region in the KLF2 promoter harboring the p53-binding site was PCR amplified. Non-immunoprecipitated HEK 293 cell chromatin was used as input (lane 1). Amplification of GAPDH in immunoprecipitates was used as a specificity control. ChIP is representative of three independent experiments.
Prior reports have suggested that p53 can modulate NF-kB expression and function 12. Given the importance NF-kB in regulating cytokine-stimulated repression of KLF2 expression 2, we examined the role of NF-kB in p53-induced down-regulation of KLF2. NF-κB activity was inhibited using a Super Repressor Inhibitor of κB pathway (SR-IκBα). Expression of SR-IκBα did not inhibit p53-stimulated repression of KLF2 promoter activity (Fig 4B). In contrast, SR-IκBα abolished TNFα-induced decrease in KLF2 promoter activity (Fig 4B), consistent with the role of NF-kB in mediating TNFα-stimulated repression of KLF2 transcription 2. We also examined the role of NF-kB in mediating p53-induced repression of KLF2 transcription by creating a KLF2 promoter-reporter construct that is devoid of putative NF-kB response elements. Four putative NF-kB sites in the 1659 bp KLF2 promoter were identified and sequentially deleted or mutated. The lack of these putative NF-kB sites did not affect p53-induced repression of the KLF2 promoter (Fig 4C). These findings demonstrate the dispensability of NF-κB in p53-induced down-regulation of KLF2 transcription.
The core KLF2 promoter region consists of a 221 bp region upstream of the transcription start site, and is highly conserved in mouse and human. This region has a myocyte enhancer factor-2 (MEF2) binding site (Fig 4D) which plays a critical role in governing KLF2 expression regulated by laminar flow 3, cytokines 2 and statins 5, 17. We examined the role of the MEF2 response element in p53-induced repression of KLF2 transcription by mutating key nucleotides in it and measuring p53-stimulated repression of the mutated and wild-type full-length promoters. Mutation of the MEF2 response element did not affect basal or p53-stimulated inhibition of promoter activity (Fig 4E). Thus, the MEF2 site is dispensable for repression of KLF2 promoter activity by p53.
Armed with the knowledge that the core 221 bp promoter is sufficient for repression of promoter activity by p53, and the MEF2 response element is dispensable for p53-stimulated trans-repression, we sought out novel p53-response elements in this region of the promoter. Examination of the promoter sequence reveals a 27 bp element resembling the p53 consensus binding site (Fig 4D). To determine if this 27 bp element functions as a p53 response element, we deleted it in the full-length promoter and measured the response to p53. Deletion of this element completely abrogated p53-stimulated repression of the KLF2 promoter (Fig 4E). We also generated two additional promoter-reporters: one that encompasses this element (−117) and one in which this 27 bp element is deleted (−88). We then compared p53-stimulated activity of the −221, −117, and −88 promoters. Compared with the −221 core promoter, p53 suppressed activity of the −117 promoter to a similar degree (Supplement Fig II). However, p53 did not inhibit activity of the −88 promoter (Supplement Fig II). Taken together, these findings show that this 27 bp sequence in the KLF2 promoter is a p53-response element, being required for p53-stimulated repression of KLF2 promoter activity.
P53 binds to the KLF2 promoter
Next, we examined whether p53 binds to this 27 bp element in the KLF2 promoter. First, we performed electrophoretic mobility shift assays (EMSA) using nuclear extract from 293 HEK cells overexpressing p53 and radiolabeled oligonucleotide with a sequence corresponding to the 27 nucleotides of this element. Nuclear extract containing p53 formed complexes with the labeled oligonucleotide, one of which was identified as containing p53 using unlabeled excess oligonucleotide and p53-specific antibody (Fig 4F). Thus, p53 binds in vitro to an oligonucleotide corresponding to the p53-response element in the KLF2 promoter.
Next, we examined occupancy by p53 of the region of the KLF2 promoter containing this element. Chromatin immunoprecipitation assays amplifying a 169 base pair region of the human KLF2 promoter, which encompasses the p53 response element, showed that p53 occupies this genomic region in the KLF2 promoter. Therefore, p53 occupies the KLF2 promoter in the genomic context (Fig 4G).
P53 induces hypoacetylation of Histone 3 on the KLF2 promoter and suppresses KLF2 transcription in a HAT-independent and HDAC-dependent manner
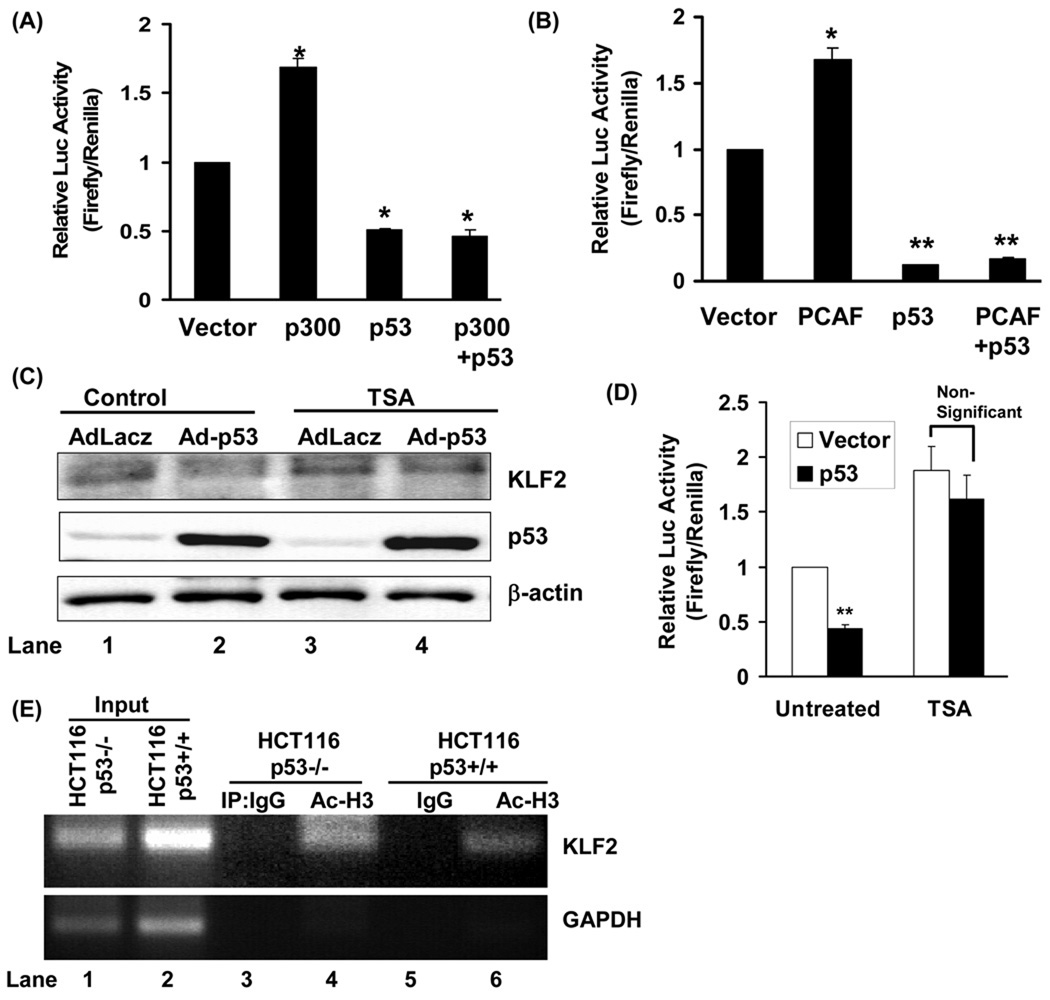
We next investigated how p53 represses KLF2 expression. P300/CBP and PCAF (p300/CBP associated factor) are histone acetyl transferases (HATs) which acetylate histone and non-histone proteins on lysine residues. Through acetylation, p300/CBP and PCAF increase the transcription of various genes and also relieve gene repression 22 such as in Smad3- induced repression of pro-inflammatory target genes 23. We considered the possibility that the p53-mediated repression of KLF2 transcription may occur through sequestration of coactivators, such as p300/CBP and PCAF. To investigate this possibility, we examined the effect of p300 and PCAF overexpression on p53-induced repression of KLF2 promoter activity. P300 independently stimulated KLF2 promoter activity, indicating that it can function as a co-activator of KLF2 transcription (Fig 5A). Similarly, PCAF stimulated KLF2 promoter activity (Fig 5B). However, neither p300 nor PCAF overexpression rescued p53-induced repression of KLF2 promoter activity (Fig 5A, 5B). Thus, sequestration of p300 or PCAF by p53 cannot explain p53-stimulated decrease in KLF2 transcription.
Figure-5. P53-mediated repression of KLF2 does is HAT-independent and HDAC-dependent.
(A, B) P53-induced repression of KLF2 promoter activity is independent of histone acetyltransferases (HAT). (A) Overexpression of the p300 HAT is unable to rescue p53-induced repression of KLF2 promoter activity. KLF2 promoter activity was measured in 293 HEK cells with and without expression of p53 and p300, alone or in combination. Normalized reporter activity is expressed relative to vector control (*p<0.05, n=3). (B) Overexpression of the PCAF HAT is unable to rescue p53-induced repression of KLF2 promoter activity. KLF2 promoter activity was measured in 293 HEK cells with and without expression of p53 and PCAF, alone or in combination. Normalized reporter activity is expressed relative to vector control (**p<0.01, n=3). (C, D) p53 inhibits KLF2 expression in a histone deacetylase (HDAC)-dependent manner. (C) p53-induced down-regulation of KLF2 expression is rescued by the class I and II HDAC inhibitor Trichostatin A (TSA). HUVECs pretreated with TSA (500 nM) for 4 hrs were infected with an adenovirus encoding p53 (Adp53) or a control adenovirus encoding LacZ (AdLacZ). Representative Immunoblots are shown (n=3) (D) p53-induced repression of KLF2 promoter activity is rescued by TSA. KLF2 promoter activity was measured in 293 HEK cells with and without expression of p53 and treatment with TSA (500 nM). Normalized reporter activity is expressed relative to vector control (**p<0.01, n=3). (E) P53 decreases acetylation of Histone H3 associated with the KLF2 promoter. ChIP assay was performed on chromatin from HCT116 p53+/+ and HCT116 p53−/− cells with Acetylated-Histone H3 (Ac-H3) antibody (lanes 4 and 6) or non-immune IgG (lanes 3 and 5), and a 169-bp region of KLF2 promoter was PCR amplified. Non-immunoprecipitated chromatin was used as an input (lanes 1 and 2). Amplification of GAPDH in immunoprecipitates was used as a specificity control. Data is representative of three experiments, and representative immunoblots are shown.
Next, we focused our attention on recruitment of transcriptional repressors by p53 to the KLF2 promoter. The Histone Deacetylases (HDACs) constitute a major category of enzymes that repress gene expression. HDACs deacetylate histone proteins leading to a compact chromatin structure and decrease in gene transcription 24. P53 associates with and recruits HDACs to various gene promoters to regulate gene expression 25. Therefore, we assessed the role of HDACs as co-repressors that are recruited by p53 to the KLF2 promoter and mediate p53-induced repression of KLF2 transcription. We first examined the effect of HDAC inhibition on p53-induced suppression of KLF2 expression. Trichostatin a (TSA), an inhibitor of class I and II HDAC abrogated p53-induced decrease in KLF2 expression in endothelial cells (Fig 5C). Moreover, TSA blocked p53-induced decline in KLF2 promoter activity (Fig 5D). Thus, inhibition of class I and II HDAC antagonizes p53-induced repression of KLF2 transcription, suggesting that one or more members of these classes of HDACs play a role in p53-induced repression of KLF2 transcription.
Wild type p53 binds to its target genes and recruits co-repressor HDACs that subsequently modify histone H3 and H4 and decrease gene expression 26, 27. Recruitment of HDACs by p53 to the KLF2 promoter would therefore be expected to lead to hypoacetylation of histones at the KLF2 promoter. Thus, we asked if p53 stimulates hypoacetylation of histones at the KLF2 promoter. Chromatin immunoprecipitation experiments were performed to determine p53-mediated change in acetylation of Histone 3 (H3) at the KLF2 promoter. Acetylation on lysine 9 of Histone H3 at the KLF2 promoter was significantly lower in p53+/+ HCT166 cells when compared to p53 −/− HCT116 cells (Fig 5E). Thus, p53 leads to hypoacetylation of H3 at the KLF2 promoter, and indicates a role for recruitment of HDACs by p53 to the promoter.
P53 promotes a pro-thrombotic endothelial cell phenotype, and impairs endothelium-dependent NO production by repressing KLF2 expression
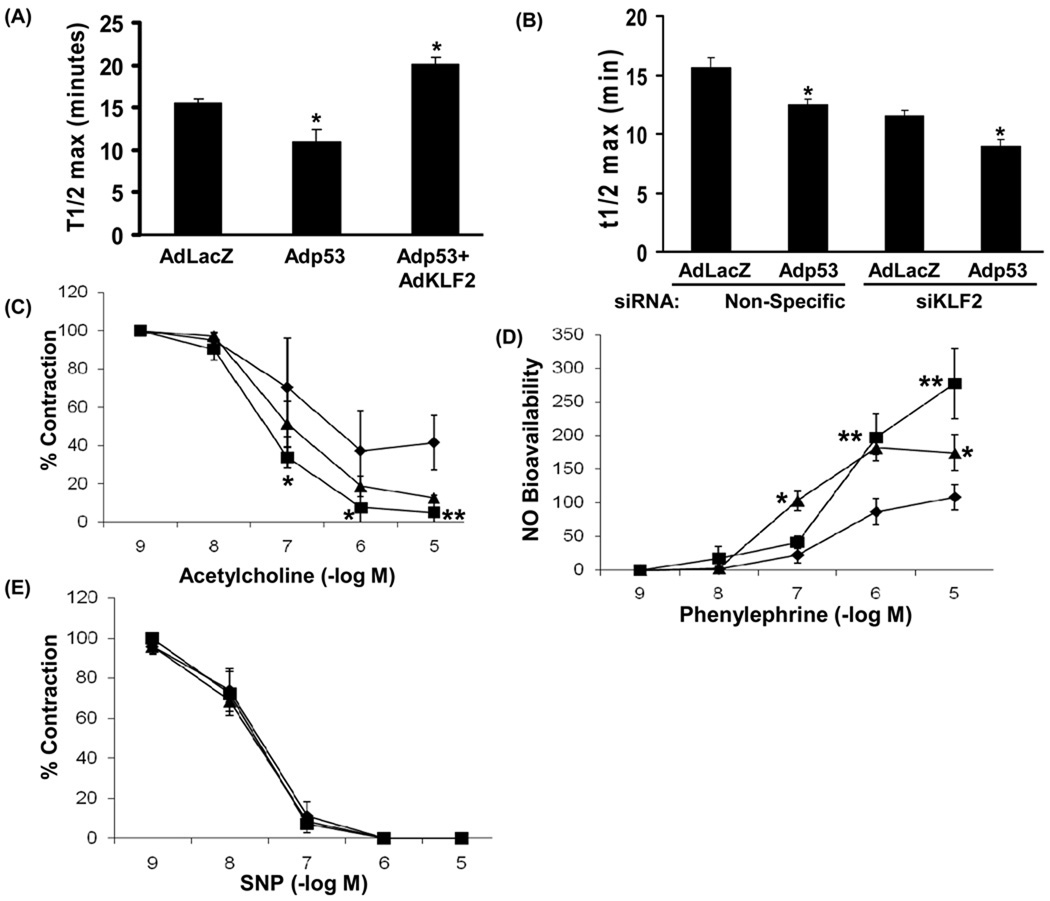
TM and PAI-1, two target genes of KLF2, are important modulators in the coagulation cascade. TM expression by the endothelium inhibits thrombosis 28, while PAI-1 expression on the endothelium promotes blood clotting 29 Therefore, the increase in endothelial PAI-1 and decrease in TM expression induced by p53 would be expected to lead to a pro-thrombotic endothelial phenotype. To test this we performed an in vitro thrombosis assay on an endothelial cell monolayer. Adenoviral overexpression of p53 in endothelial cells decreased the time to thrombosis (Fig 6A), indicating that p53 promotes a pro-thrombotic phenotype in endothelial cells. This p53-induced pro-thrombotic phenotype was rescued by forced adenoviral expression of KLF2 (Fig 6A). In contrast, siRNA-mediated knockdown of KLF2 in endothelial cells decreased the time to clot formation under basal conditions, and also potentiated p53-induced decrease in time to clot formation (Fig 6B). These findings show an important role for KLF2 repression in p53-induced pro-thrombotic phenotype in endothelial cells.
Figure-6. KLF2 down-regulation mediates P53-induced impairment of endothelial function.
(A, B) P53 promotes thrombosis on endothelial cells by suppressing KLF2 expression. (A) Overexpression of KLF2 rescues p53-induced thrombosis on endothelial cells. HUVECs infected with the indicated adenoviruses were exposed to human plasma in the presence of calcium. Time to clot formation was measured by light absorbance. Values are expressed as time to reach half-maximal absorbance (t1/2 max) *p<0.05 (n=3). (B) KLF2 down-regulation potentiates p53-induced thrombosis on endothelial cells. KLF2 was down-regulated by siRNA in HUVEC followed by adenoviral infection with AdLacZ or Adp53. Thrombosis assay was performed as described above and t1/2 max determined. *p<0.05 (n=3). (C, D, E) KLF2 rescues p53-induced impairment of endothelial-dependent vasorelaxation and decrease in bioavailability of NO. (C) Endothelium-dependent vasorelaxation (n=4), (D) bioavailable NO (n=4) and (E) endothelial-independent vasorelaxation (n=3) in rat aortic rings infected with Adp53 (◆), Adp53 plus AdFLAG-KLF2 (■) and AdLacZ (▲). Equal amounts of total virus were used for each. SNP: sodium nitroprusside. *p<0.05, and **p<0.01 compared with Adp53.
Endothelial p53 impairs endothelial-dependent vascular relaxation 11. In contrast, endothelial KLF2 increases the expression of eNOS and inhibits ET-1 expression, leading to vasodilatation 4. This prompted us to ask if impairment of endothelium-dependent vasorelaxation by p53 is also, in part, due to down-regulation of KLF2. Ex vivo vasoreactivity studies in rat aortas demonstrated that adenoviral overexpression of p53 impairs acetylcholine-induced endothelium-dependent vasorelaxation (Fig 6C), and decreases bioavailable vascular NO (Fig 6D), but does not affect endothelium-independent vasorelaxation (Fig 6E). Forced adenoviral co-expression of KLF2 rescued impairment of endothelium-dependent vasorelaxation (Fig 6C) and decreased vascular bioavailable NO (Fig 6D) induced by p53. These findings show that down-regulation of KLF2 is, in part, responsible for p53-induced impairment of endothelium-dependent production of NO and vasorelaxation.
Discussion
Endothelial dysfunction which includes impaired vasodilatation, and increased expression of adhesion molecules and pro-coagulant factors, is an early pathophysiological event in development and progression of cardiovascular diseases. P53 has been implicated in impairing endothelial function, while KLF2, by differentially regulating endothelial gene expression, is essential to the maintenance of an anti-thrombotic and vasodilatory endothelium. The major finding of the present study is that KLF2 is a gene that is targeted and repressed by p53, and this functional relationship between the two has a bearing on p53-induced impairment of normal endothelial function.
We considered several plausible mechanisms by which p53 may regulate KLF2 transcription. Prior studies have shown that NF-kB acts as a downstream effector of p53 12. Our studies with the molecular inhibitor of NF-κB (SR-IκB) and promoter deletion analysis (constructs devoid of putative NF-kB binding sites) provide evidence that the NF-κB-mediated signaling is not involved in p53-induced repression of KLF2 transcription. Further, because the element that mediates the response to p53 lies outside a region in the KLF2 promoter that includes the MEF2 binding site, our data also indicate that p53-induced KLF2 repression does not involve MEF2 factors which are known to be important for basal and cytokine-induced down-regulation of KLF2 2. As such, our findings provide an additional mechanism for transcriptional control of KLF2, showing the importance of a novel functional p53-response element in the KLF2 promoter. Examination of the p53-response element in the KLF2 promoter reveals an imperfect match with the consensus p53 binding motif which consists of two half-sites (5’-PuPuPuC(A/T)(T/A)GpyPyPy-3’), separated by 0 to 13 base pairs. Nevertheless, there are ample examples of p53-response elements that deviate from the consensus binding motif 30–32. Moreover, comparison of the human and mouse KLF2 promoter sequences shows 100% homology of this p53-response element, suggesting that p53 has an important and conserved role in regulating of KLF2 transcription via this element.
Although well known as a transcriptional activator, p53 also represses genes that are involved in various signaling pathways such as cell proliferation (c-myc, cyclin B) and apoptosis (Bcl2, surviving) 26, 33. With respect to p53-mediated gene repression, several mechanisms have been proposed. Repression may occur through displacement of co-activators by p53 from adjacent or overlapping binding sites within the promoters 25, 34. Alternatively, p53 may recruit transcriptional co-repressors to specific promoters to inhibit their activity 25, 34. Such p53-mediated repression that depends on HDAC recruitment requires additional protein(s) like mSin3a 34. Our data demonstrate that sequestration of p300/PCAF away from KLF2 promoter is not the mechanism for p53-induced repression of KLF2 transcription, but rather recruitment of HDACs as co-repressors to the KLF2 promoter mediates the effect of p53. This is supported by findings that the histone deacetylase inhibitor TSA abrogates down-regulation of KLF2 expression by p53, and p53 decreases lysine acetylation of Histone H3 at the KLF2 promoter, and is consistent with prior studies showing that p53 represses gene expression by decreasing Histone H3 acetylation through recruitment of HDAC 35
Our findings indicate that KLF2 is the principal Kruppel-like factor that mediates p53-induced dysregulation of the endothelium. However, it is highly improbable that p53-stimulated endothelial cell phenotype is solely via down-regulation of KLF2. P53 influences the expression of a vast array of genes, many of which could, directly or indirectly, mediate some of the p53-induced phenotypes in endothelial cells. It is also noteworthy that there is significant redundancy in the Kruppel-like family of transcription factors. Kruppel-like factor 4 (KLF4) is capable of conferring gene regulatory effects similar to KLF2 in endothelial cells 36. Thus, the KLF2-mediated effect of p53 may be blunted by the compensatory expression of other members of the Kruppel-like factor family.
Impaired nitric oxide production is the sine qua non of endothelial dysfunction and is considered as an early requisite step in the development of atherosclerotic vascular lesions 37. Deficiency in endothelium-derived nitric oxide leads to impaired vasodilatation, increased proliferation and migration of smooth muscle cells, and platelet activation and aggregation, predisposing to thrombogenesis 38. In recent years, though p53 has gain considerable attention for its role in atherosclerosis, this role still remains controversial, and there is a paucity of information about how p53 impacts on endothelial function. P53 expression is associated with increased apoptosis in endothelial as well as in vascular smooth muscle cells 39, and has been shown to impair endothelial-dependent vasorelaxation 11. However, whether p53 affects thrombosis is not known. By showing that p53 inhibits KLF2, a pivotal endothelial factor that is important to NO production, and one that promotes a milieu on the endothelium that inhibits thrombosis, our findings provide a unifying mechanism through which p53 in the endothelium impacts on both thrombosis and vascular tone. Finally, given the importance of p53 and KLF2 in regulating death, survival, and proliferation of many cell types, these observations may have broader implications for p53-mediated pathophysiological states.
Supplementary Material
Acknowledgements
We thank B. Vogelstein for the gift of the HCT116 cells, H.H. Ng for the KLF2 antibody, P.M. Hwang and X. Lu for the p53 expression plasmids, and A. Baldwin for the SR-IκBα plasmid.
Source of Funding
This work was supported by NIH grants HL094959, HL070929, HL065608, and HL098892 to KI, HL076754 and HL097593 to MKJ, HL087595 to ZL, and an American Heart Association BGIA 0865419D to AK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None
References
- 1.Bieker JJ. Isolation, genomic structure, and expression of human erythroid kruppel-like factor (eklf) DNA & Cell Biology. 1996;15:347–352. doi: 10.1089/dna.1996.15.347. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A, Lin Z, SenBanerjee S, Jain MK. Tumor necrosis factor alpha-mediated reduction of klf2 is due to inhibition of mef2 by nf-kappab and histone deacetylases. Mol Cell Biol. 2005;25:5893–5903. doi: 10.1128/MCB.25.14.5893-5903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parmar KM, Larman HB, Dai G, Zhang Y, Wang ET, Moorthy SN, Kratz JR, Lin Z, Jain MK, Gimbrone MA, Jr, Garcia-Cardena G. Integration of flow-dependent endothelial phenotypes by kruppel-like factor 2. J Clin Invest. 2006;116:49–58. doi: 10.1172/JCI24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, Feinberg MW, Chen Z, Simon DI, Luscinskas FW, Michel TM, Gimbrone MA, Jr, Garcia-Cardena G, Jain MK. Klf2 is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya R, Senbanerjee S, Lin Z, Mir S, Hamik A, Wang P, Mukherjee P, Mukhopadhyay D, Jain MK. Inhibition of vascular permeability factor/vascular endothelial growth factor-mediated angiogenesis by the kruppel-like factor klf2. J Biol Chem. 2005;280:28848–28851. doi: 10.1074/jbc.C500200200. [DOI] [PubMed] [Google Scholar]
- 6.Kumar A, Hoffman TA, Dericco J, Naqvi A, Jain MK, Irani K. Transcriptional repression of kruppel like factor-2 by the adaptor protein p66shc. FASEB J. 2009;23:4344–4352. doi: 10.1096/fj.09-138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine AJ. P53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 8.Ihling C, Haendeler J, Menzel G, Hess RD, Fraedrich G, Schaefer HE, Zeiher AM. Co-expression of p53 and mdm2 in human atherosclerosis: Implications for the regulation of cellularity of atherosclerotic lesions. J Pathol. 1998;185:303–312. doi: 10.1002/(SICI)1096-9896(199807)185:3<303::AID-PATH106>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, Criado LM, Klatt P, Flores JM, Weill JC, Blasco MA, Serrano M. "Super p53" mice exhibit enhanced DNA damage response, are tumor resistant and age normally. Embo J. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanz-Gonzalez SM, Barquin L, Garcia-Cao I, Roque M, Gonzalez JM, Fuster JJ, Castells MT, Flores JM, Serrano M, Andres V. Increased p53 gene dosage reduces neointimal thickening induced by mechanical injury but has no effect on native atherosclerosis. Cardiovasc Res. 2007;75:803–812. doi: 10.1016/j.cardiores.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Kim CS, Jung SB, Naqvi A, Hoffman TA, DeRicco J, Yamamori T, Cole MP, Jeon BH, Irani K. P53 impairs endothelium-dependent vasomotor function through transcriptional upregulation of p66shc. Circ Res. 2008;103:1441–1450. doi: 10.1161/CIRCRESAHA.108.181644. [DOI] [PubMed] [Google Scholar]
- 12.Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of nf-kappab in p53-mediated programmed cell death. Nature. 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 13.Zheng SJ, Lamhamedi-Cherradi SE, Wang P, Xu L, Chen YH. Tumor suppressor p53 inhibits autoimmune inflammation and macrophage function. Diabetes. 2005;54:1423–1428. doi: 10.2337/diabetes.54.5.1423. [DOI] [PubMed] [Google Scholar]
- 14.Huddleson JP, Ahmad N, Lingrel JB. Up-regulation of the klf2 transcription factor by fluid shear stress requires nucleolin. J Biol Chem. 2006;281:15121–15128. doi: 10.1074/jbc.M513406200. [DOI] [PubMed] [Google Scholar]
- 15.Huddleson JP, Ahmad N, Srinivasan S, Lingrel JB. Induction of klf2 by fluid shear stress requires a novel promoter element activated by a phosphatidylinositol 3-kinase-dependent chromatin-remodeling pathway. J Biol Chem. 2005;280:23371–23379. doi: 10.1074/jbc.M413839200. [DOI] [PubMed] [Google Scholar]
- 16.Sako K, Fukuhara S, Minami T, Hamakubo T, Song H, Kodama T, Fukamizu A, Gutkind JS, Koh GY, Mochizuki N. Angiopoietin-1 induces kruppel-like factor 2 expression through a phosphoinositide 3-kinase/akt-dependent activation of myocyte enhancer factor 2. J Biol Chem. 2009;284:5592–5601. doi: 10.1074/jbc.M806928200. [DOI] [PubMed] [Google Scholar]
- 17.Sen-Banerjee S, Mir S, Lin Z, Hamik A, Atkins GB, Das H, Banerjee P, Kumar A, Jain MK. Kruppel-like factor 2 as a novel mediator of statin effects in endothelial cells. Circulation. 2005;112:720–726. doi: 10.1161/CIRCULATIONAHA.104.525774. [DOI] [PubMed] [Google Scholar]
- 18.Yamamori T, White AR, Mattagajasingh I, Khanday FA, Haile A, Qi B, Jeon BH, Bugayenko A, Kasuno K, Berkowitz DE, Irani K. P66shc regulates endothelial no production and endothelium-dependent vasorelaxation: Implications for age-associated vascular dysfunction. J Mol Cell Cardiol. 2005;39:992–995. doi: 10.1016/j.yjmcc.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Jeon BH, Gupta G, Park YC, Qi B, Haile A, Khanday FA, Liu YX, Kim JM, Ozaki M, White AR, Berkowitz DE, Irani K. Apurinic/apyrimidinic endonuclease 1 regulates endothelial no production and vascular tone. Circ Res. 2004;95:902–910. doi: 10.1161/01.RES.0000146947.84294.4c. [DOI] [PubMed] [Google Scholar]
- 20.Rose SL, Babensee JE. Procoagulant phenotype of endothelial cells after coculture with biomaterial-treated blood cells. J Biomed Mater Res A. 2005;72:269–278. doi: 10.1002/jbm.a.30222. [DOI] [PubMed] [Google Scholar]
- 21.Liu G, Chen X. Regulation of the p53 transcriptional activity. J Cell Biochem. 2006;97:448–458. doi: 10.1002/jcb.20700. [DOI] [PubMed] [Google Scholar]
- 22.Grossman SR. P300/cbp/p53 interaction and regulation of the p53 response. Eur J Biochem. 2001;268:2773–2778. doi: 10.1046/j.1432-1327.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 23.Werner F, Jain MK, Feinberg MW, Sibinga NE, Pellacani A, Wiesel P, Chin MT, Topper JN, Perrella MA, Lee ME. Transforming growth factor-beta 1 inhibition of macrophage activation is mediated via smad3. J Biol Chem. 2000;275:36653–36658. doi: 10.1074/jbc.M004536200. [DOI] [PubMed] [Google Scholar]
- 24.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (hdacs): Characterization of the classical hdac family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, George DL. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with msin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn J, Murphy M, Kratowicz S, Wang A, Levine AJ, George DL. Down-regulation of the stathmin/op18 and fkbp25 genes following p53 induction. Oncogene. 1999;18:5954–5958. doi: 10.1038/sj.onc.1202986. [DOI] [PubMed] [Google Scholar]
- 27.Esteve PO, Chin HG, Pradhan S. Molecular mechanisms of transactivation and doxorubicin-mediated repression of survivin gene in cancer cells. J Biol Chem. 2007;282:2615–2625. doi: 10.1074/jbc.M606203200. [DOI] [PubMed] [Google Scholar]
- 28.Weiler H, Isermann BH. Thrombomodulin. J Thromb Haemost. 2003;1:1515–1524. doi: 10.1046/j.1538-7836.2003.00306.x. [DOI] [PubMed] [Google Scholar]
- 29.Gimbrone MA, Jr, Nagel T, Topper JN. Biomechanical activation: An emerging paradigm in endothelial adhesion biology. Journal of Clinical Investigation. 1997;100:S61–S65. [PubMed] [Google Scholar]
- 30.Jin YJ, Wang J, Qiao C, Hei TK, Brandt-Rauf PW, Yin Y. A novel mechanism for p53 to regulate its target gene eck in signaling apoptosis. Mol Cancer Res. 2006;4:769–778. doi: 10.1158/1541-7786.MCR-06-0178. [DOI] [PubMed] [Google Scholar]
- 31.Nemoto S, Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science. 2002;295:2450–2452. doi: 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- 32.Yugawa T, Handa K, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. Regulation of notch1 gene expression by p53 in epithelial cells. Mol Cell Biol. 2007;27:3732–3742. doi: 10.1128/MCB.02119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho J, Benchimol S. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 2003;10:404–408. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- 34.Ho JS, Ma W, Mao DY, Benchimol S. P53-dependent transcriptional repression of c-myc is required for g1 cell cycle arrest. Mol Cell Biol. 2005;25:7423–7431. doi: 10.1128/MCB.25.17.7423-7431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogden SK, Lee KC, Wernke-Dollries K, Stratton SA, Aronow B, Barton MC. P53 targets chromatin structure alteration to repress alpha-fetoprotein gene expression. J Biol Chem. 2001;276:42057–42062. doi: 10.1074/jbc.C100381200. [DOI] [PubMed] [Google Scholar]
- 36.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 37.Davignon J, Ganz P. Role of endothelial dysfunction in atherosclerosis. Circulation. 2004;109:III27–III32. doi: 10.1161/01.CIR.0000131515.03336.f8. [DOI] [PubMed] [Google Scholar]
- 38.Kinlay S, Libby P, Ganz P. Endothelial function and coronary artery disease. Curr Opin Lipidol. 2001;12:383–389. doi: 10.1097/00041433-200108000-00003. [DOI] [PubMed] [Google Scholar]
- 39.Lee CN, Cheng WF, Chang MC, Su YN, Chen CA, Hsieh FJ. Hypoxia-induced apoptosis in endothelial cells and embryonic stem cells. Apoptosis. 2005;10:887–894. doi: 10.1007/s10495-005-2946-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.