Abstract
Metastasis-related recurrence often occurs in hepatocellular carcinoma (HCC) patients who receive curative therapies. At present, it is challenging to identify patients with high risk of recurrence, which would warrant additional therapies. In this study, we sought to analyze a recently developed metastasis-related gene signature for its utility in predicting HCC survival using two independent cohorts consisting of a total of 386 patients who received radical resection. Cohort-1 contained 247 predominantly HBV-positive cases analyzed with an Affymetrix platform, while cohort-2 contained 139 cases with mixed etiology analyzed with the NCI Oligo Set microarray platform. We employed a survival risk prediction algorithm with training, test, and independent cross-validation strategies and found that the gene signature is predictive of overall and disease-free survival. Importantly, risk was significantly predicted independently of clinical characteristics and microarray platform. In addition, survival prediction was successful in patients with early disease, such as small (<5 cm in diameter) and solitary tumors, and the signature predicted particularly well for early recurrence risk (<2 years), especially when combined with serum alpha fetoprotein or tumor staging. In conclusion, we have demonstrated in two independent cohorts with mixed etiologies and ethnicity that the metastasis gene signature is a useful tool to predict HCC outcome, suggesting the general utility of this classifier. We recommend the use of this classifier as a molecular diagnostic test to assess the risk that an HCC patient will develop tumor relaps within 2 years after surgical resection, particularly for those with early stage tumors and solitary presentation.
Keywords: Early Recurrence, Prognosis, Gene Signature, HCC
INTRODUCTION
Hepatocellular carcinoma (HCC) is the most frequent malignant tumor in the liver and the third leading cause of cancer-related deaths worldwide (1). HCC is most prevalent in developing countries, but its incidence is increasing in developed countries due to chronic infection with hepatitis C virus (HCV) and resulting liver cirrhosis(2). In the United States, liver cancer has the fastest growing cancer death rate, even though, the overall cancer mortality rate has declined during the past years (3). The poor outcome of HCC patients is mainly caused by the high frequency of late-stage disease, metastasis and de novo tumor formation in the diseased liver, the so-called “field effect” (4, 5). Currently, surgery is the most effective treatment, but the recurrence rate is high, mainly due to the dissemination of malignant cells (6). Although early-stage tumors can be treated by resection, liver transplantation or local ablation, few patients present with early-stage disease and many patients still suffer from recurrence after treatment of early-stage tumors (7).
Metastasis contributes to 90% of all cancer related deaths, emphasizing the importance of metastasis risk prediction (8). Unlike other tumor types, HCC metastasis occurs mainly within the liver itself with new tumor colonies frequently invading into the major branches of the portal vein or to other parts of the liver (9–11). It is believed that de novo development of primary HCC in the remnant liver occurs with a lower frequency (12, 13). Recurrence by metastasis seems to occur mainly in an early period, i.e., within the first two years after resection, whereas recurrence due to new primary lesions often occurs after a longer period (5, 14–17). Consistently, Chen et al. found that tumors that recurred late often showed clonal origins different from the original tumors, suggesting a de novo second primary HCC (18). The comparison of early and late recurrence of HCC after hepatectomy revealed that early recurrence is associated with non-anatomical resection, microscopic vascular invasion and high alpha fetoprotein (AFP) levels (14). In contrast, late recurrence is associated with the level of chronic hepatitis, multi-nodularity, and tumor classification (14).
In a recent pilot study, we identified a metastasis signature consisting of 153 genes that could distinguish HCC patients with portal venous metastases from those without (19). This metastasis signature was developed based on cDNA microarray profiling of 20 well-defined HCC cases, of which 10 presented with tumor thrombi in the major branches of the portal vein at surgery while 10 were metastasis-free HCC patients at the time of surgery and at follow-up. In this study, we used two independent cohorts consisting of a total of 386 HCC patients to analyze the utility of this signature as a risk classifier for HCC recurrence and survival.
MATERIALS AND METHODS
Study Cohorts and Patient Characteristics
Cohort-1 hepatic tissues were obtained from the Liver Cancer Institute (LCI) with informed consent from patients who underwent radical resection between 2002 and 2003 at the Liver Cancer Institute and Zhongshan Hospital (Fudan University, Shanghai, China). The study was approved by the Institutional Review Board of the participating institutes. A total of 247 HCC patients were recruited. Cases were mainly from patients with a history of hepatitis B virus (HBV) infection or HBV-related liver cirrhosis; all were diagnosed with HCC by two independent pathologists, with detailed information on clinical presentation and pathological characteristics. For 242 patients, disease-free survival and overall survival as well as the cause of death were available.
The gene expression data of cohort-2 has been published earlier (20, 21). Briefly, gene expression profiling of cohort-2 was performed by the Laboratory of Experimental Carcinogenesis (LEC) and analyzed using NCI’s Human Array-Ready Oligo Set microarray platform (GPL1528). The microarray data is publicly available at the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) with accession numbers GSE1898 and GSE4024.
Tumor Samples and Microarray Processing
Total RNA was extracted from frozen tissues using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Only RNA samples with good RNA quality as confirmed with the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) and agarose gel electrophoresis were included in the study. For microarray profiling, tumors, and paired non-tumor tissues were profiled separately using a single channel array platform. Gene expression profiling of 22 tumor samples was carried out on Affymetrix GeneChip HG-U133A 2.0 arrays (Affymetrix, Santa Clara, CA) according to the manufacturer's protocol. The fluorescent intensities were determined with an Affymetrix GeneChip Scanner 3000, controlled by GCOS Affymetrix software. The remaining 225 tumor samples were processed on the 96 HT HG-U133A 2.0 microarray platform. The fluorescent intensities were determined with an Affymetrix GeneChip HT Array Plate Scanner, controlled by GCOS Affymetrix software. Quality controls included image inspection as well as Relative Log Expression (RLE) and Normalized Unscaled Standard Error (NUSE) implemented in the affyPLM package available at the Bioconductor (www.bioconductor.org). In accordance with Minimum Information About a Microarray Experiment (MIAME) guidelines, we deposited the microarray data and additional patient information into the GEO repository with accession number GSE14520.
Affymetrix gene expression arrays obtained from different platforms were combined with the matchprobes package in R (http://www.R-project.org)(22). Raw gene expression data were normalized using the Robust Multi-array Average (RMA) method and global median centering (23). For genes with more than one probe set, the mean gene expression was calculated.
Statistical Analysis
Class comparison and survival risk prediction of the gene expression data was performed with the BRB-Array Tools software (http://linus.nci.nih.gov/BRB-ArrayTools.html; Version 3.7.0). For survival risk prediction, we identified genes whose expression was significantly related to survival by applying univariate Cox proportional hazards regression followed by principal component analysis. Principal component analysis is a computational procedure that transforms a number of possibly correlated variables into a significantly smaller number of uncorrelated variables called principal components. This resulted in a regression coefficient (weight) related to survival time based on two principal components. Next, to compute a prognostic index, the weighted average of the principal component values was calculated, using the regression coefficients derived from the Cox regression, described above. Finally, this prognostic index was used to split samples into two groups of equal size by the median of the prognostic index. Thereby, a high value of the prognostic index corresponded to a high value of hazard of death (high risk), and consequently a relatively poor predicted survival.
Kaplan-Meier survival curves for the predicted cases to have above average risk and the cases predicted to have below average risk were plotted. In order to evaluate the predictive value of the method, 10-fold cross-validation with 1000-fold random permutation of the Cox-Mantel log-rank test was performed.
For cross-validation of the LCI and LEC cohorts, we converted the gene expression data into z-scores and then performed class prediction in BRB-Array Tools. First, we used the LEC cohort for training/testing and predicted the outcome of the LCI cohort and then, we used the LCI cohort for training/testing and predicted the outcome of the LEC cohort. Six class prediction algorithms, Support Vector Machines (SVM), Nearest Centroid (NC), 3-Nearest Neighbor (3-NN), 1-Nearest Neighbor (1-NN), Linear Discriminant Analysis (LDA) or Compound Covariate Predictor (CCP), were used to determine whether mRNA expression patterns could accurately discriminate good and poor survival HCC groups in an independent data set. The accuracy of the prediction was calculated after 1000 repetitions of this random partitioning process to control the number and proportion of false discoveries.
Kaplan-Meier survival analysis was performed using GraphPad Prism software 5.0 (GraphPad Software, San Diego, CA) and the statistical p values were generated by the Cox-Mantel log-rank test. Cox proportional hazards regression was used to analyze the effect of clinical variables on patient survival using STATA 9.2 (College Station, TX). Clinical variables included age, gender, HBV active status, pre-resection AFP, cirrhosis, alanine transferase (ALT), tumor size or size of the largest tumor when multiple tumors are present, nodular type and the HCC prognosis staging systems Barcelona Clinic Liver Cancer (BCLC), Cancer Liver Italian Program (CLIP) or Tumor Node Metastasis (TNM) classification (24–26). An AFP cutoff of 300 ng/mL, ALT of 50 U/L and tumor size of 5cm were used in Cox regression analysis and are clinically relevant values used to distinguish patient survival. A univariate test was used to examine the influence of the ‘metastasis’ gene predictor or each clinical variable on patient survival. A multivariate analysis was performed to estimate the hazards ratio of the predictor while controlling for clinical variables that were significantly associated with survival in the univariate analysis. Since tumor size and nodular type were collinear with tumor staging, these variables were not included in the multivariate analysis. It was determined that the final model met the proportional hazards assumption. Receiver operating characteristic (ROC) curves were computed by using the tumor expression level for compound covariate prediction and the ROCR package (27). The statistical significance was defined as p <0.05.
Endpoints
We analyzed the overall survival, which was defined as time from surgery to death from any disease, as well as the disease-free survival, which was defined as the time from surgery to any recurrence, distant metastasis or death from any cause. The Kaplan-Meier estimator was used to display time-to-event curves for these two endpoints.
RESULTS
Redefining the Metastasis Gene Signature
We reanalyzed the data from our pilot study on 20 well-defined HCC cases used to identify our recently published 153 gene HCC metastasis signature with the updated gene annotation, sequence data and software (19). Class comparison identified 181 differentially-expressed cDNA probes (p < 0.001, FDR < 0.05). Thirty six of the 181 probes did not have any gene annotation available in the original study (19). Alignment of the probe sequences to the human genome (NCBI BLAST) resulted in the annotation information of 8 additional genes. Therefore, 161 out of 181 probes matched to annotated genes (including all original 153 genes; Supplementary Table 1). This new 161 gene signature is referred to as a metastasis risk classifier and was used for subsequent analysis.
Predicting HCC Survival Using Two Independent Validation Cohorts
Next, we developed a strategy for testing the metastasis risk classifier by incorporating two independent patient cohorts, i.e., LCI and LEC cohorts (Figure 1A). We aimed to determine whether this classifier can predict survival, since HCC metastasis is the main causative factor for poor outcome. The recruitment criteria of the LCI cohort were based on the characteristics of the 40 original patients previously described (19). In addition to the two different microarray platforms used, the LCI and LEC cohorts differed in their patient characteristics (Table 1). The LCI cohort mainly consists of HBV positive Chinese patients (95.6%), whereas, the LEC cohort is heterogeneous, containing a mixture of Chinese, European and American patients with 41.7% HBV positive, 12.2% HCV positive and 23.0% non-viral HCC. The two cohorts also differed in gender distribution, the number of patients with underlying cirrhosis, tumor size and survival time (Table 1). The survival time of patients in the LEC cohort was significantly shorter than in the LCI cohort which was consistent with the larger tumor size of the LEC cohort.
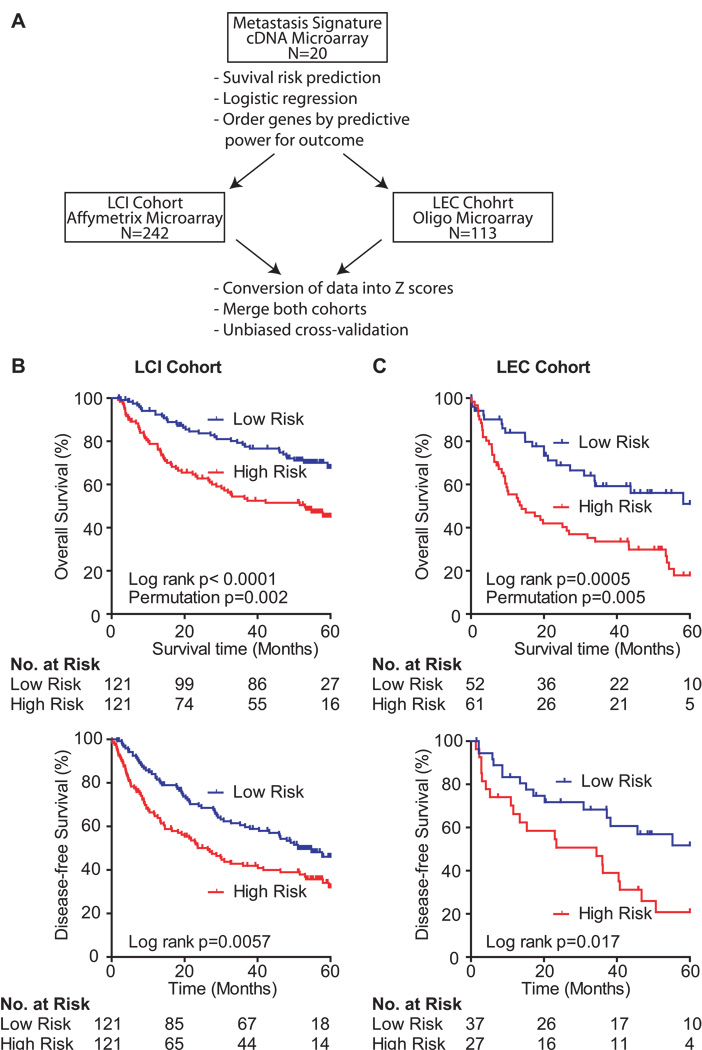
Figure 1.
Survival risk prediction analysis and application of the metastasis gene signature. (A) Schematic overview of the study design. (B) Kaplan-Meier survival curves showing the overall survival (top panel; N = 242) and the disease-free survival (bottom panel; N = 242) of the predicted high and low risk groups in the LCI cohort. (C) Kaplan-Meier survival curves showing the overall survival (top panel; N = 113) and the disease-free survival (bottom panel; N = 64) of the predicted high and low risk groups in the LEC cohort. Displayed are the Cox-Mantel log-rank, the permutation p-values and the number of patients at risk for each Kaplan-Meier survival curve.
Table 1.
Clinical Characteristics of Patients in the LCI and LEC Cohort at the Time of Surgery.
| Clinical variable | LCI (N=247) |
LEC (N=139) |
P valuea |
|---|---|---|---|
| Etiology (HBV/HCV/HBV+HCV/non-viral/NAb) | (236/ 0/ 5/ 2/ 4) | (58/ 17/ 4/ 32/ 28) | <0.0001 |
| AVR-CCc (Yes/No/NA) | 62/ 179/ 6 | NA | NA |
| Gender (Male/Female/NA) | 214/ 31/ 2 | 102/ 37/ 0 | 0.0008 |
| Age (>=50years/<50years/NA) | 136/ 109/ 2 | 92/ 47/ 0 | 0.0515 |
| AFP (>300ng/mL/<=300ng/mL/NA) | 111/ 130/ 6 | 55/73/ 11 | 0.5844 |
| ALT (>50U/L/<=50U/L/NA) | 101/ 144/ 2 | NA | NA |
| Cirrhosis (Yes/No/NA) | 224/ 21/ 2 | 69/ 70/ 0 | <0.0001 |
| Tumor size (>5cm/<=5cm /NA) | 89/ 155/ 3 | 72 /67/ 0 | 0.0038 |
| Multinodular (Yes/No /NA) | 52/ 193/ 2 | NA | NA |
| Encapsulation (No/Yes/NA) | 114/ 129/ 4 | NA | NA |
| Microscopic vascular invasion (Yes/No/NA) | 107/ 90/ 50 | NA | NA |
| BCLC staging (B-C/A-0/NA) | 53/ 174/ 20 | NA | NA |
| CLIP staging (1–5/0/NA) | 128/ 99/ 20 | NA | NA |
| TNM staging (II-III/I/NA) | 130/ 97/ 20 | NA | NA |
| Survival at 60 Months (Events/Censored/NA) | 95/ 147/ 5 | 67/ 46/ 26 | <0.0001d |
Fisher’s exact test
NA: not available
AVR-CC: active viral replication chronic carrier
Log-rank test
We tested the genes of the metastasis risk classifier for their survival association. The survival risk prediction based on 10-fold cross-validation classified patients into low and high risk groups with a significant difference in survival as analyzed by Kaplan-Meier plot, with log-rank p values of p < 0.0001 and p = 0.0005 in LCI and LEC cohorts, respectively (Figure 1B and C top panel). The cross-validated misclassification rates were significantly lower than expected by chance (permutation p < 0.01; Figure 1). Similar results were observed when disease-free survival was used as an end point (Figure 1B and C bottom panel). Thus, this signature was independently validated as a classifier to predict survival in addition to metastasis.
We performed Cox proportional hazards regression analysis to determine whether the metastasis gene signature was confounded by the underlying clinical parameters. In univariate Cox analysis, the unadjusted hazard ratio for the overall survival in the high risk versus the low risk patient groups in the LCI cohort was 2.25 (95% CI = 1.48–4.5). The Cox analysis was stratified by several clinical factors and revealed that the AFP serum levels, underlying liver cirrhosis, tumor size, microscopic vascular invasion and tumor staging such as Barcelona Clinic Liver Cancer (BCLC), Cancer Liver Italian Program (CLIP) or Tumor Node Metastasis (TNM) were associated with overall survival (Table 2) (24–26). Multivariate Cox regression analysis accounting for the prognostic clinical factors that were significant in the univariate analysis revealed that the gene signature is an independent predictor of survival (Table 2). Only limited clinical information was available for the Cox regression analysis in the LEC cohort (Table 1). Analysis of the LEC cohort showed that the gene signature was a strong prognostic factor for patient survival with a hazard ratio of 2.59 (95% CI = 1.61–4.17). The univariate Cox regression analysis of the available clinicopathological data of the LEC cohort did not result in any significant clinical factor and thus no further multivariate analysis was performed (data not shown).
Table 2.
Univariate and multivariate Cox regression analysis of clinical factors associated with overall survival of the LCI cohort (N=242)a
| Clinical variable | Hazard Ratio (95% CIb) | P value |
|---|---|---|
| Univariate Analysisc | ||
| Predictor (high vs low risk) | 2.25 (1.48–4.5) | <0.001 |
| Gender (Male vs Female) | 1.86 (0.90–3.83) | 0.094 |
| Age (>= 50 years vs < 50 years) | 0.80 (0.53–1.19) | 0.209 |
| AFP (> 300ng/mL vs <= 300ng/mL) | 1.64 (1.10–2.45) | 0.016 |
| ALT (> 50U/L vs <= 50U/L) | 1.15 (0.77–1.73) | 0.483 |
| Cirrhosis (Yes vs No) | 5.09 (1.25–20.7) | 0.023 |
| Tumor size (> 5cm vs <= 5cm) | 2.01 (1.35–3.01) | 0.001 |
| Multinodular (Yes vs No) | 1.65 (1.06–2.57) | 0.025 |
| Encapsulation (No vs Yes) | 0.76 (0.50–1.14 | 0.181 |
| Microscopic vascular invasion (Yes vs No) | 1.97 (1.26–3.09) | 0.003 |
| HBV (AVR-CC vs CC)d | 1.36 (0.85–2.16) | 0.196 |
| BCLC staging (B–C vs A-0) | 3.69 (2.38–5.73) | <0.001 |
| CLIP staging (1–5 vs 0) | 2.19 (1.38–3.48) | 0.001 |
| TNM staging (II-III vs I) | 3.08 (1.88–5.05) | <0.001 |
| Multivariate Analysise | ||
| Predictor (high vs low risk) | 1.64 (1.03–2.60) | 0.038 |
| AFP (> 300ng/mL vs <= 300ng/mL) | 1.31 (0.84–2.03) | 0.237 |
| Cirrhosis (Yes vs No) | 3.91 (0.96–16.0) | 0.058 |
| TNM staging (II-III vs I) | 2.72 (1.64–4.51) | <0.001 |
| Multivariate Analysisf | ||
| Predictor (high vs low risk) | 1.91 (1.22–2.99) | 0.005 |
| AFP (> 300ng/mL vs <= 300ng/mL) | 0.74 (0.42–1.29) | 0.289 |
| CLIP staging (1–5 vs 0) | 2.38 (1.29–4.40) | 0.006 |
| Multivariate Analysisg | ||
| Predictor (high vs low risk) | 1.79 (1.14–2.82) | 0.012 |
| AFP (> 300ng/mL vs <= 300ng/mL) | 1.16 (0.75–1.80) | 0.542 |
| BCLC staging (B–C vs A-0) | 3.43 (2.19–5.36) | <0.001 |
Bold indicates significant P values.
Analysis was performed on the entire gene expression cohort.
95% CI, 95% confidence interval.
Univariate analysis, Cox proportional hazards regression.
AVR-CC (active viral replication chronic carrier); CC (chronic carrier).
Multivariate analysis, Cox proportional hazards regression adjusting for AFP status, cirrhosis and TNM staging.
Multivariate analysis, Cox proportional hazards regression adjusting for AFP status and CLIP staging.
Multivariate analysis, Cox proportional hazards regression adjusting for AFP status and BCLC staging.
Performance of the metastasis risk classifier
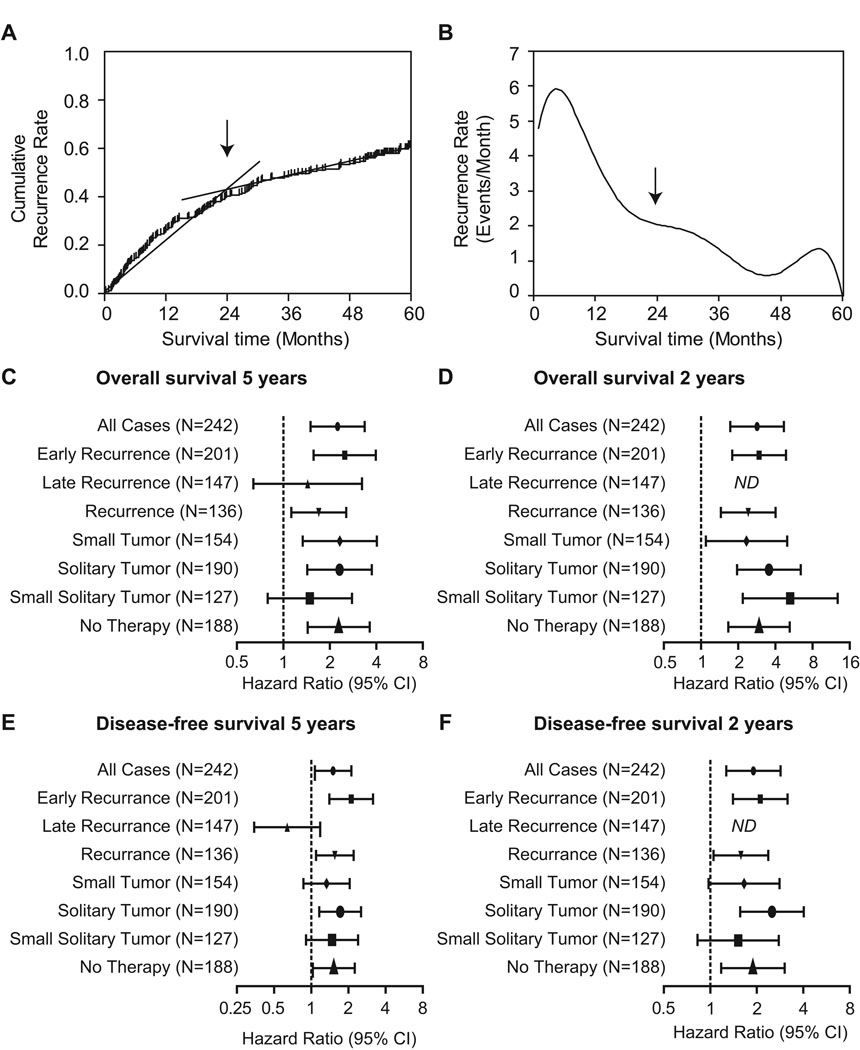
It has been suggested that there are two biologically different forms of HCC recurrence, i.e., early and late recurrence (5, 14–17). Early recurrence is believed to occur within the first two years after HCC treatment, mainly contributed by dissemination of metastatic HCC cells. In contrast, late recurrence is thought to originate de novo in the at risk liver and early recurrence is generally more common than late recurrence (18, 28). Consistently, when we analyzed the cumulative recurrence in the LCI cohort, we found that the HCC recurrence rate is biphasic (Figure 2A–B). The cumulative recurrence rate was 20.35% per year during the first two years after diagnosis, whereas, from the rate beyond two years after diagnosis decreased to 6.77% per year (Figure 2A). In agreement with these data, the recurrence rate peaked during the first year and persisted through the following years (Figure 2B). We did not analyze the LEC cohort due to the lack of sufficient recurrence data in this cohort.
Figure 2.
Analysis of the performance of the survival risk prediction dependent on HCC tumor recurrence over time after surgery. (A) Cumulative HCC recurrence rate over time. (B) Smoothed recurrence rate per month over time. (C) Forest plots showing Hazard Ratios for high risk patients in the indicated clinical groups of patients. Hazard Ratios are shown for the overall survival at 5 years, (D) the overall survival at 2 years, (E) the disease-free survival at 5 years and (F) the disease-free survival at 2 years of follow-up of the high risk subgroup as compared with the low risk group. Hazard ratios above 1.0 indicate significantly worse outcome. ND, not determined.
To study the prognostic capacity of the metastasis risk classifier with respect to the time of recurrence, we compared the hazards ratios of patient groups with early and late recurrence. The metastasis risk classifier significantly predicted overall survival and disease-free survival only in patients with early, but not with late recurrence (Figure 2C–F). In addition, the classifier was not affected by postoperative adjuvant therapy and was able to predict overall survival within the first two years in patients with small solitary tumors (tumor size ≤ 5cm; Figure 2D). These results are consistent with the hypothesis that early and late recurrence differ in their gene expression profiles and indicate that the metastasis risk classifier is only applicable to metastasis-related relapse and can be used to classify early HCC recurrence.
Independent Cross-Validation and Analysis of Sensitivity and Specificity
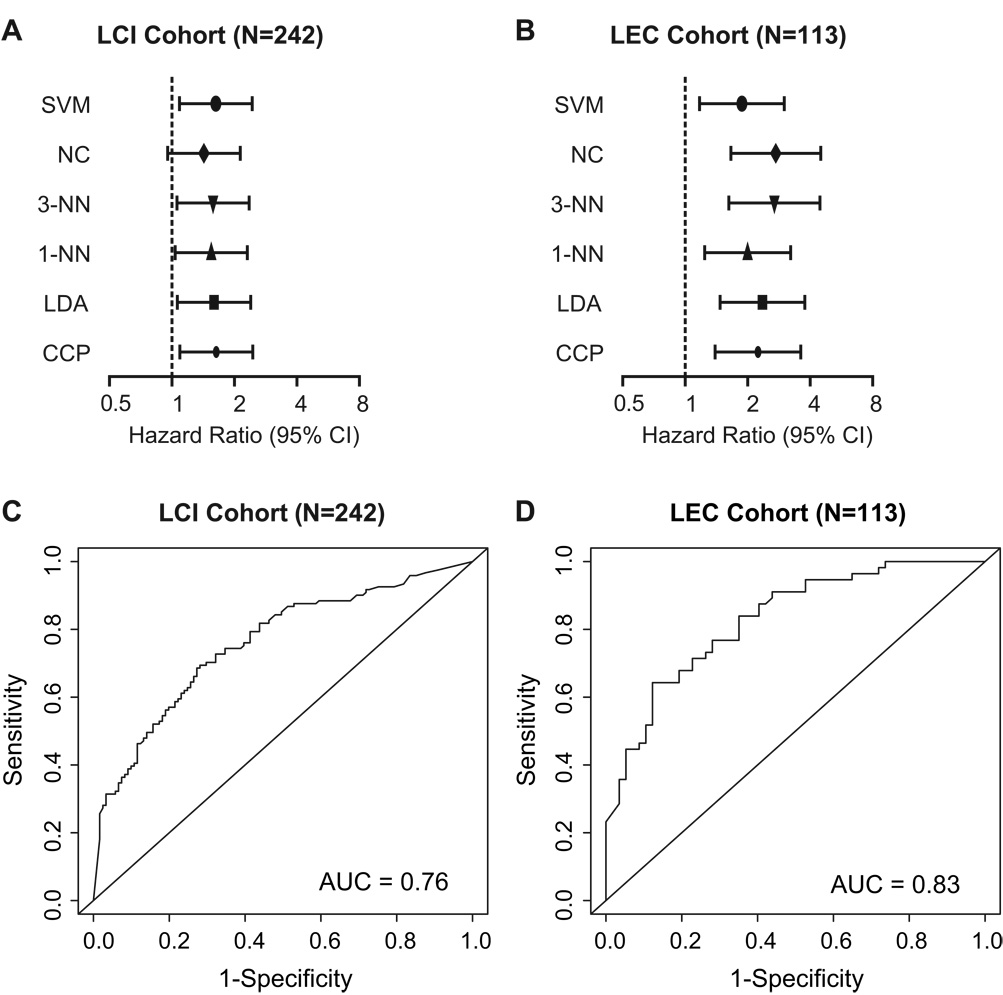
In order to determine if the signature has any practical measure, we performed a new sample assignment/prediction simulation strategy by independently cross-validating the two cohorts. We converted the gene expression data of both cohorts into z-scores. The resulting survival risk prediction was then used for unbiased cross-validation of both cohorts. We used six class prediction algorithms, Support Vector Machines (SVM), Nearest Centroid (NC), 3-Nearest Neighbor (3-NN), 1-Nearest Neighbor (1-NN), Linear Discriminant Analysis (LDA) or Compound Covariate Predictor (CCP), to predict good and poor survival HCC subgroups. To assess outcome prediction, we used one of the cohorts as a template and the second cohort as an independent validation set and vice versa. After using the LEC cohort as the template cohort and the LCI cohort as the validation cohort, Cox proportional hazards regression analysis showed that five out of the six prediction algorithms were able to significantly predict outcome (Figure 3A). Next, we used the LCI cohort as template and the LEC cohort as validation cohort and found that all six prediction algorithms significantly predicted the patient outcome (Figure 3B). Therefore, even though the two cohorts differed in their patient characteristics and were analyzed on two different microarray platforms, the metastasis risk classifier was consistently able to predict the survival. Of note, both cohorts could prospectively serve as templates for patient classification in the future. Receiver operating characteristic (ROC) curves showed that the predictive accuracy of the Compound Covariate Predictor had high sensitivity (i.e. a low probability of falsely classifying a patient as low risk; The sensitivity is 0.760 and 0.839 for the LCI and LEC cohort, respectively.) and good specificity (i.e. a low probability of falsely classifying a patient as high risk; The specificity is 0.603 and 0.649 for the LCI and LEC cohort, respectively.) in both cohorts (Figure 3C and D).
Figure 3.
Unbiased cross-validation of the survival risk prediction and analysis of the sensitivity and specificity by Receiver Operating Characteristic (ROC) curves. (A) Six class prediction algorithms, i.e., Support Vector Machines (SVM), Nearest Centroid (NC), 3-Nearest Neighbor (3-NN), 1-Nearest Neighbor (1-NN), Linear Discriminant Analysis (LDA) or Compound Covariate Predictor (CCP) were used to predict good and poor survival HCC groups in the independent validation data set. Forest plots show Hazard Ratios for high risk patients in clinical groups of patients. Hazard Ratios are shown for the overall survival for the LCI cohort at 5 years using the LEC cohort as a training/test set and predicting outcome in the LCI cohort. (B) Hazard Ratios of the LEC cohort are shown using LCI as the training/test set and prediction of the LEC cohort are depicted. (C) ROC curve of the LCI cohort and (D) ROC curve of the LEC cohort applying the compound covariate predictor. AUC; area under the curve. SVM, Support Vector Machines, NC, Nearest Centroid, 3-NN, 3-Nearest Neighbor, 1-NN, 1-Nearest Neighbor, LDA, Linear Discriminant Analysis. CCP, Compound Covariate Predictor.
Improving Prediction by Combining Clinical Prognostic Factors and the Metastasis Risk Classifier
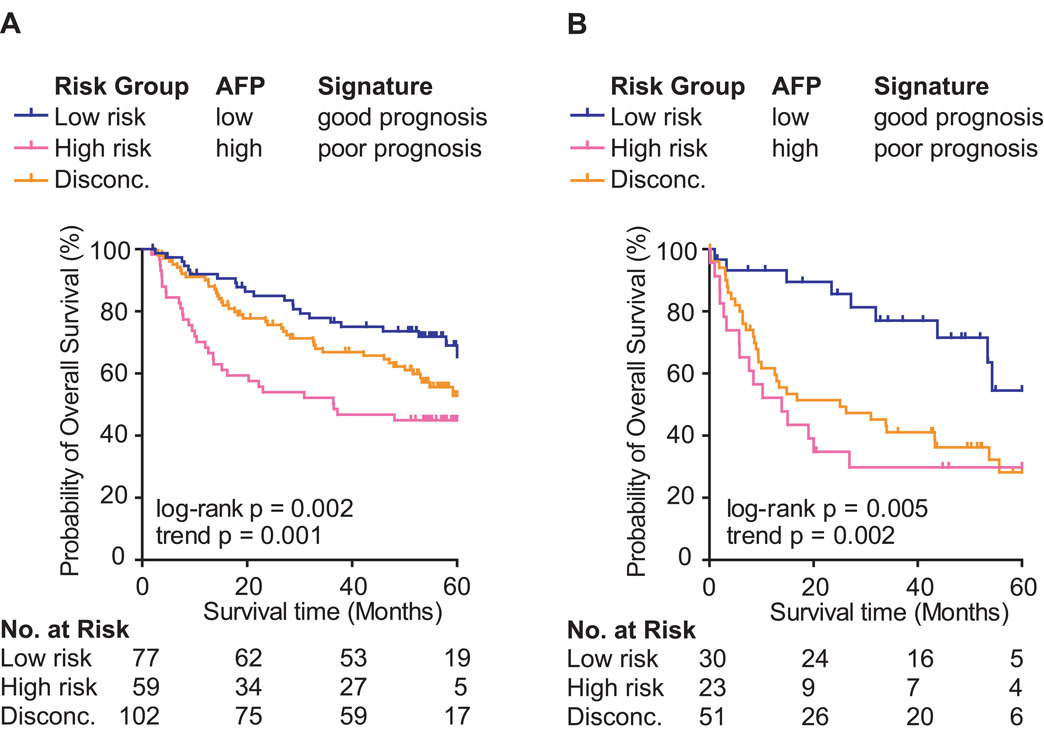
Currently, the only clinically available marker for HCC is alpha fetoprotein (AFP), whose serum levels have been linked to HCC prognosis (20, 29) (Table 2 and Figure S1). We sought to determine whether prognostic prediction of the LCI cohort (Figure 4A) and the LEC cohort (Figure 4B) could be improved by combining AFP and the metastasis risk classifier. We divided patients into subgroups based on an AFP level cutoff of 300 ng/mL and the survival risk determined by the metastasis risk classifier (Figure 4). This resulted in three outcome groups (low risk, high risk and discordant). While the low and high risk patients were both classified into the same outcome groups by AFP and the gene classifier, it appeared that there was a subset of patients misclassified by both methods (discordant cases, i.e., high risk according to the metastasis risk classification and low risk prediction by AFP or vice versa). Kaplan-Meier survival analysis showed that patients with discordant risk prediction have poorer outcome than low risk patients and therefore, might benefit from more rigid therapies. Stratification of discordant cases revealed that neither the gene signature nor AFP is a stronger predictor but that the combination of the gene signature classifier with AFP may improved prediction outcome (Figure S2).
Figure 4.
Combination of survival risk prediction applying the Compound Covariate Predictor (CCP) and AFP (300 ng/mL cutoff) to stratify patient subgroups. (A) The Kaplan-Meier curves show overall survival of the LCI cohort (N = 238) and (B) LEC cohort (N = 104) sub-grouped by survival risk prediction and AFP. Disconc.: cases with discordant risk assessments, i.e., high risk according to the metastasis risk classification and low risk prediction by AFP, i.e. AFP less than 300 ng/mL.
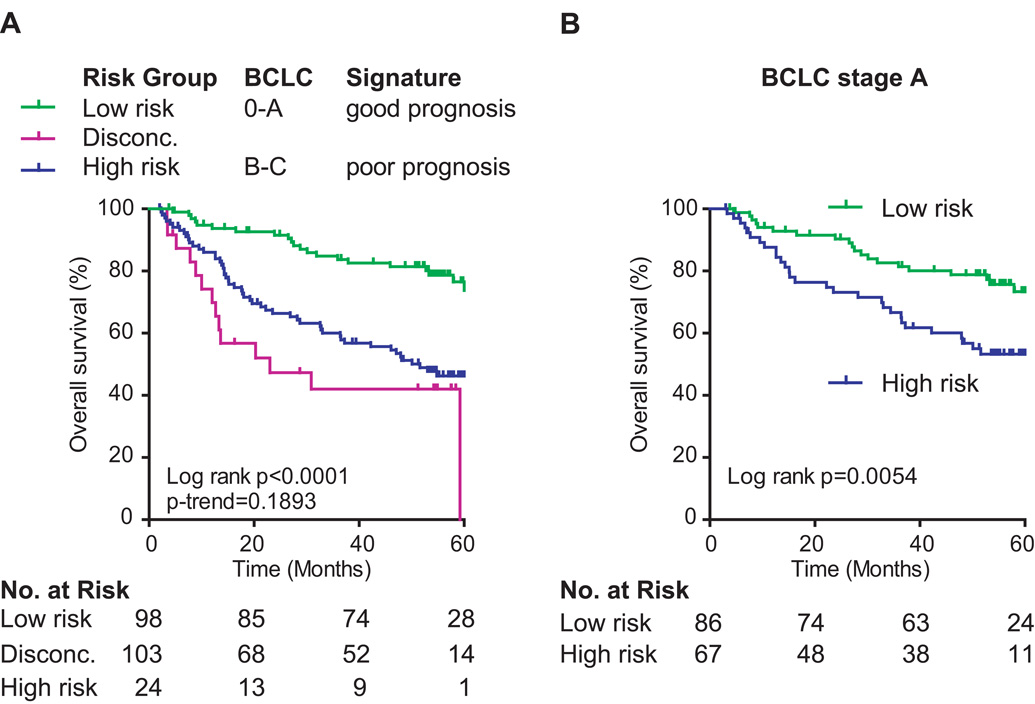
We also sought to determine if the gene classifier can improve BCLC staging as both were independent predictors of HCC survival. BCLC staging, which includes tumor size and liver function, is frequently used in the clinic to determine treatment options. BCLC stage A includes early stage HCC patients with single tumors or three tumors smaller than 3 cm and Child-Pugh class A–B. Patients with BCLC stage A are suitable for radical therapies such as resection, transplantation or percutaneous treatments. We only performed analyses on the LCI cohort since the LEC cohort lacks BCLC staging data. Similarly to the results obtained with AFP, we found that the metastasis risk classifier improved survival prediction when combined with BCLC (Figure 5A). Importantly, the gene signature was capable of significantly stratifying patients into low and high risk groups, especially among those with early stage HCC as defined by BCLC stage A (Figure 5B and Figure S3). Therefore, these results confirmed that the gene signature can significantly improve BCLC recurrence risk assessment. Taken together, combination of the recurrence-risk classifier with clinical staging as a molecular diagnostic test might be clinically useful to improve recurrence risk prediction and to determine treament modality, particularly for those with early stage tumors and solitary presentation.
Figure 5. A.
Patient stratification using survival risk prediction and BCLC staging. (A) Kaplan-Meier curves are showing overall survival of the LCI cohort (N = 225) by sub-grouping according to CCP class prediction of good or poor prognosis and BCLC stage 0-A or B–C. (B) Kaplan-Meier curves of patients with BCLC staging A (N = 153) stratified by CCP survival risk prediction. Disconc.: cases with discordant risk assessments, i.e., high risk according to the metastasis risk classification and early stage prediction by BCLC.
DISCUSSION
Recurrence is a common post-surgical event contributing to the poor prognosis of HCC patients. Currently there are few effective therapeutic options to reduce metastasis-related recurrence. This is due, in part, to our inability to identify in advance the subgroup of HCC patients that are at high risk of developing metastatic disease. Risk stratification is particularly important for those patients with early stage of HCC who do not have vascular invasion and regional tumor cell dissemination at the time of diagnosis. This problem has hindered our ability to identify a specific therapeutic regimen that could improve the outcome of HCC since no ‘one-size-fits-all’ therapeutic strategy has been shown to be effective. Recent findings from two phase III randomized control trials on the use of Sorafenib as a therapeutic agent for advanced HCC are encouraging, but the survival benefit appears modest and its value in the prevention and treatment of postoperative metastatic recurrence are still under investigation (30, 31). There is an urgent need to develop genetic profiling tools to stratify patients with respect to prognosis and response to therapy, an essential step towards personalized medicine-based cancer management. For this purpose, we recently identified miR-26 as a biomarker to predict HCC survival and response to adjuvant IFN therapy (32).
The traditional tumor evolution model suggests that a primary tumor is initially benign and over time acquires mutations that give a few tumor cells the ability to metastasize (8, 33). If a tumor is detected and treated before it spreads, the chances of long-term survival should be increased (34). Therefore, early detection is crucial to improve patient outcome. However, recent publications show that even if tumors are detected early, they might have already completed most of the steps on their way to metastasis (35). For example, genome analyses of primary colon tumors and paired metastases suggest that the genetic machinery that causes metastases may be hard-wired into the tumor from the beginning (36). Similarly, copy number analysis of prostate cancers and their metastasis revealed that lethal metastatic prostate cancer is of monoclonal origin and that most metastatic cancers arise from a single cell (37). Consistently, our recent studies revealed that global gene expression patterns are very similar between primary HCCs and their paired metastases (19). These results provide a rationale for profiling primary tumors to predict patient prognosis.
In this study, we have validated our recently identified metastasis risk classifier by profiling primary HCC tissues in two independent cohorts with mixed etiologies as a tool to predict recurrence and survival attributed to metastatic HCC. Multivariate analyses including various clinical risk factors and clinical staging indicate that the molecular classifier is an independent prognostic predictor, especially applicable to early recurrence, a poor prognostic factor mainly associated with metastatic dissemination of HCC cells, but not late recurrence, an outcome contributed mainly by high carcinogenic activities of diseased livers. These results indicate that early and late recurrences differ in their molecular profile. Importantly, the gene classifier could predict poor outcome in patients with small solitary tumors, which has been traditionally viewed as having low risk for tumor recurrence. Therefore, the metastasis risk classifier adds independent prognostic value to the recurrence risk assessment, especially in early stage HCC patients where current clinical staging fails to provide an accurate assessment. The ability to identify patients with high risk for recurrence in advance would reduce unnecessary economic burden and side effects for those low risk patients who may not benefit from these treatments.
Since our molecular signature is independent of other prognostic clinical factors, we also tested whether an improved prediction can be achieved by combining the signature with clinically relevant serum AFP or tumor staging (38, 39). Our data confirmed this hypothesis in two independent cohorts when the gene signature was combined with AFP. Encouraging results were also obtained in the LCI cohort in which the gene classifier improves HCC survival prediction when combined with BCLC staging, especially for those with early stage HCC. These data require further validation in additional cohorts as tumor staging data was not available in the LEI cohort. As the combination of the metastasis risk classifier and either AFP or BCLC staging leads to the identification of discordant cases which have poorer outcome than low risk cases we suggest that patients with discordant risk prediction should receive more rigid therapies.
It should be noted that osteopontin (OPN) was the top ranked gene in our classifier (i.e. the most highly over expressed in metastatic HCC) (19). Further studies indicated that OPN may be a potential therapeutic target for metastatic HCC as inhibition of OPN by neutralizing antibody, small peptides or lentivirus-mediated RNA interference can block HCC cell invasion in vitro and inhibited pulmonary metastasis in mice (19, 40, 41). Further studies are warranted to determine whether the application of the metastasis risk classifier in combination with novel agents such as inhibitors of OPN can improve HCC outcome.
For other cancer types, there are already gene classifiers used in the clinic. For breast cancer, there are two commercial reference laboratory tests based on gene-expression profiling (MammaPrint® and Oncotype DX®) that are either agency-approved or widely-accepted by the oncology community (42–44). The Oncotype DX®assay measures the expression of 16 genes by qRT-PCR and requires a routinely processed formalin-fixed paraffin embedded tumor tissue block. The MammaPrint® assay measures the expression of 70 genes with a microarray and requires snap frozen tumor tissue or fresh tumor tissue procured in a special buffer. Therefore, we suggest that the metastasis gene signature, similarly to the MammaPrint® assay, can be used as an assay in the clinic since we showed that the metastasis gene signature can be applied to different microarray platforms.
In conclusion, we have validated the metastasis risk classifier as a tool to predict HCC outcome in two independent cohorts with mixed etiologies and ethnicity, suggesting the general utility of this classifier. In addition, the gene classifier was able to predict disease-free survival and early recurrence. In combination with serum AFP levels or BCLC staging, the gene classifier may improve survival risk prediction. Thus, we recommend the use of this classifer as a molecular diagnostic test to assess the recurrence risk of HCC patients, particularly those with early stage after curative resection.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the microarray core at the NCI-SAIC for help on high-through-put microarray analysis; Curtis Harris for critical reading of the manuscript; Dr. Richard Simon for statistical advice; Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team.; the NIH Fellows Editorial Board for editing the manuscript; Karen MacPherson for bibliographic assistance.
Financial Support: This work was supported by the intramural Research Program of the Center for Cancer Research, the US National Cancer Institute (Z01-BC 010313 and Z01-BC 010876), and by China National Key Projects for Infectious Disease (2008ZX10002-021) and the State Key Basic Research Program of China (2009CB521701).
Abbreviations
- HCC
hepatocellular carcinoma
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- AFP
alpha fetoprotein
- LEC
Laboratory of Experimental Carcinogenesis
- LCI
Liver Cancer Institute
- TNM
Tumor Node Metastasis
- CLIP
Cancer Liver Italian Program
- BCLC
Barcelona Clinic Liver Cancer
- SVM
Support Vector Machines
- NC
Nearest Centroid
- 3-NN
3-Nearest Neighbor
- 1-NN
1-Nearest Neighbor
- LDA
Linear Discriminant Analysis
- CCP
Compound Covariate Predictor
- OPN
osteopontin
- HR
hazard ratio
- CI
confidence intervals
- ROC
receiver operating characteristic
- AUC
area under the curve
Footnotes
Potential conflict of interest: Nothing to report.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular Carcinoma Incidence, Mortality, and Survival Trends in the United States From 1975 to 2005. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 4.Libbrecht L, Craninx M, Nevens F, Desmet V, Roskams T. Predictive value of liver cell dysplasia for development of hepatocellular carcinoma in patients with non-cirrhotic and cirrhotic chronic viral hepatitis. Histopathology. 2001;39:66–73. doi: 10.1046/j.1365-2559.2001.01172.x. [DOI] [PubMed] [Google Scholar]
- 5.Sherman M. Recurrence of hepatocellular carcinoma. N Engl J Med. 2008;359:2045–2047. doi: 10.1056/NEJMe0807581. [DOI] [PubMed] [Google Scholar]
- 6.Tang ZY. Hepatocellular carcinoma-Cause, treatment and metastasis. World J Gastroenterol. 2001;7:445–454. doi: 10.3748/wjg.v7.i4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–1236. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 9.Cha C, Fong Y, Jarnagin WR, Blumgart LH, DeMatteo RP. Predictors and patterns of recurrence after resection of hepatocellular carcinoma. J Am Coll Surg. 2003;197:753–758. doi: 10.1016/j.jamcollsurg.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 10.El Assal ON, Yamanoi A, Soda Y, Yamaguchi M, Yu L, Nagasue N. Proposal of invasiveness score to predict recurrence and survival after curative hepatic resection for hepatocellular carcinoma. Surgery. 1997;122:571–577. doi: 10.1016/s0039-6060(97)90130-6. [DOI] [PubMed] [Google Scholar]
- 11.Kanematsu T, Matsumata T, Takenaka K, Yoshida Y, Higashi H, Sugimachi K. Clinical management of recurrent hepatocellular carcinoma after primary resection. Br J Surg. 1988;75:203–206. doi: 10.1002/bjs.1800750305. [DOI] [PubMed] [Google Scholar]
- 12.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, et al. Gene Expression in Fixed Tissues and Outcome in Hepatocellular Carcinoma. N Engl J Med. 2008 doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoshida Y, Villanueva A, Llovet JM. Molecular profiling to predict hepatocellular carcinoma outcome. Expert Rev Gastroenterol Hepatol. 2009;3:101–103. doi: 10.1586/egh.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imamura H, Matsuyama Y, Tanaka E, Ohkubo T, Hasegawa K, Miyagawa S, et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol. 2003;38:200–207. doi: 10.1016/s0168-8278(02)00360-4. [DOI] [PubMed] [Google Scholar]
- 15.Portolani N, Coniglio A, Ghidoni S, Giovanelli M, Benetti A, Tiberio GA, et al. Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann Surg. 2006;243:229–235. doi: 10.1097/01.sla.0000197706.21803.a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poon RT, Fan ST, Ng IO, Lo CM, Liu CL, Wong J. Different risk factors and prognosis for early and late intrahepatic recurrence after resection of hepatocellular carcinoma. Cancer. 2000;89:500–507. [PubMed] [Google Scholar]
- 17.Poon RT. Differentiating early and late recurrences after resection of HCC in cirrhotic patients: implications on surveillance, prevention, and treatment strategies. Ann Surg Oncol. 2009;16:792–794. doi: 10.1245/s10434-009-0330-y. [DOI] [PubMed] [Google Scholar]
- 18.Chen YJ, Yeh SH, Chen JT, Wu CC, Hsu MT, Tsai SF, et al. Chromosomal changes and clonality relationship between primary and recurrent hepatocellular carcinoma. Gastroenterology. 2000;119:431–440. doi: 10.1053/gast.2000.9373. [DOI] [PubMed] [Google Scholar]
- 19.Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–423. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 20.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 21.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 22.R Development Core Team. R. A language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 23.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 24.The Cancer of the Liver Italian Program (CLIP) investigators. A new prognostic system for hepatocellular carcinoma: a retrospective study of 435 patients: the Cancer of the Liver Italian Program (CLIP) investigators. Hepatology. 1998;28:751–755. doi: 10.1002/hep.510280322. [DOI] [PubMed] [Google Scholar]
- 25.Llovet JM, Bru C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 26.International Union Against Cancer (UICC) TNM Classification of Malignant Tumours. 6th Edition. Hoboken, NJ: John Wiley & Sons; 2002. [Google Scholar]
- 27.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 28.Wu JC, Huang YH, Chau GY, Su CW, Lai CR, Lee PC, et al. Risk factors for early and late recurrence in hepatitis B-related hepatocellular carcinoma. J Hepatol. 2009;51:890–897. doi: 10.1016/j.jhep.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 30.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 31.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 32.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 34.Etzioni R, Penson DF, Legler JM, di Tommaso D, Boer R, Gann PH, et al. Overdiagnosis due to prostate-specific antigen screening: lessons from U.S. prostate cancer incidence trends. J Natl Cancer Inst. 2002;94:981–990. doi: 10.1093/jnci/94.13.981. [DOI] [PubMed] [Google Scholar]
- 35.Dong F, Budhu AS, Wang XW. Translating the metastasis paradigm from scientific theory to clinical oncology. Clin Cancer Res. 2009;15:2588–2596. doi: 10.1158/1078-0432.CCR-08-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wildi S, Pestalozzi BC, McCormack L, Clavien PA. Critical evaluation of the different staging systems for hepatocellular carcinoma. Br J Surg. 2004;91:400–408. doi: 10.1002/bjs.4554. [DOI] [PubMed] [Google Scholar]
- 39.Cillo U, Bassanello M, Vitale A, Grigoletto FA, Burra P, Fagiuoli S, et al. The critical issue of hepatocellular carcinoma prognostic classification: which is the best tool available? J Hepatol. 2004;40:124–131. doi: 10.1016/j.jhep.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 40.Sun BS, Dong QZ, Ye QH, Sun HJ, Jia HL, Zhu XQ, et al. Lentiviral-mediated miRNA against osteopontin suppresses tumor growth and metastasis of human hepatocellular carcinoma. Hepatology. 2008;48:1834–1842. doi: 10.1002/hep.22531. [DOI] [PubMed] [Google Scholar]
- 41.Takafuji V, Forgues M, Unsworth E, Goldsmith P, Wang XW. An osteopontin fragment is essential for tumor cell invasion in hepatocellular carcinoma. Oncogene. 2007;26:6361–6371. doi: 10.1038/sj.onc.1210463. [DOI] [PubMed] [Google Scholar]
- 42.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 43.Van de Vijver MJ, He YD, Van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 44.Kim C, Paik S. Gene-expression-based prognostic assays for breast cancer. Nat Rev Clin Oncol. 2010;7:340–347. doi: 10.1038/nrclinonc.2010.61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.