Abstract
The most common chromosomal abnormalities associated with autism are 15q11–q13 duplications. Maternally derived or inherited duplications of 15q pose a substantial risk for an autism phenotype, while paternally derived duplications may be incompletely penetrant or result in other neurodevelopmental problems. Therefore, the determination of maternal versus paternal origin of this duplication is important for early intervention therapies and for appropriate genetic counseling to the families. We adapted a previous single-reaction tube assay (high-resolution melting curve analysis) to determine the parent of origin of 15q duplications in 28 interstitial duplication 15q samples, one family and two isodicentric subjects. Our method distinguished parent origin in 92% of the independent samples as well as in the familial inherited duplication and in the two isodicentric samples. This method accurately determines parental origin of the duplicated segment and measures the dosage of these alleles in the sample. In addition, it can be performed on samples where parental DNA is not available for microsatellite analysis. The development of this single-tube assay will make it easier for genetic testing laboratories to provide parent-of-origin information and will provide important information to clinical geneticists about autism risk in these individuals.
Introduction
The most common chromosomal abnormalities associated with autism are duplications of the proximal region of chromosome 15q encompassing the imprinted Prader-Willi/Angelman syndrome (PWS/AS) locus. As array comparative genomic hybridization (arrayCGH) methods have become more commonplace in the clinical setting, the frequency of detection of submicroscopic interstitial 15q duplications continues to increase (Wang et al., 2004; Depienne et al., 2009). The PWS/AS locus contains an imprinting center that displays parent-of-origin-specific methylation at a CpG island in the promoter of the SNRPN gene (Glenn et al., 1996). This regulation restricts expression of the HBII-85 C/D box small nucleolar RNA cluster on the maternal allele (Sahoo et al., 2008), while the UBE3A gene is silenced on the paternal allele (Horsthemke and Wagstaff, 2008). As with many other loci throughout the human genome, this imprinted locus is flanked by low copy repeats that can recombine during nonhomologous allelic recombination events resulting in deletions that cause either PWS or AS and duplications that can result in an autism phenotype, particularly when maternally derived.
For patients with segmental aneuploidies of chromosome 15q11.2–q13, either deletions or duplications, parent of origin of the involved segment is of critical importance because dosage of the imprinted PWS/AS critical region is affected. In PWS and AS, assays are available to identify the methylation state of the deleted chromosome; however, it is frequently not feasible to clearly determine parent of origin for patients with duplication using molecular assays available clinically. This is an important limitation to the current methodologies because there is tremendous variation in phenotypic risks for individuals with maternal versus paternal chromosome 15q duplications. There is strong evidence to suggest that maternally derived or inherited duplications of 15q pose a substantial risk for an autism spectrum disorder (ASD) phenotype, while paternally derived duplications may be incompletely penetrant in association with neurodevelopmental symptoms or in some cases are benign. The determination of maternal versus paternal imprinting status of this duplication is therefore clinically important in terms of both early intervention therapies for autism and also for appropriate genetic counseling of unaffected individuals who may carry a paternally derived or inherited 15q interstitial duplication.
Currently, for duplications, parent-of-origin testing is done in a research setting only and may require some degree of trial and error, which is not acceptable in a clinical testing lab. Current methods for parent-of-origin detection include the use of methylation-specific polymerase chain reaction (PCR) of the SNRPN exon α locus or amplification of DNA polymorphisms from the proband and parents. These methods are semiquantitative and/or require parental DNA samples, which are not always available. A variety of methods have been developed to identify deletions and methylation defects associated with PWS and AS, including the use of 21 differentially methylated CpG dinucleotides at the SNRPN locus as a diagnostic test for PWS (Kubota et al., 1996). Recently, a method was described using high-resolution melting (HRM) curve analysis to detect and distinguish the PWS/AS deletion as well as uniparental disomy (UPD) cases (White et al., 2007). Here we have applied this methodology to develop a single-reaction tube assay to determine the parent of origin of 15q duplications. We show that this method will accurately detect interstitial as well as isodicentric duplications of 15q, including stably inherited duplications, and can be performed on samples where parental DNA is not available.
Materials and Methods
DNA samples
All experiments on human samples were performed in compliance with the University of Tennessee Health Science Center Institutional Review Board (IRB). De-identified 15q duplication samples (n = 28) including cases of inherited 15q duplication were obtained from Signature Genomic Laboratories (under a protocol approved by the Spokane IRB). Additional samples from familial inherited interstitial 15q duplication and isodicentric samples (n = 4) were provided by E.H.C. and N.C.S. An AS Class I deletion sample (one allele with no methylation), PWS maternal UPD sample (both alleles methylated), and nonduplicated healthy subjects (n = 15) (one allele methylated and one allele not methylated) were used as controls for comparison to our 15q duplication samples. All 15q duplication samples were previously tested by arrayCGH and in some cases fluorescence in situ hybridization (FISH) to confirm the presence of either interstitial or isodicentric duplications of chromosome 15q encompassing the AS/PWS locus.
HRM analysis
Five hundred nanograms of patient genomic DNA was sodium bisulfite treated using the EZ DNA Methylation Kit (Zymo Research), in accordance to the manufacturer's protocol. After bisulfite treatment ∼50 ng of DNA was amplified, using primers specific for bisulfite-converted DNA and conditions from White et al. (2007). PCR was performed in 12.5 μL using the LightCycler 480 High Resolution Melting Master (Roche), with 2.5 mM MgCl2 and 2 pmol primers. PCR and methylation sensitive high resolution melting (MS-HRM) were performed in the LightCycler 480 (Roche Molecular Diagnostics) machine. Each reaction was performed in duplicate. The cycling conditions were as follows: 1 cycle of 95°C for 10 min; 40 cycles of 95°C for 10 s, 55°C for 14 s, and 72°C for 24 s, followed by an HRM step of 95°C for 1 min, 40°C for 1 min, 60°C for 0.01 s, and continuous acquisition to 95°C at 25 acquisitions per 1°C. Data were analyzed with LightCycler software version 1.5 using the melting curve analysis option followed by normalization from the gene scanning options.
Results
HRM curve assay design and controls
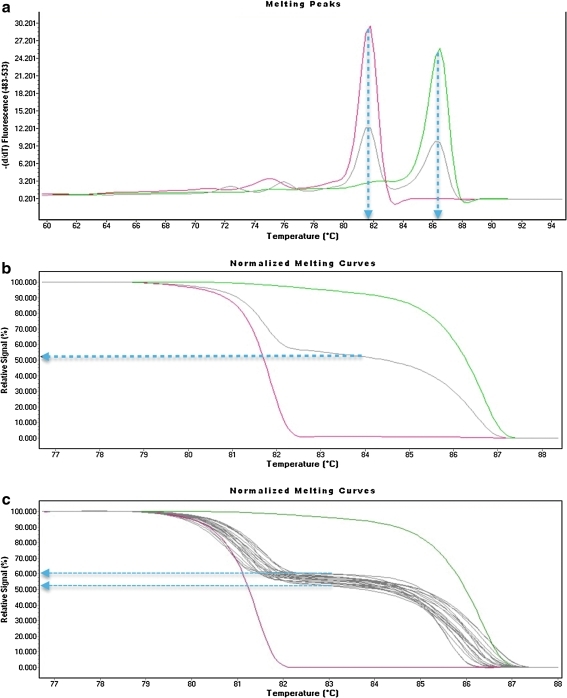
The basis of our methylation-sensitive HRM curve assay is a study by White et al. (2007) demonstrating that in the case of either deletion or UPD for the SNRPN gene, it would be possible to determine the parent of origin of the remaining homoallelic region using a primer set that amplifies the bisulfite-converted residues resulting in a single but distinct melting curve for AS and PWS. Since the reciprocal interstitial duplication of 15q should produce three alleles (one from the nonduplicated chromosome and two from the duplicated chromosome), we first needed to determine if both of these chromosomes could be detected in normal control samples. Indeed, two peaks of approximately equal fluorescent intensity could be detected in normal controls (Fig. 1a). These peaks matched the melting point curves for the paternal (methylated) DNA from an AS deletion sample and the maternal (unmethylated) DNA from a PWS UPD sample (Fig. 1a). The midpoint for the melting curves were ∼81.7°C for the paternal allele and ∼86.2°C for the maternal allele. Normalization of the overall fluorescent intensity for both peaks reveals that ∼52% of the control signal originates from the maternal allele and ∼48% of the signal from the paternal allele (Fig. 1b). It should be noted that only in control samples did we also observe two small melting curve peaks at ∼74°C and 76°C. HRM analysis of an additional 14 healthy nonduplication controls revealed a tight clustering of control normalized signal intensity between 52% and 60% (Fig. 1c).
FIG. 1.
SNRPN quantitative HRM analysis in control samples. The colors indicate HRM analysis in AS deletion (red), PWS UPD (dark green), and a nonduplication control sample (gray). (a) Melting peaks indicate one peak at ∼81.7°C for the AS deletion sample that has no maternal allele and one peak at ∼86.2°C for PWS UPD that has no paternal allele. The normal control shows two peaks of approximate equal intensity at ∼81.7°C for the paternal allele and ∼86.2°C for the maternal allele. (b) The normalized melting profiles indicate that 100% of the relative signal intensity comes from the ∼81.7°C melting curve in the AS deletion sample, while 100% of the signal comes from the ∼86.2°C melting curve in the PWS UPD. A blue line drawn from the midpoint between the two melt curves in the nonduplication control sample indicates that ∼52% of the signal comes from the paternal melting curve (∼86.2°C) and 48% of the signal comes from the maternal melting curve (∼81.7°C). (c) A quantitative HRM analysis of multiple nonduplication control samples is shown, and although some variation in profiles could be detected, there was a very tight range of relative signal intensity in 15 nonduplication control samples between 52% and 60% (blue arrows). The individual melting curves ranged between 80.5°C and 82.3°C (average 81.4°C) for the paternal allele and 85°C–87°C (average 86°C) for the maternal allele. These values essentially match the peak intensity melt temperatures observed in (a) for the control. HRM, high-resolution melting; PWS/AS, Prader-Willi/Angelman syndrome; UPD, uniparental disomy.
HRM method distinguishes parent of origin in 92% of independent 15q duplication samples
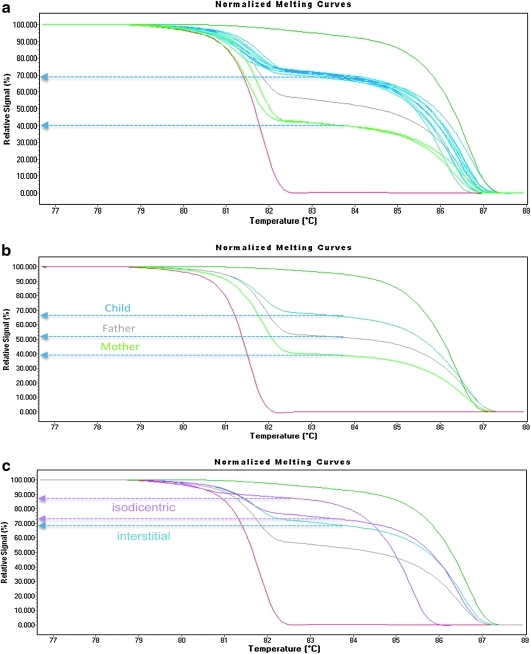
We obtained 28 DNA samples from individuals with an interstitial duplication of 15q as determined by targeted arrayCGH analysis from Signature Genomics Laboratories. Methylation-sensitive HRM analysis of these samples revealed the parent of origin clearly in 26/28 samples tested (Fig. 2a). The majority of these duplications were of maternal origin (24) with a maternal-to-paternal allele ratio of 2.2:1 (Fig. 2a). Two samples showed an absolute signal intensity that indicates the signal from the maternal allele melting curve to be at a lower ratio than the paternal allele 0.67:1. These two samples represent paternally methylated duplications and are less likely to display the ASD phenotype (although phenotypic data were unavailable for analysis). Two samples were shifted to the left in comparison to the others samples, and although the signal intensity matched normal control samples, we did not assign them to a category (data not shown). In practice, the measure of absolute percent signal intensity is indicative of a maternal methylation state of the duplication when the midpoint between the two DNA melting curves is 66% or higher and a paternal duplication when the midpoint is 40% or lower.
FIG. 2.
Quantitative identification of maternal and paternal 15q duplications. Arrow heads indicate relative signal intensity between the two melting curves. (a) Quantitative HRM determination of parental origin using normalized melt profiles. Bisulfite-treated DNA samples from 28 individuals with interstitial duplications of unknown origin were analyzed by quantitative HRM and compared to AS deletion (red) and PWS-UPD (dark green) controls. One nonduplicated control DNA sample is indicated in gray at ∼50% relative signal intensity. Normalized melting profiles revealed a clear grouping of 26 maternal duplication samples (blue) at an average relative signal intensity of ∼66% and a grouping of 2 paternal duplication samples (green) at a relative signal intensity of ∼40%. (b) Detection of parental origin in stable inherited interstitial 15q duplications. HRM normalization analysis in a family, where the child (blue = 66% relative signal intensity) has inherited a maternal duplication and his mother (green = 39% relative signal intensity) a duplication of paternal origin. As expected, the father, who does not have a chromosomal duplication, showed a relative signal intensity of 52%, within the 52%–60% range found in normal controls. (c) Quantitative HRM can determine both parental origin and distinguish between interstitial and isodicentric duplications on 15q. The colors indicate HRM analysis in AS deletion (red), PWS UPD (dark green), nonduplication control samples (gray), and maternal interstitial duplication (blue). The two known isodicentric duplication samples are indicated in pink. One sample had an HRM relative signal intensity of 73%, indicating a 2.7:1 ratio of maternal to paternal signal, while the other had a signal intensity of 87%, consistent with a 6.7:1 maternal-to-paternal ratio. Both isodicentric 15q duplication samples showed relative signal intensities >66%, the observed interstitial duplication relative signal intensity.
Stably inherited interstitial 15q duplications
Although duplications of 15q are de novo in the majority of cases reported (Hogart et al., 2008), there have been documented cases of a paternally derived interstitial duplication chromosome passing through the maternal germline and resulting in an ASD phenotype in the child (Cook et al., 1997). We wanted to determine if stable, inherited interstitial duplications could also be easily distinguished using the HRM method since this duplicated sequence would presumably be identical in the mother and the child with the exception of epigenetic modification of the differentially methylated SNRPN locus. A family with a stable maternally inherited interstitial duplication was used for HRM analysis (Fig. 2b). All three individuals in the family displayed distinct HRM patterns as predicted. The child, who has an ASD phenotype, inherited the interstitial duplication maternally (66% relative signal intensity), while the mother has a duplication of paternal origin (39% signal intensity) and does not have autism (Fig. 2b). As expected, the father has two normal chromosome 15s and thus shows a relative signal intensity of 52%. The maternal inheritance of this duplication was confirmed by microsatellite analysis using polymorphic markers within the duplicated region (Supplemental Fig. S1, available online at www.liebertonline.com). These data suggest that the changes in methylation state and not the primary sequence of the region being analyzed result in changes in the observed HRM patterns.
Isodicentric as well as interstitial duplications can be distinguished using HRM
Although this method works quite well for determining the parent of origin in interstitial duplication samples, we were curious if it would also work on samples with greater than three copies of the SNRPN locus as is the case in the more severe isodicentric duplication 15q syndrome cases (Battaglia, 2005). Two isodicentric 15q duplication samples were analyzed by HRM and both were found to produce relative signal intensities greater than the 66% signal for the maternal allele observed in the interstitial duplication samples (Fig. 2c). One sample had a predicted ratio of 2.7:1 maternal to paternal copies of the region, as would be expected for a typical isodicentric 15q duplication that contains two normal alleles plus two copies of this locus on the isodicentric marker chromosome. The other sample had a ratio of 6.7:1 for maternal to paternal signal. The relative signal intensity for the paternal allele was as low as 12.5% of the total signal, but a paternal peak could clearly be observed in the raw signal chromatogram (data not shown). This subject has a complex supernumerary der(15) that results in hexasomy for the PWS/AS critical region, with five maternally methylated copies of the region (Mann et al., 2004). These data suggest that the HRM method will also work for determining the parent of origin of the duplication in more complex idic duplications of 15q as well, although it is not intended to be a substitute for interphase FISH analysis when establishing the number of copies of the region in an idic (15q) duplication sample.
Discussion
In the era of high-resolution whole-genome oligonucleotide arrayCGH, there is no question that the detection of chromosome 15q duplications encompassing the PWS/AS locus combined with confirmation by FISH analysis has become standard practice in the genetics clinic for the investigation of ASD phenotypes. Here we have developed a method that supplements this cytogenetic analysis with critical information on the parental origin of these duplications that has a direct impact on appropriate genetic counseling for these individuals. Although other methods have been developed to determine the parent of origin of the PWS/AS locus using methylation-specific PCR, methylation-sensitive single-nucleotide primer extension, methylation-specific restriction enzyme digestion, and methylation-specific multiplex-ligation-dependent probe amplification, there are technical difficulties applying these approaches in the clinical setting because they were designed to detect deletions or UPD and thus are less reliable when used in a condition where both alleles are present. In addition, these methods require a two-step procedure that separates amplification from analysis, while in the quantitative HS-HRM method, both amplification and analysis occur in the real-time PCR machine, eliminating the possibility for error or contamination of DNA samples. This is an important feature of this assay because methods with fewer transfer steps result in higher quality control and thus are better suited to meet Clinical Laboratory Improvement Act (CLIA)–approved guidelines (wwwn.cdc.gov/clia/regs/toc.aspx).
Early intervention therapies for autism clearly improve the quality of life and eventual outcome in these individuals (Landa, 2007; Dawson, 2008; Eldevik et al., 2009). Although paternally derived interstitial duplications of 15q may result in neurological effects (Bolton et al., 2004; Veltman et al., 2005), they are less likely to be associated with an autism phenotype, while maternal interstitial duplications much more substantially increase risk for a phenotype on the autism spectrum (reviewed in [Hogart et al., 2008]).
This method not only determines parental origin of the duplicated segment, but also simultaneously measures the dosage of these alleles in the sample. Although there is some indication that methylation at the SNRPN locus may be anywhere between 95% and 100% of the CpG sites, this did not complicate our ability to clearly distinguish maternal from paternal duplications in 26 independent samples (Fig. 2a) where parental DNA was not available. The possibility, however unlikely, that duplications of 15q may also harbor imprinting center defects that result in incomplete or inappropriate methylation of the SNRPN locus is a caveat for all methods that utilize this differential methylation to establish the parental origin of the duplication. In these cases parental DNA would be required for microsatellite analysis, and this is not always possible. Also, this method worked both on inherited interstitial duplications that differed only in methylation state and on samples with known isodicentric duplications of the region.
One limitation of this method lies in the ability to determine the number of copies of the duplicated region using quantitative fluorescent intensity. For example, in one of the idic 15q duplication samples the ratio of maternal to paternal alleles was found to be 6.7:1. Although adjusting the normalization parameters within LightCycler software can compensate to some extent, it may not be possible to obtain accurate quantification of samples at the extremely high or low ends of the HRM curve. In addition, the ratios even for interstitial duplication samples were often less than the expected 2:1 for this type of chromosomal rearrangement. It must be pointed out, however, that the method was designed only to determine which chromosome 15 (maternal or paternal) is duplicated, and not to supplant current methods used to determine the number of copies of the duplicated region such as interphase FISH analysis.
Supplementary Material
Acknowledgments
We are grateful to the subjects and families, and the clinicians who collected DNA samples. We also appreciate the additional samples provided by J. Rosenfeld of Signature Genomic Laboratories, S. Skinner of Greenwood Genetic Center, and E. Dykens and T.J. Bischell of Vanderbilt University, and the technical support from the University of Tennessee Health Science Center Molecular Resource Center. This work was funded by a grant from the Herbert and Mary Shainberg Neuroscience Research Fund to L.T.R.
Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Battaglia A. The inv dup(15) or idic(15) syndrome: a clinically recognisable neurogenetic disorder. Brain Dev. 2005;27:365–369. doi: 10.1016/j.braindev.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Bolton PF. Veltman MW. Weisblatt E, et al. Chromosome 15q11-13 abnormalities and other medical conditions in individuals with autism spectrum disorders. Psychiatr Genet. 2004;14:131–137. doi: 10.1097/00041444-200409000-00002. [DOI] [PubMed] [Google Scholar]
- Cook EH., Jr. Lindgren V. Leventhal BL, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60:928–934. [PMC free article] [PubMed] [Google Scholar]
- Dawson G. Early behavioral intervention, brain plasticity, and the prevention of autism spectrum disorder. Dev Psychopathol. 2008;20:775–803. doi: 10.1017/S0954579408000370. [DOI] [PubMed] [Google Scholar]
- Depienne C. Moreno-De-Luca D. Heron D, et al. Screening for genomic rearrangements and methylation abnormalities of the 15q11-q13 region in autism spectrum disorders. Biol Psychiatry. 2009;66:349–359. doi: 10.1016/j.biopsych.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Eldevik S. Hastings RP. Hughes JC, et al. Meta-analysis of early intensive behavioral intervention for children with autism. J Clin Child Adolesc Psychol. 2009;38:439–450. doi: 10.1080/15374410902851739. [DOI] [PubMed] [Google Scholar]
- Glenn CC. Saitoh S. Jong MT, et al. Gene structure, DNA methylation, and imprinted expression of the human SNRPN gene. Am J Hum Genet. 1996;58:335–346. [PMC free article] [PubMed] [Google Scholar]
- Hogart A. Wu D. Lasalle JM, et al. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis. 2010;38:181–191. doi: 10.1016/j.nbd.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthemke B. Wagstaff J. Mechanisms of imprinting of the Prader-Willi/Angelman region. Am J Med Genet A. 2008;146A:2041–2052. doi: 10.1002/ajmg.a.32364. [DOI] [PubMed] [Google Scholar]
- Kubota T. Sutcliffe JS. Aradhya S, et al. Validation studies of SNRPN methylation as a diagnostic test for Prader-Willi syndrome. Am J Med Genet. 1996;66:77–80. doi: 10.1002/(SICI)1096-8628(19961202)66:1<77::AID-AJMG18>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Landa R. Early communication development and intervention for children with autism. Ment Retard Dev Disabil Res Rev. 2007;13:16–25. doi: 10.1002/mrdd.20134. [DOI] [PubMed] [Google Scholar]
- Mann SM. Wang NJ. Liu DH, et al. Supernumerary tricentric derivative chromosome 15 in two boys with intractable epilepsy: another mechanism for partial hexasomy. Hum Genet. 2004;115:104–111. doi: 10.1007/s00439-004-1127-5. [DOI] [PubMed] [Google Scholar]
- Sahoo T. del Gaudio D. German JR, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman MW. Thompson RJ. Craig EE, et al. A paternally inherited duplication in the Prader-Willi/Angelman syndrome critical region: a case and family study. J Autism Dev Disord. 2005;35:117–127. doi: 10.1007/s10803-004-1039-1. [DOI] [PubMed] [Google Scholar]
- Wang NJ. Liu D. Parokonny AS, et al. High-resolution molecular characterization of 15q11-q13 rearrangements by array comparative genomic hybridization (array CGH) with detection of gene dosage. Am J Hum Genet. 2004;75:267–281. doi: 10.1086/422854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HE. Hall VJ. Cross NC. Methylation-sensitive high-resolution melting-curve analysis of the SNRPN gene as a diagnostic screen for Prader-Willi and Angelman syndromes. Clin Chem. 2007;53:1960–1962. doi: 10.1373/clinchem.2007.093351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.