Abstract
The enzymes involved in metabolism and signaling are regulated by posttranslational modifications that influence their catalytic activity, rates of turnover, and targeting to subcellular locations. Most prominent among these has been phosphorylation/dephosphorylation, but now a distinct class of modification coming to the fore is a set of versatile redox modifications of key cysteine residues. Here we review the chemical, structural, and regulatory aspects of such redox regulation of enzymes and discuss examples of how these regulatory modifications often work in concert with phosphorylation/dephosphorylation events, making redox dependence an integral part of many cell signaling processes. Included are the emerging roles played by peroxiredoxins, a family of cysteine-based peroxidases that now appear to be major players in both antioxidant defense and cell signaling. Antioxid. Redox Signal. 14, 1065–1077.
Introduction
Enzymes are the primary catalysts that promote chemical reactions fundamental to biological processes, yet without regulatory mechanisms to ensure the proper expression and activation state at the appropriate time and place for such enzymes, cells would be unable to function properly or respond to their environment. Recognition is increasing that one level of posttranslational control exerted on enzymes involved in metabolism, cell signaling, and oxidative stress protection is by modulation of their redox state either at the catalytic site or at distinct regulatory sites. Cysteine residues are the predominant targets of redox-linked regulation and are the focus of this review.
As incorporated into proteins, cysteinyl residues bear a thiol (sulfhydryl) group that represents the most reduced state of sulfur in proteins. Conversion of these groups to oxidized or alkylated species occurs postranslationally and can act as a switch, changing the catalytic properties of an enzyme. The word “switch” can be used in many different contexts, including the noncovalent binding of a substrate to a cooperative enzyme that leads to a steep response curve over a very limited concentration range. Although reversibility is considered a requirement in some discussions of cell signaling switches, this review includes as potential switches any modification, reversible or irreversible, which changes the SH state of cysteines in enzymes and leads to a change in activity or other functional properties. In this way we can take a relatively comprehensive view of how cellular redox changes can be sensed by enzymes and reflected in changes in their properties. In addition, catalytic cycles that involve cysteine redox state changes are discussed given that reactivation within the normal cycle often reflects action of a reductant, which could itself exert a level of control on metabolite flux through a cysteine-dependent enzymatic process.
The Chemistry of Cysteinyl Residues Within Proteins
As a basis for the consideration of redox effects on cysteine-containing enzymes, we begin with a summary of the various chemical reactions in which thiol groups are found to participate within biological systems. Due to the unique chemistry of the sulfur-containing sidechain that can be strongly influenced by the microenvironment of the surrounding protein, cysteine residues are important players both in enzyme catalytic sites and in regulatory aspects of enzyme function. Enzymes with active-site cysteine residues typically rely on the thiolate (deprotonated) form of the cysteine for activity, and reactivity toward substrates (and oxidants) is therefore enhanced by a microenvironment that perturbs the normally high pKa (∼8.5) of cysteine thiols to a value at or lower than neutral pH. In the case of the disulfide-bond oxidoreductase DsbA, the pKa drops as low as 3.5 (51, 86). Thus, although the vast majority of cysteine residues within cytoplasmic proteins are in the protonated form at physiological pH, the small subset within enzyme catalytic or regulatory sites are largely or fully ionized due to their low pKa values. Aside from shifts in pKa, additional features that enhance reactivity are relatively poorly defined and under intense investigation (see later in this section), but may include the presence of acid–base catalysts (21) and specialized substrate-docking sites (47).
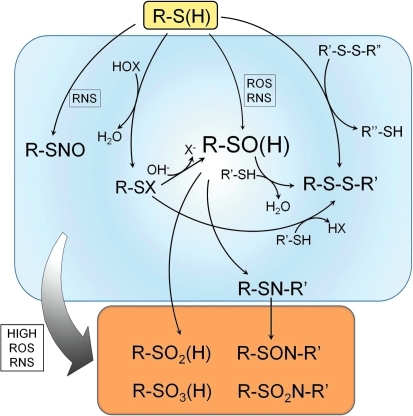
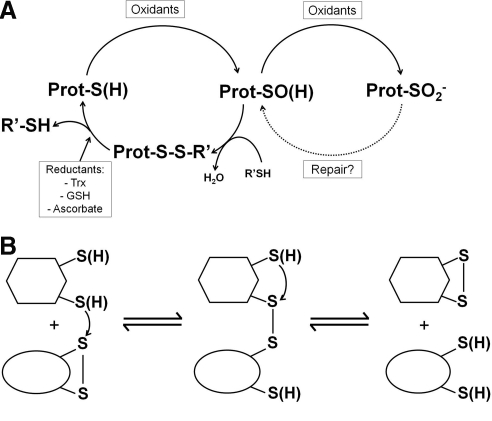
Thiol groups in proteins can undergo both one- and two-electron chemistry to generate more oxidized products. With one-electron oxidation, thiol groups are converted to thiyl radicals (R-S•), species that can participate in free radical chain reactions and can go on to form all of the various oxidized species generated by 2-electron chemistry and summarized in Figure 1 [see ref. (86) for details]. These species also have the potential to react directly with another radical, NO•, to form S-nitrosylated thiol groups and can participate in oxygen-dependent pathways that produce superoxide (86). Thiolate reactions with H2O2, organic hydroperoxides, and peroxynitrite are two-electron processes that form the reactive oxidation product cysteine sulfenic acid, a metastable intermediate that is readily transformed into other oxidative products such as disulfide bonds (with other protein cysteines or glutathione) and sulfenamides. Sulfenic acids are also intermediates in the formation of more extensively oxidized and generally irreversible oxidative and nitrosative products (i.e., sulfinic acids, sulfinamides, sulfonic acids, and sulfonamides; Fig. 1). Myeloperoxidase-generated oxidants such as hypohalous acids (hypochlorous acid and hypobromous acid) and N-chloramines also produce sulfenic acids and other reversibly or irreversibly oxidized products of cysteine residues (23, 58, 66, 80). While sulfenic acid, sulfenamide, S-nitrosothiol, and disulfide products are readily reduced to thiols by the prominent thioredoxin- and glutathione-dependent redox systems, sulfinamides and sulfonamides are characterized as crosslinks that are resistant to such reduction (23, 66). Sulfinic acids in one family of proteins, the peroxiredoxins (Prxs), can be repaired through an ATP-dependent process catalyzed by sulfiredoxins, but are likely to be irreversibly oxidized products in most proteins (14, 67). There is no known biological repair pathway for sulfonic acids. Similar to enzyme regulation through reversible phosphorylation, the oxidation of a protein to form a sulfenic acid or other reversibly oxidized product may elicit a functional change through a conformational change or steric blockage, while subsequent reduction (akin to dephosphorylation effected by phosphatases) reverses the effect (Fig. 2A). Although sulfinic acid formation is usually irreversible, it provides an additional level of oxidative regulation (Fig. 2A). As one example, when matrix metalloproteinases are oxidized to a sulfinic acid at a regulatory cysteinyl site, they become activated through enhanced autoproteolytic activity (22, 27).
FIG. 1.
Biological modifications of cysteine thiols. Reactive cysteine thiols (R-SH), typically in their ionized, thiolate form (R-S−), are oxidized by hydrogen peroxide, organic hydroperoxides, hypohalous acids (HOX), and peroxynitrite to form sulfenic acids, which may be stabilized or go on to form other reversible (disulfides [R-SS-R′] or sulfenamides [R-SN-R′]) or irreversible oxidation products (sufinic acids [R-SO2H], sulfonic acids [R-SO3H], sulfinamides [R-SON-R′], and sulfonamides [R-SO2N-R′]). Reversible S-nitrosocysteine modifications are also observed in cells exposed to RNS. Both ROS and RNS promote these oxidations. Modifications within the blue box are considered reversible (by cellular reductants like thioredoxin and glutathione), whereas the species in the orange box are not. Although sulfinic and sulfonic acids are shown here as irreversible modifications, recent discoveries show that sulfinic acid forms of some Prxs can be recovered through action of specialized sulfinic acid reductases (sulfiredoxins). Prx, peroxiredoxin; RNS, reactive nitrogen species; ROS, reactive oxygen species. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
FIG. 2.
Two potential loops of oxidative regulation (A) and the chemical steps of thiol–disulfide interchange reactions (B). (A) Oxidants and particularly H2O2 modify specific reactive cysteine residues within proteins to sulfenic acids (R-SOH), with a corresponding alteration in protein function. The sulfenic acid is also susceptible to further modification by oxidants to form sulfinic acid (R-SO2−), a modification that could have distinct effects on functional properties of enzymes. This species in some Prxs may be repaired by sulfiredoxins to recover the unmodified protein, but typically the sulfinic acid is an irreversible oxidation state in the cell. (B) Reversible thiol-disulfide interchange reactions among proteins proceed via mixed disulfide intermediates and can lead to migration of disulfide bonds to other locations in the same or separate protein(s).
Because of their reactivity toward thiol groups, the primary fate of sulfenic acids is the formation of disulfide bonds with other cysteine residues within the same or another protein, or with glutathione. As will be described in more detail below in the examples of protein tyrosine phosphatases (PTPs) and Prxs, the presence of a proximal resolving cysteine or accessibility toward glutathione that leads to facile disulfide bond formation offers a more stable oxidized species for reductive recycling and, in doing so, prevents the generation of further oxidized products. Once disulfide bonds are formed, they are susceptible to migration to other sites or other proteins through thiol–disulfide interchange reactions depicted in Figure 2B. Such thiol–disulfide interchange is in fact the mechanism through which reduction occurs in most thiol-containing proteins through the resultant oxidation of molecules such as thioredoxin and glutathione. Reduction of cysteinyl centers in thioredoxin and glutathione occurs through the action of specialized flavoproteins, thioredoxin reductase and glutathione reductase; the reducing equivalents are provided through the oxidation of NADPH with flavin adenine dinucleotide (FAD) and a protein redox-active disulfide mediating the electron transfer.
It is well established that only a subset of cysteine residues in proteins are susceptible to oxidation or alkylation, illustrating the specificity of these reactions (40, 45). In this light, it would be of great value to the scientific community if enough were known about the nature of regulatory cysteine sites to allow prediction of such sites in proteins. This would require the knowledge of what the structures around such sites had in common for activating these residues, if anything, and the ability to find and score such features in other proteins. While we are still early in the process of identifying and looking for commonalities among reactive cysteine sites selectively modified by particular oxidants and electrophiles, initial studies have identified relatively few characteristic features among the rather small numbers of reactive sites identified to date (15, 47, 69, 70), suggesting that developing successful predictions will be challenging. Even at this early stage it seems that a single predictive set of rules will not identify all reactive cysteines, but a great deal remains to be discovered regarding the identity of these sites and that will help inform the bioinformatics analyses so that it may be possible to develop not a single, but a series of predictive tools to recognize such sites.
Thiol Modifications Among Enzymes That Directly Use Cysteine for Catalysis
A number of enzymes involved in metabolic, cell signaling, and antioxidant defense pathways utilize activated cysteinyl residues at their active sites. Because this cysteine typically changes redox state during the course of reaction and/or may be inherently reactive toward oxidants and electrophiles given the protein microenvironment that supports their catalytic reactivity, these enzymes may also be subject to regulation through modification at these sites. In fact, simply the need to reductively recycle enzymes following catalysis with an oxidizing substrate, as for ribonucleotide reductase and Prxs, makes these enzymes dependent on availability of sufficient levels of an appropriate electron donor in its reduced state for the cell to support multiple turnovers.
As described above, enzymes with low pKa cysteine residues at their active sites tend to be more susceptible to oxidation. Given their obligate role in catalysis, modification of such active-site cysteines inhibits the enzymes that possess them. The most well-known example of regulatory oxidation occurring at the active site of cell signaling proteins is in the case of PTPs, proteins which possess a low pKa cysteine (pKa 4–6.5) that attacks the phosphorylated protein substrate to dephosphorylate it and generate a cysteinyl phosphate intermediate within the PTP enzyme that is then hydrolyzed (17). The common step of regulatory oxidation at this catalytic cysteine of PTPs is the formation of a sulfenic acid via H2O2-mediated oxidation. Evidence for this sulfenic acid intermediate has been obtained for PTP family members such as PTP1, vaccinia H1-related phosphatase, mitogen-activated protein kinase phosphatase 3, SH2 domain-containing protein tyrosine phosphatase (SHP)-1, SHP-2, and cell division cycle (Cdc) 25 phosphatases B and C (18, 48, 74, 75).
Accumulating evidence suggests that various PTPs go on to form one of a number of reversible oxidation products. Disulfide bond formation with a resolving cysteine located nearby within the same protein, sometimes referred to as a backdoor cysteine, has been observed in several PTPs, including the low-molecular-weight PTPs, the cell cycle Cdc phosphatases, SHP-1, SHP-2, and phosphatase and tensin homolog (PTEN) (7, 8, 42, 75); this modification serves to protect these proteins by preventing further, irreversible oxidation of the sulfenic acid. The conformational change that may accompany disulfide bridge formation can also aid access to the site by reductants like thioredoxin (Trx) or glutaredoxin (Grx) (83). Thiol–disulfide exchange with other cysteinyl residues in the same or different proteins can also allow the disulfide bond to migrate, as was recently observed for SHP-1 and SHP-2, where the formation of an intrasubunit disulfide bond between two (proximal and distal) backdoor cysteines provides a rapid path for PTP reactivation (8).
In PTPs lacking a nearby resolving cysteine, one known avenue of further modification is a glutathionylation of the active-site cysteine that may facilitate reductive recycling of reversibly inhibited PTPs like PTP1B (21). Another avenue of modification revealed by crystallographic analyses of PTP1B and PTPα is for the sulfenic acid formed at the active site to undergo attack by the peptide amide nitrogen of an adjacent serine residue to form water and a 5-membered ring structure with a sulfenamide linkage (68, 91). Formation of this cyclic sulfenamide, which is reversible, significantly alters the active-site architecture, bringing buried residues to the surface and presumably, as with disulfide bond formation, enhancing access of reductants to the site.
Another group of proteins containing an oxidation-susceptible, low-pKa cysteinyl residue at the active site is the thiol proteases, including papain, caspases, and cathepsins. These proteins catalyze proteolysis through a mechanism analogous to that of the serine proteases, except that a nucleophilic thiolate of cysteine rather than an activated serine hydroxyl acts as the attacking nucleophile. Some of the earliest data directly linking the oxidation of cysteine residues to the regulation of activity were with papain, where sulfenic acid formation following H2O2 treatment caused a reversible inhibition in activity (1). Similarly, caspases and cathepsins have been shown to be sensitive to reversible oxidative inhibition in vitro by H2O2, nitric oxide donors, and/or protein hydroperoxides through similar mechanisms (15, 29, 30, 32, 60).
Glyceraldehyde-3-phosphate dehydrogenase (GapDH) is an additional example of a protein recognized early on to form a sulfenic acid at the active-site cysteine, modulating its activity (1). This glycolytic enzyme produces 1,3-bisphosphoglycerate via the oxidative phosphorylation of glyceraldehyde-3-phosphate in the presence of NAD+ and inorganic phosphate through formation of a thiohemiacetal intermediate at the essential cysteine. Oxidative modification of this cysteine to sulfenic acid not only inhibits its dehydrogenase activity, but also switches on acyl phosphatase activity (1, 35, 72); other oxidized forms of this cysteine do not impart acyl phosphatase activity. GapDH is also susceptible to S-glutathionylation and S-nitrosation by various reactive oxygen and nitrogen species (3). GapDH is frequently discovered as a modified protein in various redox proteomics studies, suggesting that it is readily modified in vivo (6, 15).
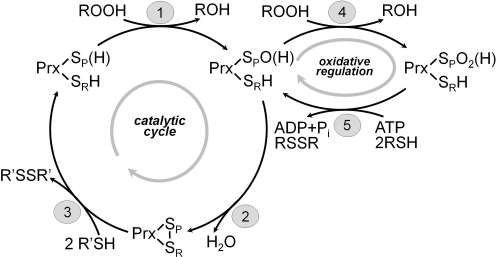
Another set of cysteine-based peroxidase proteins that includes the Prxs, cysteine-containing glutathione peroxidase (Gpx) homologs, and the organic hydroperoxide resistance protein is oxidized at an active-site cysteine by reaction with hydroperoxide or peroxynitrite substrates as part of their normal catalytic cycle (Fig. 3, reactions 1 through 3). Considerable evidence has now accumulated that demonstrates the susceptibility of a number of eukaryotic Prxs to inactivation during turnover with high levels of peroxide substrate, showing that the sulfenic acid intermediate generated during normal catalysis can also operate as a redox switch in these enzymes (described in more detail below; Figs. 2A and 3, reactions 4 and 5).
FIG. 3.
Mechanisms of catalysis by Prxs. The three main reactions universal to the catalytic cycle of Prxs are [1] peroxidation, [2] resolution, and [3] recycling. During reaction 2, a local unfolding event occurs to facilitate the formation of disulfide bonds, with SP and SR designating the sulfur atoms of the peroxidatic and resolving cysteines, respectively. For the typical 2-Cys Prxs, the resolving cysteine, CR, is located in the C-terminus of the protein and the CR from one subunit forms a disulfide bond with the peroxidatic cysteine, CP, on its partner subunit. For atypical 2-Cys Prxs, the CR is found in an alternate location, frequently in the same subunit as the corresponding CP. 1-Cys Prxs do not contain a CR and are presumably recycled by a low-molecular-weight thiol or are directly reduced by thioredoxin. 2 R′SH in reaction 3 represents a thioredoxin-like protein or domain. Overoxidation of CP (reaction 4) and ATP-dependent reduction of Cys-SPO2H by sulfiredoxin (reaction 5) depict redox regulation and repair occurring in some eukaryotic typical 2-Cys Prxs.
Enzymes with Regulatory Cys Not at the Active Site
While this area is less well developed than the examples above where oxidation affects catalytic cysteine residues, there are now an increasing number of examples of enzymes with relatively well-characterized redox switches that are not at the active site but are, nevertheless, important in controlling their biological functions. We suspect that there are many more such sites that have not yet been recognized and that we have a long way to go before the scope of this type of regulation is fully appreciated.
There are many anecdotal references in the literature to enzymes for which the inclusion of dithiothreitol (DTT) in the assay mixture is necessary for maximal activity, and it is likely that for many of these proteins, the DTT requirement reflects the inhibitory effect of regulatory oxidation at one or more cysteinyl residues. In one such case, the human mitochondrial branched chain aminotransferase, this DTT effect was mapped to two cysteinyl residues within a Cys-Xaa Xaa-Cys motif that form a regulatory disulfide bond upon H2O2-mediated sulfenic acid formation at the more C-terminal residue of the pair (11). Another enzyme frequently used in a variety of research applications, Escherichia coli alkaline phosphatase, has an opposite requirement. It is normally secreted into the periplasmic space and requires for its activity the disulfide bonds formed upon translocation across the plasma membrane. In a clever application, the enzyme was engineered to prevent translocation, allowing its activity to be used to assess the redox status of thioredoxin proteins within the cytoplasm of mutant bacteria (77).
As examples relevant to cell signaling, some protein kinases are regulated by redox modifications at Cys sites not directly involved in catalysis. Unlike for the PTPs, oxidative modifications of kinases are inhibitory in some cases and stimulatory in others, and relatively few mechanisms are well defined. This diversity of the effects of oxidation parallels the fact that some phosphorylation events also positively regulate signal transduction pathways, while others have an inhibitory effect. Here we describe several of the best molecularly characterized effects of oxidation on kinase activity, with biological settings under which these redox modifications are observed highlighted in the following section.
Some of the best mapped redox effects are for protein kinase A (PKA) and cGMP-dependent protein kinase (PKG)Iα, which appear to respond in opposite ways to oxidative modifications (13, 86). Following treatment of cells with diamide, PKA is glutathionylated, on a Cys residue adjacent to the activation loop, and this promotes enzyme inactivation through the dephosphorylation of a nearby phosphothreonine (33). In contrast, H2O2 treatment of PKGIα causes an intermolecular disulfide bond to form between two Cys42 residues (in different subunits) within a regulatory region; this activates the enzyme by increasing its affinity for substrate (5).
Two upstream kinases in the c-Jun N-terminal kinase (JNK) pathway, MAPK/extracellular signal-regulated kinase (ERK) kinase kinase 1 (MEKK1) and apoptosis signal-regulating kinase 1 (Ask1), are also affected in opposite ways by regulatory oxidation (13). JNK itself also appears to be redox regulated, although this effect seems to be through an inhibitory interaction with a redox-sensitive protein, glutathione S-transferase π, which releases JNK for activation when oxidized (13). For MEKK1, glutathionylation at a Cys residue within the ATP-binding domain inhibits its kinase activity (12). The activating effect of reactive oxygen species (ROS) on Ask1 occurs via Trx1; the kinase is inhibited by an interaction with reduced Trx1 that is disrupted by Trx oxidation. Recent evidence suggests that there is also an alternative or additional direct redox sensitivity of at least one Cys in the Ask1 protein, but this is not yet well characterized (59, 86). Interestingly, Trx1 itself possesses a regulatory dithiol that is distinct from the active-site redox center, adding to the redox modifications that can influence signaling through stress-activated kinase pathways (85).
For the tyrosine kinase cellular sarcoma kinase (cSrc), oxidation at two cysteinyl residues is activating when combined with dephosphorylation of a C-terminal Tyr on the protein (25, 67). Interestingly, for cSrc and other fibroblast growth factor receptor family members, recent studies have also demonstrated that formation of an intermolecular disulfide bond between two Cys277 residues from different subunits inhibits the kinase activity (81).
Two other signaling kinase families, RAC-alpha serine/threonine-protein kinase or protein kinase B (Akt) (also known as protein kinase B) and protein kinase C (PKC), also exhibit redox-sensitive kinase activity. Akt exposed to oxidative stress conditions forms an inhibitory, intramolecular disulfide bond that inhibits its activity by promoting the dephosphorylation of Akt by protein phosphatase 2A (50). For PKC, effects can be inhibitory or stimulatory depending on the isoform and site of oxidation. Although not well defined at the molecular level, an oxidation within the kinase domain can inhibit PKC activity, whereas disulfide bond formation in the regulatory subunit can activate PKC activity through the removal of autoinhibition (13, 26).
Linking Biological Function to Thiol Switches in Enzymes
As we gather more and more specifics regarding reactive cysteines that regulate protein function in vitro, the expanded question of where (within the cell) and under what conditions such regulatory modifications occur in biological systems must also be asked. This issue is a significant challenge to address as it is widely recognized that there are only rather limited approaches available for measuring and mapping such sites and determining the molecular bases underlying redox sensitivity of particular enzymes or pathways (13). Also challenging to achieve is the goal to freeze the redox status of all proteins at an instantaneous point in time in a cell before detection of the modifications (37). There are some powerful tools emerging for such analyses (e.g., OxICAT and various other trapping and detection methods for sulfenic acids, S-nitrosothiols, glutathionylated proteins, and the total reversibly oxidized thiol pool) (2, 31, 45, 52, 59, 64), but even when such modified proteins are identified, complementary in vitro work will be needed to unravel the functional effects of the modification(s). Finally, a full causal analysis to establish the role of regulatory cysteines in biological processes often relies on the ability to express or knock down expression of proteins of interest and, as an optimal outcome, demonstrate loss of redox sensitivity upon mutation of specific cysteinyl residues. This is a slow and laborious task that can be carried out on only a few targeted proteins. Although this approach is not applicable to enzymes in which cysteine residues are directly involved in their activity, the link between modifications of such cysteines and activity is inherently better defined. An additional complication is that some proteins' activities may rely on binding partners that are redox sensitive, and these may be even more difficult to track down.
For bacteria, redox signaling is generally considered to be one aspect of an oxidative stress response that allows these unicellular organisms to respond to oxidative threats in their immediate environment, which can change rapidly and dramatically. For eukaryotes, and particularly multicellular organisms, there is an additional redox signaling phenomenon that has diverged to be quite distinct from oxidative stress-related signaling. At the extremes, such oxidative stress-related and nonstress-related redox-linked signaling are quite distinct, although a strong case can be made for considerable overlap between the two, in part because cellular protein modifications are likely a continuum of low to high levels of modification and the spectrum of modifications changes under various conditions (3, 14).
In accord with the long precedence for considering oxidation as a damaging influence (76), protein oxidation observed as a result of oxidative stress is an accepted phenomenon, involving the expected modifications at thiol residues in addition to the oxidation of methionines, as well as carbonylation, adduction, and deamination and other types of damage to various protein sidechains. Damage or stress sensing is the context in which such pathways and proteins as Kelch-like ECH-associated protein-1 (Keap1)/NF-E2-related factor-2 (Nrf2) and tumor protein 53 (p53) are activated through thiol modifications that may include alkylations as well as oxidations (3, 14, 15, 31), and there is clearly much overlap between these signals. In comparison with oxidative stress-related signaling, the concept of nonstress-related redox signaling (hereafter simply referred to as redox signaling) is still somewhat controversial, in part because molecular details of protein oxidation affecting signaling pathways are few, and the community is still wrestling with issues, including how the known chemistry and rates of reaction between potentially oxidized proteins and hydrogen peroxide, in particular, can be reconciled with the rates and levels of hydrogen peroxide generation and diffusion in the cell (see below) (21, 78, 86). Having said that, evidence for the involvement of ROS-dependent thiol switches in evoking or modulating nonstress-related signaling events has been emerging and is gaining clarity in terms of mechanisms involved. Specifics of such pathways have been outlined in a number of recent reviews (3, 13, 24, 59), a few of which are summarized below.
Before illustrating specific examples of thiol switches thought to be involved in redox signaling, it should be mentioned that the response of a particular protein, pathway, or cell may vary dramatically depending on the growth conditions, cell type, and stimulant. Even where the actual direct cause-and-effect of ROS generation and subsequent thiol oxidation in one or more specific protein(s) involved in signaling may be quite difficult to establish, one would expect the redox status of the components within and around a cell to be an important backdrop influencing how well and in what way a cell can respond to given stimuli. Indeed, along with changing protein expression and basal phosphorylation levels that influence the wiring of the system and the priming of the component sensors, redox status may be one of the most critical aspects of the differing context through which signal transduction occurs in normal and dysfunctional cells. An illustration of this latter point in a phosphorylation pathway is the cooperativity that derives from the dual phosphorylation required on ERK for downstream signaling in Xenopus oocytes (20). Because MEK can only act on the ERK sites one at a time, two MEK-ERK binding/catalysis events must occur to activate signaling. Thus, at lower fluxes through the pathway that on average generate only singly phosphorylated ERK, this ERK population is inactive but primed for a second wave of activation to more easily generate doubly phosphorylated ERK and send the signal. This type of cooperative behavior is said to be more switchlike. While this paradigm is one oversimplified aspect of the biochemistry of ERK signaling (38), the example provides a good analogy for how the oxidation status of proteins or complexes in a cell could strongly influence how readily they are activated, further modified, or translocated under certain redox conditions in response to upstream signals.
As mentioned above, one of the most discussed oxidative modifications thought to influence signal transduction pathways is that occurring at the active-site cysteine of PTPs in which the inhibitory modification would serve to enhance and prolong the phosphotyrosine signals generated by receptor and nonreceptor tyrosine kinases. PTP oxidation has been observed to occur as a result of various cell stimuli, including growth factor (platelet-derived growth factor, epithelial growth factor, and insulin) or cytokine (tumor necrosis factor alpha [TNFα]) treatment, B-cell and T-cell activation, and ADP-stimulation of macrophages (21, 64, 67, 83). Localization of the target proteins of ROS to sites of ROS production (the “where” of “where and when”) is also an important aspect of the selective modification of signaling-relevant cysteines. In an article emphasizing the need for localization of PTP1B near activated Nox4 complexes in the endoplasmic reticulum, Keaney and colleagues demonstrated the intermediacy of H2O2 in the phosphatase inhibition required to activate epithelial growth factor signaling in human aortic endothelial and COS-7 cells (9). Localization of NADPH oxidase components with PTPs and other targeting proteins at lamellipodial leading edges and focal adhesions is also thought to help organize the necessary components for cell adhesion and migration (84). It is notable in this situation that integrin and growth factor stimulation, which invoke independent activation of distinct ROS sources (NADPH oxidases for growth factors and mitochondria and 5-lipoxygenase for integrins), act synergistically to promote these signaling events. While reversible PTP oxidation has been observed in many signal transduction cascades, a recent finding that cancer cell lines have a significant pool (∼40%) of irreversibly hyperoxidized PTPs suggests that a one-way switch may also be operational in certain biological systems (3, 46). This provides an illustration of the overlap discussed above that inherently exists between oxidative stress and normal redox signaling-linked processes.
Although cases clearly exist in which PTP oxidation occurs and modulates cell signaling processes, the cellular mechanism through which this oxidation occurs is less clear. It is frequently argued that the modest rate of oxidation of PTPs by H2O2, at 2–160 M−1 s−1, is insufficient to explain the in vivo oxidation of these proteins (21, 78, 86). Although the answer to this dilemma is not yet known, factors that could, nonetheless, promote such oxidations could include enzymatic catalysis of the PTP oxidation, modulation of oxidation sensitivity via interactions with substrates or other proteins or additional posttranslational modifications, or mechanisms through which tight control of the production and breakdown of H2O2 can allow it to reach very high local levels as suggested by the floodgate hypothesis described below.
The ability to modulate the proteolytic activity of caspases through oxidative modifications of the active-site cysteine is an intriguing way in which apoptotic pathways may be regulated by redox modifications. Given that oxidation inhibits caspase activity, however, this would not explain the link between oxidative stress and the promotion of apoptosis. A recent study by Pan and Berk clearly demonstrated the inhibitory effect of caspase-3 glutathionylation on caspase cleavage and apoptosis in vivo. Expression of active Grx1, which catalyzes the deglutathionylation reaction, was essential for TNFα-mediated apoptosis, although at least part of this effect was apparently due to glutathionylation at sites remote from the active site, where they interfere with cleavage of the caspase-3 by caspase-8 (55). While caspase-3 exhibits a relatively slow rate of oxidation when exposed to H2O2, its apparent susceptibility to oxidation in vivo may in part be explained by the finding that the interaction of caspase-3 with substrates enhances its oxidant sensitivity by as much as 40-fold (29). This demonstrates that redox sensitivity can indeed be strongly dependent on interactions with substrates or other proteins.
In addition to GapDH, mentioned above, a number of metabolic enzymes, including creatine kinase, are regulatable by redox-sensitive thiol switches, a subject that has been reviewed elsewhere (3, 93). Under oxidative stress conditions imposed by addition of oxidants or inactivation of the Trx or glutathione pathways, redox proteomic analyses demonstrated disulfide bond formation in GapDH between the active-site Cys and a neighboring Cys in both E. coli and yeast cells (40, 45). GapDH also forms intermolecular disulfide bonds under both nitrosative and oxidative conditions that lead to aggregate formation and correlates with stress-induced apoptosis (3). S-nitrosylation of GapDH may also trigger cell death through association of the modified protein with, and stabilization of, an E3-ubiquitin ligase, Siah1 (3). Interestingly, inhibition of glycolysis through oxidative inhibition of GapDH leads to a shift in metabolic flux that promotes the pentose phosphate pathway, a major provider of NADPH in eukaryotes. This would in turn provide electrons for reductive recycling of both Trx and glutathione to help restore the normal redox balance within oxidatively challenged cells (3).
Redox-dependent modulation of signaling kinase activities has been demonstrated in many cases through the treatment of cells with exogenous oxidants like H2O2, menadione, and diamide. Although application of these reagents is of only marginal physiological significance at best, they do indicate ways in which cellular oxidative stress can influence redox signaling. The activating effects of ROS on Ask1 and PKGIα, as well as the inhibitory effects of ROS on MEKK1 and Akt, have all been shown to occur after such treatments. More physiological stimuli have also been linked to the direct redox-mediated activation of key signaling kinases, including TNFα (Ask1), insulin (PKGIα), extracellular matrix (Src), and glucose deprivation (Ask1, through redox-sensitive Grx1 interaction) (13, 25).
Redox Regulation and the Involvement of Thiol-Based Peroxidases in Cell Signaling
Multiple aspects of cysteine-based peroxidase function are important to the redox control exerted over cell signaling functions; not only are there various modes of regulation among these thiol-dependent peroxidases, but these clearly vary to some extent across biology based on our current knowledge. Simply the removal of peroxides, including peroxynitrite, to lower ROS can account for much of the observed influence of these proteins on signaling (10, 41, 67). The accompanying oxidation of the peroxidase enzymes to sulfenic acid and/or disulfide forms and consequent oxidation of Trx or Trx-like proteins during normal turnover with peroxide substrates must also have an oxidizing effect on the protein thiol pool. Oxidant sensing by some Prxs where peroxide levels are high and turnover is sustained also leads to hyperoxidation, which switches them off (Figs. 2A and 3, reaction 4). The structural origin of the sensitivity of Prxs toward inactivation during turnover was found to be due to the presence of a C-terminal helix within sensitive Prxs, which packs over the active site and impedes the next step of cataly-sis, disulfide bond formation (reaction 2 in Fig. 3), thereby promoting hyperoxidation (reaction 4 in Fig. 3).
The idea that this redox switch is of functional significance within the cell is suggested by the fact that the evolution of these structural features that support hypersensitivity of eukaryotic Prxs has coincided with evolution of a repair system, sulfiredoxin, to recover them. Nonetheless, the biological role for this hyperoxidation sensitivity, particularly in mammals, remains rather unclear and may also vary significantly among organisms. We briefly review here the key findings that link Prxs and thiol-containing glutathione peroxidase homologs to cell signaling processes and the mechanisms that have been proposed for their involvement. For more extensive discussions of the salient aspects of cellular redox regulation linked to thiol peroxidase function, the reader is referred to other recent reviews in this area (14, 21, 28, 86).
Unicellular organisms
For bacteria the transcriptional regulator OxyR is directly oxidized by hydrogen peroxide in a very rapid process (with a half-time of about 30 s in the presence of 100 nM H2O2), then more slowly reduced back to its resting state by Grx1 to deactivate transcription of the OxyR regulon (34). This regulator is therefore like a very slow peroxidase enzyme, not optimized for removal of peroxide but rather for a rapid sensing and slower turn off to achieve transient transcriptional activation of antioxidant expression. In a somewhat complementary sense, the highly active, thiol-containing peroxidase alkyl hydroperoxide reductase peroxidase component (AhpC), the first bacterial Prx discovered, is an abundant protein that reacts rapidly with hydrogen peroxide (k = 3.2 × 107 M−1 s−1 at pH 7), and is recycled in a considerably more efficient manner (than OxyR) by its own reductant, AhpF, to effectively scavenge, with a kcat as high as ∼55 s−1, endogenously or exogenously produced hydrogen peroxide (53, 57, 73). By virtue of this activity, AhpC levels and redox status affect OxyR activation indirectly, but profoundly, through the enzyme's ability to control hydrogen peroxide concentrations (90). Interestingly, expression of AhpC, AhpF, and Grx1 proteins is under the control of OxyR, providing regulatory feedback mechanisms. Although enzymatic peroxide removal and transcriptional control of gene expression appear to be distinct processes in prokaryotes, there is still cross-talk between the two systems to promote ROS removal and cell survival.
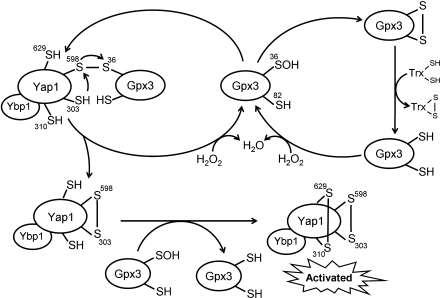
For yeast, the issues of peroxide removal and transcriptional regulation are far more intricately linked. Both Saccharomyces cerevisiae and Schizosaccharomyces pombe use thiol peroxidases as redox sensors that communicate with transcriptional regulators through formation of intermolecular disulfide bonds (Figs. 4 and 5) (14). In S. cerevisiae, Gpx3 (a thiol-containing Gpx-like enzyme, also known as oxidant receptor peroxidase-1) and yeast activator protein-1 (Yap1) constitute an H2O2 sensing relay, whereby sulfenic acid formation at Cys36 in Gpx3 is followed by intermolecular disulfide bond formation with Yap1, a transcription factor that controls antioxidant gene expression (Fig. 4). The intermolecular disulfide is resolved through thiol–disulfide interchange with another cysteine in Yap1, activating Yap1 and restoring the reduced state of Gpx3. Thus, Gpx3 acts as the peroxide sensor protein for oxidizing Yap1. A second disulfide bond also forms within Yap1, presumably through thiol–disulfide exchange and a second oxidizing cycle with Gpx3. Because a resolving cysteine (Cys82) is already present in Gpx3 that participates in the normal catalytic cycle and competes with Cys598 of Yap1 for disulfide bond formation with the sulfenic acid of Cys36, a kinetic pause (presumably imparted by the requirement for a conformational change) that hinders intrasubunit disulfide bond formation in Gpx3 is required to allow redox interaction with Yap1 to compete effectively (63). As an alternative or additional mechanism, this redox relay from Gpx3 to Yap1 may be facilitated by an additional player in the system, the Yap-1 binding protein (14). Thus, Gpx3 oxidation occurring concomitant with peroxide reduction can be sensed by the cell either through oxidation of the Trx pool as part of the normal catalytic cycle or by activation of a transcription factor, Yap1.
FIG. 4.
The mechanism of Saccharomyces cerevisiae Gpx3–Yap1 redox interaction. Gpx3 reacts with H2O2 at its peroxidatic cysteine (Cys36), which becomes oxidized to a sulfenic acid. The Cys36 sulfenic acid then reacts either with its own resolving cysteine, Cys82, or with Cys598 in the C-terminal region of Yap1 to form an intermolecular disulfide bond. Subsequent thiol–disulfide interchange with Cys303 in Yap1 completes the transfer of the oxidation state from Gpx3 to Yap1. A second disulfide bond (Cys310–Cys629) of Yap1 may be introduced by thiol–disulfide interchange with the first and/or reaction with another oxidized Gpx3. Yap1 binding protein, Ybp1, may be involved in bringing oxidized Gpx3 Cys36 close to Yap1 Cys598 and/or preventing its condensation with Gpx3 Cys82. The intramolecular disulfide bond of Gpx3 (Cys36 and Cys82) can be reduced by Trx. Gpx, glutathione peroxidase; Trx, thioredoxin; Yap1, yeast activator protein-1; Ybp1, Yap1 binding protein.
FIG. 5.
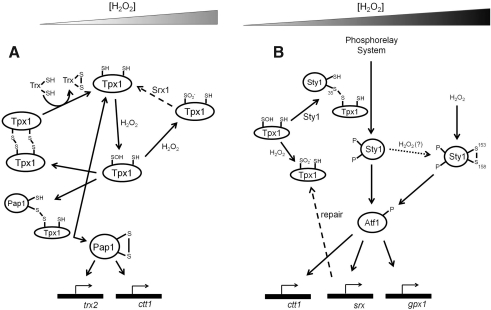
Model of two interconnected Schizosaccharomyces pombe H2O2-responsive pathways. (A) Mechanism for the redox relay between Tpx1 and Pap1. Partitioning of the sulfenic acid of Tpx1 between condensation with the normal resolving cysteine of the catalytic cycle or a thiol group from Pap1 proceeds in a similar way as the Gpx3–Yap1 pathway shown in Figure 4. Activation of Pap1 regulates expression of a thioredoxin (from trx2) and cytosolic catalase-1 (from ctt1). At high concentrations of H2O2, Tpx1 is hyperoxdized to sulfinic acid, which can be reversed by sulfiredoxin. (B) The Sty1 pathway. Sty1 (also known as Spc1), a relative of the c-Jun N-terminal kinase/p38 stress-activated protein kinases in higher organisms, is activated independently by a phosphorelay system that is promoted by formation of a mixed disulfide between Cys35 on Sty1 and the peroxidatic Cys of Tpx1. Activated Sty1 activates the bZIP-type transcription factor Atf1, leading to upregulation of antioxidant gene expression (including Gpx from gpx1 and sulfiredoxin from srx1). An alternative mode of activating Sty1 may be formation of an internal disulfide bond that is induced by high H2O2 concentrations. Pap1, Schizosaccharomyces pombe activator protein-1; Sty1, stress-activated protein kinase in fission yeast.
S. pombe has two interconnected H2O2-responsive pathways, with each responding to different levels of peroxide; the Yap1 homolog Schizosaccharomyces pombe activator protein-1 (Pap1) responds to low H2O2 (e.g., below 0.2 mM) and is inactivated at peroxide levels above 1 mM, whereas the stress-activated protein kinase in fission yeast (Sty1) is responsive to high H2O2 concentrations (Fig. 5) (14). The Prx thiol peroxidase 1 (Tpx1) seems to participate in redox relay systems with both of these transcriptional regulators. Its interaction with Pap1 is directly analogous to the Gpx3-Yap1 system of S. cerevisiae, except that high H2O2 concentrations lead to inactivation of both Tpx1 and Pap1, processes that are reversed by sulfiredoxin expression under the control of Sty1 (Fig. 5). Sty1 is activated independently by a bacterial-like His phosphorelay system (with the Ser/Thr protein kinase Wis1 ultimately phosphorylating Sty1) through a mechanism that seems to be promoted by formation of a mixed disulfide between Cys35 on Sty1 and the peroxidatic Cys of Tpx1, leading to cAMP-dependent transcription factor 1 activation and the consequent upregulation of antioxidant gene expression (14). Although high H2O2 concentrations also promote hyperoxidation of Tpx1, potentially taking away its ability to participate in this relay, an alternative mode of activating Sty1 may arise from formation of an internal disulfide bond that is induced by high H2O2 concentrations (16).
An additional way in which a redox signal alters thiol peroxidase enzyme function in yeast is in the appearance of a chaperone-like heat stress protection activity acquired by hyperoxidized S. cerevisiae Tpx1 and Tpx2 (3, 36, 67). This modification to sulfinic acid at the active site promotes aggregation to yield higher molecular weight forms with ATP-independent chaperone activity, implicating this redox-dependent process as a switch between peroxidase and chaperone activity invoked by oxidative stress. That this type of chaperone activity may be important to at least some multicellular organisms is supported by the observed role of peroxiredoxin 2 in Caenorhabditis elegans in protecting these organisms from heat stress (54).
Mechanisms for thiol peroxidase redox switches in higher organisms
The needs for signal transduction in complex, multicellular organisms are quite distinct from those of free living unicellular organisms since a significant portion of the response to the external environment must be devoted to the coordination of responses among cell types within various tissues and organs. The process of experimentally defining the exact role(s) that a given redox switch has on protein function directly affecting biological processes is also a much more demanding and complex process.
The case for the involvement of Prx proteins in cell signaling within mammalian systems is strong, although there is still much to learn about the mechanistic details (14, 67, 86). Although this discussion centers around redox regulation, it is notable that mammalian Prxs have also been shown to be regulated by phosphorylation, C-terminal truncation, and acetylation (56, 87–89), and it is likely that there is a great deal of crosstalk between the different types of posttranslational modifications influencing activity. Modulation of hyperoxidation sensitivity is just one way in which the different modifications can affect activity, since C-terminal truncations or alterations are known to desensitize Prxs toward hyperoxidation (39, 71). In addition to the peroxide-mediated redox regulation discussed below, there is also evidence for redox-linked modifications, including glutathionylation and S-nitrosylation of Prx enzymes, which may play roles in redox regulation of these enzymes in vivo (19, 79). Interestingly, Prxs are surprisingly insensitive to oxidation by hypochlorous acid and chloramines generated by phagocytes, in spite of their high reactivity toward the considerably less reactive oxidant H2O2 (61).
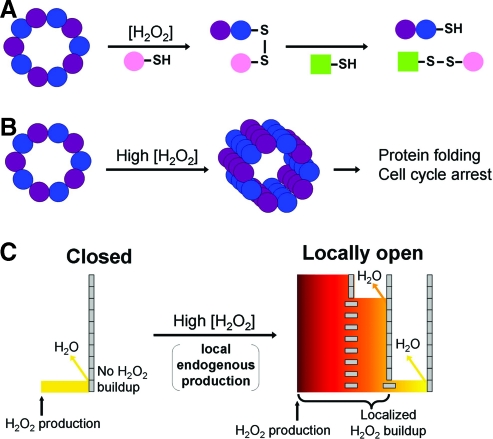
Regulation of the antioxidant activity of Prxs will clearly have an impact on cellular redox status, with the inhibition of activity leading to an increase in peroxide levels and greater flux through the Gpx and even catalase enzymes. This would be an important shift in oxidant control because Prxs are so abundant and exhibit high reactivity with hydroperoxides, suggesting that they normally serve as the primary scavenger of H2O2 (28, 86). Because the redox switch of hyperoxidation would be expected to be thrown around specialized sites of ROS generation, the buffering effect of Prxs would be lost upon localized hyperoxidation. This logic has led our group to propose the floodgate model for Prx involvement in redox signaling (Fig. 6C) (28, 88), which posits that Prx sensitivity toward peroxide-mediated inactivation allows zones of high peroxide to be established, promoting oxidation of other localized redox sensors that would otherwise be protected by the highly efficient scavenging activity of Prxs. Such highly localized effects—redox signaling is well known to be a highly localized phenomenon—would support a very high gradient of peroxide concentrations around sites of ROS generation, but prevent the flood. Whether or when this floodgate effect would be operational is as yet unclear; while a number of reports link H2O2 treatment of cells with Prx hyperoxidation (21, 62, 65, 92), there are only a few publications that describe physiological stimuli that induce significant amounts of Prx hyperoxidation [e.g., during TNFα treatment of Leydig cells to induce apoptosis, and by 6-hydroxydopamine stimulation of dopaminergic neurons used as a model of Parkinson's disease (44, 65)]. If the effect is indeed highly local, however, it may be that only very small amounts of Prx hyperoxidation are needed, amounts that are difficult to detect within the large pool of Prxs. Another significant point that is raised is that very active Gpx proteins are often also present (although amounts are quite variable), and these would dampen or buffer any Prx inactivation effects (21). Of course, this is also helpful to the cell as the Gpx would help control the flood. It may be that redox regulation is also observed within the Gpx pool surrounding ROS sources that contributes to the augmentation of redox signaling. It is interesting that an analogous argument has recently been made that membrane-localized Prx phosphorylation, another phenomenon that serves to inhibit the antioxidant function of Prxs, also promotes redox signaling through a floodgate-like mechanism (82, 87).
FIG. 6.
Three proposed roles for Prxs in peroxide signaling. (A) Thiol–disulfide interchange between Prxs and other proteins resulting in the transfer of the Prx disulfide to an acceptor protein or complex (depicted by the disulfide bond between pink and green proteins or other molecules). The reactivation of the Prx occurs by thiol–disulfide interchange with the reductant (green). (B) The chaperone model, represented by the formation of higher order oligomers of overoxidized Prxs. This is involved in stress-related signaling and requires sensitivity of Prxs toward hyperoxidation. In A and B, the Prxs are represented as purple and blue decamers under normal cellular conditions. (C). The floodgate model is an unproven mechanism. Prxs are represented as tall barriers made up of gray rectangles—vertical for active, horizontal for overoxidized and inactive. The multiple barriers on the right reflect the cell-wide Prx distribution; Prxs that are close to the peroxide generation site (marked by an arrow) are overwhelmed and inactivated, whereas those at increasing distances away are not. This creates a steep peroxide gradient and allows for localized peroxide build-up after endogenous peroxide generation. The level of hydrogen peroxide is represented by both color gradient and height. This proposed role may be involved in both stress- and nonstress-related signaling and requires that the Prxs be sensitive to hyperoxidation. Reprinted by permission from Hall et al. (28). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Additional possibilities for the role of hyperoxidation in cell signaling processes must also be considered and may work in concert with floodgate control. While the heat shock protection offered by the aggregated forms of Tpx1 and Tpx2 in yeast is not directly relevant to mammals, the chaperone activity that could assist in recovering oxidatively damaged, unfolded proteins may be (Fig. 6B). Interestingly, Prx1 chaperone activity seems to be greater than that of Prx2 due to its extra Cys residue (Cys83) that participates in intersubunit disulfide bond formation, stabilizing higher order aggregates (43, 49). Another intriguing possibility is that the hyperoxidized and/or aggregated Prx protein may itself serve as a signal. Such a function was suggested for hyperoxidized Prx2 aggregates that accumulated in mouse C10 lung epithelial cells with relatively low, continuous exposure to H2O2. The appearance of these aggregates correlated with cell cycle arrest, and recovery of the active protein after removal of the H2O2 correlated with a resumption of the normal cell cycle (62).
In terms of other recognized influences of the Prx redox state on cell signaling, it is clear from work by Hampton and coworkers that Prx3, the most highly expressed Prx in mitochondria, plays an important role in modulating apoptosis (4). One idea that is gaining momentum is that Prx, by virtue of its rapid reaction with peroxides, may itself be a major redox sensor that transmits its oxidation signal not only to the Trx pool, but also to other target proteins (Fig. 6A). It is notable in this regard that potentially regulatory interactions of Prx proteins with such targets as glutathione S-transferase/JNK, cellular myelocytomatosis oncogene (cMyc), cellular abelson (cAbl) kinase, and platelet-derived growth factor receptors have been reported, although a direct link between Prx redox status and activity of a signaling protein is not yet clear (10, 21). Speculation that the activation of Ask1 as promoted by Trx oxidation is a consequence of ROS-stimulated, Trx-dependent Prx activity is an intriguing possibility for the facilitated oxidation model of Prx function (86).
We note that none of these models yet provides a physiological role for the observation that many of the abundant Prx proteins designated as typical 2-Cys Prxs are stabilized as decamers when in their reduced or hyperoxidized states, whereas oxidation promotes their dissociation to dimers (28). One might therefore speculate that dimers would be more capable of interacting with other proteins than would decamers or higher order aggregates, and that disulfide bond formation, unlike reduction or hyperoxidation, would promote these interactions. This is yet another possible mode through which Prx proteins could modulate cell signaling functions in a redox-sensitive manner.
Conclusions
While cysteine residues often play critical roles in enzyme catalysis, they also act as redox switches in many enzymes, allowing for communication between the global or local cellular redox properties and enzymatic function. While much remains to be learned regarding the landscape of biological conditions over which these transformations are important, evidence points to redox control of metabolism and signaling being much more widespread and important than has been appreciated. Also, there is clearly an element of crosstalk between redox-controlled signaling and the phosphorylation cascades that constitute a much better understood component of signal transduction. Thiol-based peroxidase functions are clearly linked to ROS control and in some organisms directly mediate transcriptional responses. The molecular details of how redox chemistry functions in mammalian signaling is the subject of much debate and this promises that we are in for an exciting era of discovery of the specific molecular targets involved in ROS-dependent signaling.
Abbreviations Used
- AhpC
alkyl hydroperoxide reductase peroxidase component
- Akt
RAC-alpha serine/threonine-protein kinase or protein kinase B
- Ask1
apoptosis signal-regulating kinase 1
- Atf1
bZIP-type cAMP-dependent transcription factor
- cAbl
cellular abelson kinase
- Cdc
cell division cycle
- cMyc
cellular myelocytomatosis oncogene
- cSrc
cellular sarcoma kinase
- DTT
dithiothreitol
- ERK
extracellular signal-regulated kinase
- FAD
flavin adenine dinucleotide
- GapDH
glyceraldehyde-3-phosphate dehydrogenase
- Gpx
glutathione peroxidase
- Grx
glutaredoxin
- JNK
c-Jun N-terminal kinase
- Keap1
Kelch-like ECK-associated protein 1
- MEKK
MAPK/ERK kinase kinase
- Nrf2
NF-E2-related factor-2
- Pap1
Schizosaccharomyces pombe activator protein-1
- PKA
protein kinase A
- PKC
protein kinase C
- PKG
cGMP-dependent protein kinase
- Prx
peroxiredoxin
- PTEN
phosphatase and tensin homolog
- PTP
protein tyrosine phosphatase
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SHP
SH2 domain-containing protein tyrosine phosphatase
- Sty1
stress-activated protein kinase in fission yeast
- TNFα
tumor necrosis factor alpha
- Tpx1
thiol peroxidase 1
- Trx
thioredoxin
- Yap1
yeast activator protein-1
- Ybp1
Yap1 binding protein
Acknowledgments
Support from the National Institutes of Health to L.B.P. (RO1 GM050389, with a subcontract to P.A.K.; and R33 CA126659) is acknowledged. Additional support from the National Science Foundation (MCB-0517343) and from the NSF-NIGMS Program in Mathematical Biology (RO1 GM075304) to Jacquelyn S. Fetrow with L.B.P. as a coinvestigator is also acknowledged. Kimberly Nelson is acknowledged for editorial contributions to the final article.
References
- 1.Allison WS. Formation and reactions of sulfenic acids in proteins. Acc Chem Res. 1976;9:293–299. [Google Scholar]
- 2.Bechtold E. Reisz JA. Klomsiri C. Tsang AW. Wright MW. Poole LB. Furdui CM. King SB. Water-soluble triarylphosphines as biomarkers for protein S-nitrosation. ACS Chem Biol. 2010;5:405–414. doi: 10.1021/cb900302u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandes N. Schmitt S. Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown KK. Eriksson SE. Arner ES. Hampton MB. Mitochondrial peroxiredoxin 3 is rapidly oxidized in cells treated with isothiocyanates. Free Radic Biol Med. 2008;45:494–502. doi: 10.1016/j.freeradbiomed.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 5.Burgoyne JR. Madhani M. Cuello F. Charles RL. Brennan JP. Schroder E. Browning DD. Eaton P. Cysteine redox sensor in PKGIa enables oxidant-induced activation. Science. 2007;317:1393–1397. doi: 10.1126/science.1144318. [DOI] [PubMed] [Google Scholar]
- 6.Butterfield DA. Hardas SS. Lange ML. Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer's disease: many pathways to neurodegeneration. J Alzheimers Dis. 2010;20:369–393. doi: 10.3233/JAD-2010-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caselli A. Marzocchini R. Camici G. Manao G. Moneti G. Pieraccini G. Ramponi G. The inactivation mechanism of low molecular weight phosphotyrosine-protein phosphatase by H2O2. J Biol Chem. 1998;273:32554–32560. doi: 10.1074/jbc.273.49.32554. [DOI] [PubMed] [Google Scholar]
- 8.Chen CY. Willard D. Rudolph J. Redox regulation of SH2-domain-containing protein tyrosine phosphatases by two backdoor cysteines. Biochemistry. 2009;48:1399–1409. doi: 10.1021/bi801973z. [DOI] [PubMed] [Google Scholar]
- 9.Chen K. Kirber MT. Xiao H. Yang Y. Keaney JF., Jr. Regulation of ROS signal transduction by NADPH oxidase 4 localization. J Cell Biol. 2008;181:1129–1139. doi: 10.1083/jcb.200709049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi MH. Lee IK. Kim GW. Kim BU. Han YH. Yu DY. Park HS. Kim KY. Lee JS. Choi C. Bae YS. Lee BI. Rhee SG. Kang SW. Regulation of PDGF signalling and vascular remodelling by peroxiredoxin II. Nature. 2005;435:347–353. doi: 10.1038/nature03587. [DOI] [PubMed] [Google Scholar]
- 11.Conway ME. Poole LB. Hutson SM. Roles for cysteine residues in the regulatory CXXC motif of human mitochondrial branched chain aminotransferase enzyme. Biochemistry. 2004;43:7356–7364. doi: 10.1021/bi0498050. [DOI] [PubMed] [Google Scholar]
- 12.Cross JV. Templeton DJ. Oxidative stress inhibits MEKK1 by site-specific glutathionylation in the ATP-binding domain. Biochem J. 2004;381:675–683. doi: 10.1042/BJ20040591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross JV. Templeton DJ. Regulation of signal transduction through protein cysteine oxidation. Antioxid Redox Signal. 2006;8:1819–1827. doi: 10.1089/ars.2006.8.1819. [DOI] [PubMed] [Google Scholar]
- 14.D'Autreaux B. Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 15.Dalle-Donne I. Rossi R. Colombo G. Giustarini D. Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Day AM. Veal EA. Hydrogen peroxide-sensitive cysteines in the Sty1 MAPK regulate the transcriptional response to oxidative stress. J Biol Chem. 2010;285:7505–7516. doi: 10.1074/jbc.M109.040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denu JM. Dixon JE. Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Curr Opin Chem Biol. 1998;2:633–641. doi: 10.1016/s1367-5931(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 18.Denu JM. Tanner KG. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 19.Fang J. Nakamura T. Cho DH. Gu Z. Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson's disease. Proc Natl Acad Sci USA. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrell JE., Jr. Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 21.Forman HJ. Maiorino M. Ursini F. Signaling functions of reactive oxygen species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu X. Kassim SY. Parks WC. Heinecke JW. Hypochlorous acid oxygenates the cysteine switch domain of pro-matrilysin (MMP-7). A mechanism for matrix metalloproteinase activation and atherosclerotic plaque rupture by myeloperoxidase. J Biol Chem. 2001;276:41279–41287. doi: 10.1074/jbc.M106958200. [DOI] [PubMed] [Google Scholar]
- 23.Fu X. Mueller DM. Heinecke JW. Generation of intramolecular and intermolecular sulfenamides, sulfinamides, and sulfonamides by hypochlorous acid: a potential pathway for oxidative cross-linking of low-density lipoprotein by myeloperoxidase. Biochemistry. 2002;41:1293–1301. doi: 10.1021/bi015777z. [DOI] [PubMed] [Google Scholar]
- 24.Gallogly MM. Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Giannoni E. Buricchi F. Raugei G. Ramponi G. Chiarugi P. Intracellular reactive oxygen species activate Src tyrosine kinase during cell adhesion and anchorage-dependent cell growth. Mol Cell Biol. 2005;25:6391–6403. doi: 10.1128/MCB.25.15.6391-6403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopalakrishna R. Jaken S. Protein kinase C signaling and oxidative stress. Free Radic Biol Med. 2000;28:1349–1361. doi: 10.1016/s0891-5849(00)00221-5. [DOI] [PubMed] [Google Scholar]
- 27.Gu Z. Kaul M. Yan B. Kridel SJ. Cui J. Strongin A. Smith JW. Liddington RC. Lipton SA. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- 28.Hall A. Karplus PA. Poole LB. Typical 2-Cys peroxiredoxins—structures, mechanisms and functions. FEBS J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hampton MB. Stamenkovic I. Winterbourn CC. Interaction with substrate sensitises caspase-3 to inactivation by hydrogen peroxide. FEBS Lett. 2002;517:229–232. doi: 10.1016/s0014-5793(02)02629-7. [DOI] [PubMed] [Google Scholar]
- 30.Headlam HA. Gracanin M. Rodgers KJ. Davies MJ. Inhibition of cathepsins and related proteases by amino acid, peptide, and protein hydroperoxides. Free Radic Biol Med. 2006;40:1539–1548. doi: 10.1016/j.freeradbiomed.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 31.Held JM. Danielson SR. Behring JB. Atsriku C. Britton DJ. Puckett RL. Schilling B. Campisi J. Benz CC. Gibson BW. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol Cell Proteomics. 2010;9:1400–1410. doi: 10.1074/mcp.M900643-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Z. Pinto JT. Deng H. Richie JP., Jr Inhibition of caspase-3 activity and activation by protein glutathionylation. Biochem Pharmacol. 2008;75:2234–2244. doi: 10.1016/j.bcp.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphries KM. Deal MS. Taylor SS. Enhanced dephosphorylation of cAMP-dependent protein kinase by oxidation and thiol modification. J Biol Chem. 2005;280:2750–2758. doi: 10.1074/jbc.M410242200. [DOI] [PubMed] [Google Scholar]
- 34.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishii T. Sunami O. Nakajima H. Nishio H. Takeuchi T. Hata F. Critical role of sulfenic acid formation of thiols in the inactivation of glyceraldehyde-3-phosphate dehydrogenase by nitric oxide. Biochem Pharmacol. 1999;58:133–143. doi: 10.1016/s0006-2952(99)00060-x. [DOI] [PubMed] [Google Scholar]
- 36.Jang HH. Lee KO. Chi YH. Jung BG. Park SK. Park JH. Lee JR. Lee SS. Moon JC. Yun JW. Choi YO. Kim WY. Kang JS. Cheong GW. Yun DJ. Rhee SG. Cho MJ. Lee SY. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Klomsiri C. Nelson KJ. Bechtold E. Soito L. Johnson LC. Lowther WT. Ryu S-E. King SB. Furdui CM. Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection; evaluation of conditions affecting probe incorporation into redox-sensitive proteins. Methods Enzymol. 2010;473:77–94. doi: 10.1016/S0076-6879(10)73003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolch W. Calder M. Gilbert D. When kinases meet mathematics: the systems biology of MAPK signalling. FEBS Lett. 2005;579:1891–1895. doi: 10.1016/j.febslet.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Koo KH. Lee S. Jeong SY. Kim ET. Kim HJ. Song K. Chae H-Z. Regulation of thioredoxin peroxidase activity by C-terminal truncation. Arch Biochem Biophys. 2002;397:312–318. doi: 10.1006/abbi.2001.2700. [DOI] [PubMed] [Google Scholar]
- 40.Le Moan N. Clement G. Le Maout S. Tacnet F. Toledano MB. The Saccharomyces cerevisiae proteome of oxidized protein thiols: contrasted functions for the thioredoxin and glutathione pathways. J Biol Chem. 2006;281:10420–10430. doi: 10.1074/jbc.M513346200. [DOI] [PubMed] [Google Scholar]
- 41.Lee JY. Jung HJ. Song IS. Williams MS. Choi C. Rhee SG. Kim J. Kang SW. Protective role of cytosolic 2-cys peroxiredoxin in the TNF-alpha-induced apoptotic death of human cancer cells. Free Radic Biol Med. 2009;47:1162–1171. doi: 10.1016/j.freeradbiomed.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 42.Lee SR. Yang KS. Kwon J. Lee C. Jeong W. Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 43.Lee W. Choi KS. Riddell J. Ip C. Ghosh D. Park JH. Park YM. Human peroxiredoxin 1 and 2 are not duplicate proteins: the unique presence of CYS83 in Prx1 underscores the structural and functional differences between Prx1 and Prx2. J Biol Chem. 2007;282:22011–22022. doi: 10.1074/jbc.M610330200. [DOI] [PubMed] [Google Scholar]
- 44.Lee YM. Park SH. Shin DI. Hwang JY. Park B. Park YJ. Lee TH. Chae HZ. Jin BK. Oh TH. Oh YJ. Oxidative modification of peroxiredoxin is associated with drug-induced apoptotic signaling in experimental models of Parkinson disease. J Biol Chem. 2008;283:9986–9998. doi: 10.1074/jbc.M800426200. [DOI] [PubMed] [Google Scholar]
- 45.Leichert LI. Gehrke F. Gudiseva HV. Blackwell T. Ilbert M. Walker AK. Strahler JR. Andrews PC. Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci USA. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lou YW. Chen YY. Hsu SF. Chen RK. Lee CL. Khoo KH. Tonks NK. Meng TC. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. FEBS J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- 47.Marino SM. Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michalek RD. Nelson KJ. Holbrook BC. Yi JS. Stridiron D. Daniel LW. Fetrow JS. King SB. Poole LB. Grayson JM. The requirement of reversible cysteine sulfenic acid formation for T cell activation and function. J Immunol. 2007;179:6456–6467. doi: 10.4049/jimmunol.179.10.6456. [DOI] [PubMed] [Google Scholar]
- 49.Moon JC. Hah YS. Kim WY. Jung BG. Jang HH. Lee JR. Kim SY. Lee YM. Jeon MG. Kim CW. Cho MJ. Lee SY. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- 50.Murata H. Ihara Y. Nakamura H. Yodoi J. Sumikawa K. Kondo T. Glutaredoxin exerts an antiapoptotic effect by regulating the redox state of Akt. J Biol Chem. 2003;278:50226–50233. doi: 10.1074/jbc.M310171200. [DOI] [PubMed] [Google Scholar]
- 51.Nelson JW. Creighton TE. Reactivity and ionization of the active site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry. 1994;33:5974–5983. doi: 10.1021/bi00185a039. [DOI] [PubMed] [Google Scholar]
- 52.Nelson KJ. Klomsiri C. Codreanu SG. Soito L. Liebler DC. Rogers LC. Daniel LW. Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection; methods to visualize and identify labeled proteins. Methods Enzymol. 2010;473:95–115. doi: 10.1016/S0076-6879(10)73004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nelson KJ. Parsonage D. Hall A. Karplus PA. Poole LB. Cysteine pKa values for the bacterial peroxiredoxin AhpC. Biochemistry. 2008;47:12860–12868. doi: 10.1021/bi801718d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olahova M. Taylor SR. Khazaipoul S. Wang J. Morgan BA. Matsumoto K. Blackwell TK. Veal EA. A redox-sensitive peroxiredoxin that is important for longevity has tissue- and stress-specific roles in stress resistance. Proc Natl Acad Sci USA. 2008;105:19839–19844. doi: 10.1073/pnas.0805507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan S. Berk BC. Glutathiolation regulates tumor necrosis factor-alpha-induced caspase-3 cleavage and apoptosis: key role for glutaredoxin in the death pathway. Circ Res. 2007;100:213–219. doi: 10.1161/01.RES.0000256089.30318.20. [DOI] [PubMed] [Google Scholar]
- 56.Parmigiani RB. Xu WS. Venta-Perez G. Erdjument-Bromage H. Yaneva M. Tempst P. Marks PA. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc Natl Acad Sci USA. 2008;105:9633–9638. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parsonage D. Youngblood DS. Sarma GN. Wood ZA. Karplus PA. Poole LB. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 2005;44:10583–10592. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pattison DI. Davies MJ. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem Res Toxicol. 2001;14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 59.Paulsen CE. Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Percival MD. Ouellet M. Campagnolo C. Claveau D. Li C. Inhibition of cathepsin K by nitric oxide donors: evidence for the formation of mixed disulfides and a sulfenic acid. Biochemistry. 1999;38:13574–13583. doi: 10.1021/bi991028u. [DOI] [PubMed] [Google Scholar]
- 61.Peskin AV. Low FM. Paton LN. Maghzal GJ. Hampton MB. Winterbourn CC. The high reactivity of peroxiredoxin 2 with H2O2 is not reflected in its reaction with other oxidants and thiol reagents. J Biol Chem. 2007;282:11885–11892. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- 62.Phalen TJ. Weirather K. Deming PB. Anathy V. Howe AK. van der Vliet A. Jönsson TJ. Poole LB. Heintz NH. Oxidation state governs structural transitions in peroxiredoxin II that correlate with cell cycle arrest and recovery. J Cell Biol. 2006;175:779–789. doi: 10.1083/jcb.200606005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poole LB. Karplus PA. Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 64.Poole LB. Nelson KJ. Discovering mechanisms of signaling-mediated cysteine oxidation. Curr Opin Chem Biol. 2008;12:18–24. doi: 10.1016/j.cbpa.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rabilloud T. Heller M. Gasnier F. Luche S. Rey C. Aebersold R. Benahmed M. Louisot P. Lunardi J. Proteomics analysis of cellular response to oxidative stress. Evidence for in vivo overoxidation of peroxiredoxins at their active site. J Biol Chem. 2002;277:19396–19401. doi: 10.1074/jbc.M106585200. [DOI] [PubMed] [Google Scholar]
- 66.Raftery MJ. Yang Z. Valenzuela SM. Geczy CL. Novel intra- and inter-molecular sulfinamide bonds in S100A8 produced by hypochlorite oxidation. J Biol Chem. 2001;276:33393–33401. doi: 10.1074/jbc.M101566200. [DOI] [PubMed] [Google Scholar]
- 67.Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 68.Salmeen A. Barford D. Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxid Redox Signal. 2005;7:560–577. doi: 10.1089/ars.2005.7.560. [DOI] [PubMed] [Google Scholar]
- 69.Salsbury FR., Jr. Knutson ST. Poole LB. Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17:299–312. doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanchez R. Riddle M. Woo J. Momand J. Prediction of reversibly oxidized protein cysteine thiols using protein structure properties. Protein Sci. 2008;17:473–481. doi: 10.1110/ps.073252408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sayed AA. Williams DL. Biochemical characterization of 2-Cys peroxiredoxins from Schistosoma mansoni. J Biol Chem. 2004;279:26159–26166. doi: 10.1074/jbc.M401748200. [DOI] [PubMed] [Google Scholar]
- 72.Schmalhausen EV. Nagradova NK. Boschi-Muller S. Branlant G. Muronetz VI. Mildly oxidized GAPDH: the coupling of the dehydrogenase and acyl phosphatase activities. FEBS Lett. 1999;452:219–222. doi: 10.1016/s0014-5793(99)00627-4. [DOI] [PubMed] [Google Scholar]
- 73.Seaver LC. Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seth D. Rudolph J. Redox regulation of MAP kinase phosphatase 3. Biochemistry. 2006;45:8476–8487. doi: 10.1021/bi060157p. [DOI] [PubMed] [Google Scholar]
- 75.Sohn J. Rudolph J. Catalytic and chemical competence of regulation of cdc25 phosphatase by oxidation/reduction. Biochemistry. 2003;42:10060–10070. doi: 10.1021/bi0345081. [DOI] [PubMed] [Google Scholar]
- 76.Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 77.Stewart EJ. Aslund F. Beckwith J. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J. 1998;17:5543–5550. doi: 10.1093/emboj/17.19.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stone JR. Yang S. Hydrogen peroxide: a signaling messenger. Antioxid Redox Signal. 2006;8:243–270. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- 79.Sullivan DM. Wehr NB. Fergusson MM. Levine RL. Finkel T. Identification of oxidant-sensitive proteins: TNF-alpha induces protein glutathiolation. Biochemistry. 2000;39:11121–11128. doi: 10.1021/bi0007674. [DOI] [PubMed] [Google Scholar]
- 80.Summers FA. Morgan PE. Davies MJ. Hawkins CL. Identification of plasma proteins that are susceptible to thiol oxidation by hypochlorous acid and N-chloramines. Chem Res Toxicol. 2008;21:1832–1840. doi: 10.1021/tx8001719. [DOI] [PubMed] [Google Scholar]
- 81.Sun G. Kemble DJ. To C or not to C: direct and indirect redox regulation of Src protein tyrosine kinase. Cell Cycle. 2009;8:2353–2355. doi: 10.4161/cc.8.15.9225. [DOI] [PubMed] [Google Scholar]
- 82.Toledano MB. Planson AG. Delaunay-Moisan A. Reining in H2O2 for Safe Signaling. Cell. 2010;140:454–456. doi: 10.1016/j.cell.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 83.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 84.Ushio-Fukai M. Compartmentalization of redox signaling through NADPH oxidase-derived ROS. Antioxid Redox Signal. 2009;11:1289–1299. doi: 10.1089/ars.2008.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watson WH. Pohl J. Montfort WR. Stuchlik O. Reed MS. Powis G. Jones DP. Redox potential of human thioredoxin 1 and identification of a second dithiol/disulfide motif. J Biol Chem. 2003;278:33408–33415. doi: 10.1074/jbc.M211107200. [DOI] [PubMed] [Google Scholar]
- 86.Winterbourn CC. Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 87.Woo HA. Yim SH. Shin DH. Kang D. Yu DY. Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 88.Wood ZA. Poole LB. Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 89.Wood ZA. Schröder E. Harris JR. Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 90.Yamamoto Y. Ritz D. Planson AG. Jonsson TJ. Faulkner MJ. Boyd D. Beckwith J. Poole LB. Mutant AhpC peroxiredoxins suppress thiol-disulfide redox deficiencies and acquire deglutathionylating activity. Mol Cell. 2008;29:36–45. doi: 10.1016/j.molcel.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang J. Groen A. Lemeer S. Jans A. Slijper M. Roe SM. den Hertog J. Barford D. Reversible oxidation of the membrane distal domain of receptor PTPalpha is mediated by a cyclic sulfenamide. Biochemistry. 2007;46:709–719. doi: 10.1021/bi061546m. [DOI] [PubMed] [Google Scholar]
- 92.Yang KS. Kang SW. Woo HA. Hwang SC. Chae HZ. Kim K. Rhee SG. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem. 2002;277:38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 93.Ziegler DM. Role of reversible oxidation-reduction of enzyme thiols-disulfides in metabolic regulation. Annu Rev Biochem. 1985;54:305–329. doi: 10.1146/annurev.bi.54.070185.001513. [DOI] [PubMed] [Google Scholar]