Abstract
Background and Aims
Peripheral populations of plant species are often characterized by low levels of genetic diversity as a result of genetic drift, restricted gene flow, inbreeding and asexual reproduction. These effects can be exacerbated where range-edge populations are fragmented. The main aim of the present study was to assess the levels of genetic diversity in remnant populations of Hypopitys monotropa (syn. Monotropa hypopitys; yellow bird's nest) at the edge of the species' European range in Northern Ireland, since these remnant populations are small and highly fragmented.
Methods
Every plant found through surveys of 21 extant populations was genotyped for eight microsatellite loci to estimate levels and patterns of genetic diversity and clonality.
Key Results
Levels of genetic diversity were relatively high in the populations studied, and the incidence of clonal reproduction was generally low, with a mean of only 14·45 % of clonal individuals. Clones were small and highly spatially structured. Levels of inbreeding, however, were high.
Conclusions
The observed low levels of clonality suggest that the majority of genets in the populations of H. monotropa studied are fertile and that reproduction is predominantly sexual. As the species is highly self-compatible, it is likely that the high levels of inbreeding observed in the populations in the present study are the result of self-pollination, particularly given the small numbers of individuals in most of the patches. Given this extent of inbreeding, further genetic monitoring would be advisable to ensure that genetic diversity is maintained.
Keywords: Clonality, conservation, distribution range, fragmentation, inbreeding, Hypopitys monotropa, Monotropoideae, Pyrolaceae
INTRODUCTION
In plants, contemporary factors determining levels of within and between population genetic variation are often associated with differences in mating systems (sexual vs. asexual, selfing vs. outcrossing) and since many species of plant display flexibility in reproductive strategy, differing modes of reproduction under a variety of environmental conditions directly affect levels of diversity. Clonal reproduction is widespread in plants and can occur via vegetative spread, resulting in the creation of one or more genetically identical ramets, which have grown from an original progenitor plant (genet). Clonal growth has long been viewed as a mechanism to allow an individual to persist in adverse conditions, and factors causing plants to make the switch from sexual to clonal reproduction are often correlated with suboptimal environmental conditions (Tybjerg and Vestergaard, 1992; Eckert, 2002; Honnay and Bossuyt, 2005; Silvertown, 2008). Advantages of clonal reproduction in such marginal habitats include the ability to produce new individuals in the absence of a mate and relative speed of development and hardiness of ramets relative to seedlings.
Habitat fragmentation and loss have also been implicated in causing plant populations to allocate more resources to vegetative reproduction (Smith et al., 2003; Lhullier et al., 2006). Population fragmentation is a common occurrence at the edge of a species' ecological range, due to a lack of suitable habitat (Soulé, 1973; Shumaker and Babble, 1980; Brown, 1984; Caughley et al., 1988). These peripheral populations tend to be characterized by low levels of within-population genetic diversity combined with high levels of genetic differentiation between populations (Vucetich and Waite 2003; Eckert et al. 2008), and these could be compounded by asexual reproduction or processes such as geitonogamy, i.e. cross-fertilization between ramets of the same genet. It has also been suggested, however, that these range edge populations may be more likely to harbour some degree of adaptive potential that the species may ultimately need to survive the changing conditions arising from present-day global warming (Lesica and Allendorf, 1995; Booy et al., 2000; Hampe and Petit, 2005).
Hypopitys monotropa (Ericaceae; syn. Monotropa hypopitys), commonly referred to as yellow bird's nest or pinesap, is a herbaceous perennial plant found in temperate regions within Europe, Asia and North America. Throughout its range the species is considered scarce (Wallace, 1975), and within the UK most of the extant populations are mainly restricted to the south-eastern counties of England. In Northern Ireland, the species exists at the western edge of its distribution range in Europe and is extremely rare due to the limited occurrence of its preferred temperate woodland habitat, and it now occurs only in two regions, namely around the Lower Lough Erne region in Co. Fermanagh, and in a small stretch of woodland along the Co. Antrim coast at Straidkilly (Fig. 1). The species is a self-compatible hermaphrodite and plants can reproduce both sexually and clonally. Hypopitys monotropa is epiparasitic, persisting underground for the majority of the year and producing aerial spikes from late July until late September. Insect pollinators have been noted visiting the flowers (Wallace, 1977), and when seed production does occur, persistence of seedlings is dependent on the presence of mycorrhizal fungi of the genus Tricholoma, as is the case with many members of the Ericaceae (Bidartondo and Bruns, 2001; Leake et al., 2004).
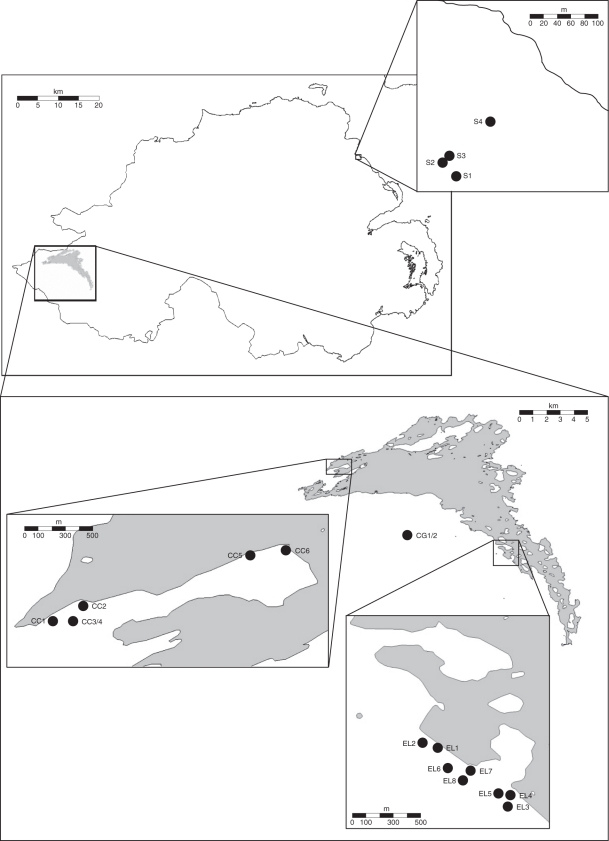
Fig. 1.
Map showing locations of populations analysed in this study. Inset maps show Straidkilly populations (S1–S4), Castle Caldwell populations (CC1–CC6) and Ely Lodge populations (EL1–8). The Knockninny population is not shown.
The main aim of the present study was to assess the levels of genetic diversity in the remaining populations of H. monotropa in Northern Ireland. A previous study on another member of the Monotropoideae, Orthilia secunda, which also exists as a series of small, highly fragmented populations at the edge of its range in Northern Ireland, revealed extremely high levels of clonality (Beatty et al., 2008). Each population comprised a single clone, with a complete lack of within-population genetic variation. Consequently, we were particularly interested in determining whether these peripheral populations of H. monotropa were also characterized by high levels of clonal growth, since extant populations of both species are restricted to the same two areas and since such data is important for the formation of rational, sustainable conservation measures for these threatened populations.
MATERIALS AND METHODS
Surveys and study populations
Surveys were carried out in 2007 and 2008 for Co. Fermanagh populations, and 2009 and 2010 for Co. Antrim populations. All sites where Hypopitys monotropa had previously been recorded were visited and the numbers of plants present were recorded and their positions in each population mapped (Fig. 1 and Table 1). Errigal Banks, Co. Londonderry, was also surveyed in 2007 and 2008, since the species had previously been recorded there, although it had not been found in recent years.
Table 1.
Number of plants (N) found in each population of H. monotropa over successive years
|
N |
Both years |
|||||||
|---|---|---|---|---|---|---|---|---|
| County | Location | Grid Ref. | Population | Year 1* | Year 2† | MLGs | % in both‡ | Population size§ |
| Co. Antrim | Straidkilly | D297156 | S1 | 76 (59) | 87 (82) | 95 | 40 | 127 |
| D297156 | S2 | 49 (35) | 42 (34) | 44 | 57 | 48 | ||
| D297156 | S3 | 9 (8) | 3 (3) | 8 | 38 | 8 | ||
| D297156 | S4 | 1 | 1 | 1 | 100 | 1 | ||
| Co. Fermanagh | Castle Caldwell | H013603 | CC1 | 1 | – | – | – | NC |
| H018605 | CC2 | 6 (6) | 9 (7) | 9 | 44 | 11 | ||
| H017605 | CC3 | 10 (8) | 8 (5) | 9 | 44 | 10 | ||
| H017605 | CC4 | 2 (2) | – | – | – | NC | ||
| H024606 | CC5 | 5 (4) | 8 (5) | 5 | 80 | 5 | ||
| H025607 | CC6 | 2 (2) | 1 | 2 | 50 | 2 | ||
| Correl Glen | H074546 | CG1 | 1 | 9 (4) | 4 | 25 | 9 | |
| H075546 | CG2 | 1 | – | – | – | NC | ||
| Ely Lodge | H176526 | EL1 | 26 (23) | 11 (9) | 24 | 33 | 26 | |
| H176520 | EL2 | 16 (15) | 4 (4) | 15 | 27 | 15 | ||
| H181515 | EL3 | 5 (5) | 19 (13) | 15 | 20 | 22 | ||
| H184514 | EL4 | 13 (13) | 14 (11) | 13 | 85 | 13 | ||
| H184514 | EL5 | 2 (2) | – | – | – | NC | ||
| H178518 | EL6 | – | 2 (1) | – | – | NC | ||
| H181517 | EL7 | – | 1 | – | – | NC | ||
| H180515 | EL8 | – | 2 (2) | – | – | NC | ||
| Knockninny | H272303 | K1 | 2 (1) | – | – | – | NC | |
Figures in parentheses indicate number of distinct multi-locus genotypes (MLGs) in populations with more than a single individual.
NC, Not calculated.
* 2007 for Co. Fermanagh populations, 2009 for Co. Antrim populations.
† 2008 for Co. Fermanagh populations, 2010 for Co. Antrim populations.
‡ Percentage of total MLGs found in both years.
§ Based on capture–recapture calculation (see Materials and methods).
Sampling and DNA extraction
Hypopitys monotropa is protected under Schedule 8 of the Wildlife (Northern Ireland) Order (1985) and, as such, it is an offence to pick, uproot or destroy the plant. Consequently, two scales were taken from each plant and stored in silica gel for transportation to the laboratory. DNA was extracted from one scale per individual using the Qiagen DNeasy Plant Mini Kit, after an initial 3-min grinding at 30 Hz using a Retsch MM300 mixer mill. DNA was quantified visually on 1 % agarose gels stained with ethidium bromide and diluted to a concentration of 50 ng μL−1 for subsequent PCR.
Microsatellite genotyping
Individuals were genotyped for five H. monotropa microsatellite loci previously described in Klooster et al. (2009): Mono02, Mono15, Mono20, Mono21 and Mono22. Three additional loci developed for this study using the ISSR-cloning technique outlined in Provan and Wilson (2007) were also used (Table 2). Forward primers were modified by the addition of a 19-bp M13 tail (5′-CACGACGTTGTAAAACGAC-3') and reverse primers were modified by the addition of a 7-bp tail (5'-GTGTCTT-3'). PCR was carried out in a total volume of 10 µL containing 100 ng genomic DNA, 10 pmol of dye-labelled M13 primer (6-FAM or HEX), 1 pmol of tailed forward primer, 10 pmol reverse primer, 1× PCR reaction buffer, 200 µm each dNTP, 2·5 mm MgCl2 and 0·25 U GoTaq Flexi DNA polymerase (Promega). PCR was carried out on a MWG Primus thermal cycler using the conditions described in Klooster et al. (2009) and genotyping was carried out on an AB3730xl capillary genotyping system. Allele sizes were scored in GeneMapper V4·1 (Applied Biosystems) using LIZ-500 size standards and were checked by comparison with previously sized control samples.
Table 2.
Hypopitys monotropa microsatellite primers developed for this study
| Locus | Repeat | Primers | Size range |
|---|---|---|---|
| MHNSSR108 | (GA)8 | ACATTTGGGAAAATGGGAGA | 130–146 bp |
| TTCAATGGCACGTCTTACACA | |||
| MHNSSR119 | Complex (GA) | GGAAGTTTCTCCATCCAGGTT | 146–180 bp |
| AGCAATCAAAACCAGGACCA | |||
| MHNSSR135 | (AG)8 | CGGTTTCAGGAAACAAAACC | 126–148 bp |
| TTGTCCGGGAATTCTCTCTC |
Data analysis
As H. monotropa possesses the capacity for clonal reproduction, clonemates were identified by calculating the probability (PGEN) of each multi-locus genotype (MLG) arising through sexual as opposed to clonal reproduction following the method of Parks and Werth (1993):
where h is the number of loci at which the genotype is heterozygous, x1i is the allele frequency of the first allele in the genotype at locus i, x2i is the allele frequency of the second allele in the genotype at locus i. The observation of heterozygosity allows the differentiation between genetic identity due to clonal propagation and identity due to inbreeding or selfing, which would lead to an increase in homozygosity. Samples which shared MLGs with PGEN < 0·05 were considered clonemates and identified as such on the maps, and duplicate genotypes were removed from subsequent analyses. PGEN values were calculated using the Genclone software package (V2·0; Arnaud-Haond and Belkhir, 2007). Levels of expected heterozygosity (HE) based on nuclear microsatellite allele frequencies were calculated using the Arlequin software package (V3·01; Excoffier et al., 2005) for populations with a sample size of N ≥ 5. The significance of differences in values of HE between years was estimated using two-tailed paired t-tests, and a Spearman's rank correlation coefficient was calculated to test for any association between population size and genetic diversity. Levels of clonal diversity were estimated by calculating genotypic richness (R), defined as (G – 1)/(N – 1), where G is the number of MLGs and N is the number of plants in the population. Where samples were located in successive years, an estimate of population size was derived using a simple capture–recapture formula (number of MLGs in year 1 × number of MLGs in year 2 ÷ number of MLGs identified in both years). Inbreeding coefficients (FIS) were estimated using the Genepop software package (V4.0.1.0; Raymond and Rousset 1995). To test for genetic differences in populations between successive years, analyses of molecular variation (AMOVA) were carried out using the Arlequin software package (V3·01; Excoffier et al., 2005). AMOVAs were also carried out to determine the levels of genetic differentiation between populations within a location.
RESULTS
The present occurrence of Hypopitys monotropa in Northern Ireland would appear to be restricted to four locations in Co. Fermanagh (Castle Caldwell, Correl Glen, Ely Lodge and Knockninny) and one location in Co. Antrim (Straidkilly), although the Knockninny population was not found in the second year of surveying. The population recorded from Errigal Banks, Co. Londonderry was not found in either of the years surveyed and is most likely now extinct. Populations tended to be very small, generally with less than ten spikes per patch, although two large populations were found in Straidkilly in both 2009 and 2010 (Table 1). Some notable changes in population size were observed over successive years, including growth from a single spike to nine spikes in Correl Glen population CG1 and from five to 19 spikes in Ely Lodge population EL3, and a decrease from 16 to four spikes in Ely Lodge population EL2.
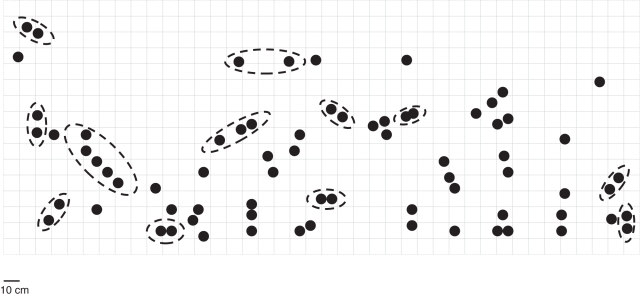
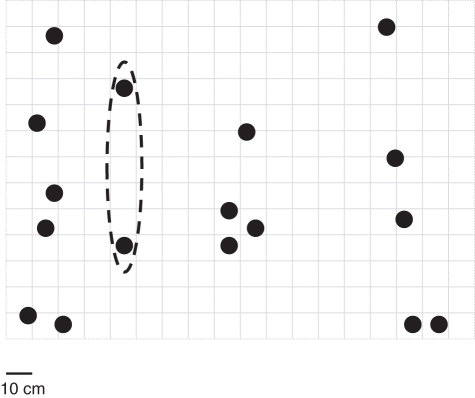
Between ten and 23 alleles were detected at the eight microsatellite loci analysed (mean = 14·75). All identical MLGs had PGEN values of <0·001, confirming that they arose via asexual reproduction. Identification of clonemates based on identical MLGs indicated that the levels of clonal reproduction were generally low, with a mean of only 14·45 % of clonal individuals (Table 1). Identical MLGs were always highly spatially structured and generally small (Figs 2 and 3; also see Figs S1–S25 in Supplementary data available online). Usually, these involved pairs of clonemates within 10 cm of each other, although larger clones were detected [e.g. five ramets spanning 40 cm (Straidkilly population S1, 2009; Fig. 2) and two identical MLGs separated by 60 cm with no intervening spikes (Ely Lodge population EL2, 2007; Fig. 3)]. In total, 38 MLGs were represented by two ramets, 12 by three ramets and two each by four and five ramets. The percentage of MLGs persisting in a population across both years of study (in cases where N was greater than 1) ranged from 20 % (Ely Lodge population EL3) to 85 % (Ely Lodge population EL4; Table 1).
Fig. 2.
Distribution of individuals in the Straidkilly S1, 2009 population. Dashed lines enclose multi-locus genotypes which are identical. Each square represents 10 cm.
Fig. 3.
Distribution of individuals in the Ely Lodge EL2, 2007 population. Dashed lines enclose multi-locus genotypes which are identical. Each square represents 10 cm.
Levels of within-population expected heterozygosity (HE) calculated for populations with N ≥ 5 (Table 3) ranged from 0·309 (Castle Caldwell population CC2, 2007) to 0·672 (Straidkilly population S3, 2009), with mean values of 0·507 and 0·492 in both years. Levels of HE calculated across both years ranged from 0·336 (Castle Caldwell population CC2) to 0·672 (Straidkilly population S3), with a mean value of 0·521. Levels of clonal diversity (R) ranged from 0·571 (Castle Caldwell populations CC3 and CC5) to 1·000 (Castle Caldwell population CC2 and Ely Lodge populations EL3 and EL4), with mean values of 0·870 and 0·737 in both years. Levels of R calculated across both years ranged from 0·333 (Castle Caldwell population CC5) to 0·737 (Ely Lodge population EL2), with a mean value of 0·552. The only significant difference in HE between successive years was a drop from HE = 0·596 to HE = 0·417 in Castle Caldwell population CC3 (two-sided paired t-test P = 0·020). The Spearman's rank correlation coefficient revealed no association between population size (N) and levels of HE (rs = 0·289; P = 0·122). Population sizes based on the capture–recapture calculation from MLGs across both years ranged from 1 (Straidkilly population S4) to 127 (Straidkilly population S1). Inbreeding coefficients (FIS) ranged from 0·114 (Ely Lodge population EL1, 2008) to 0·813 (Castle Caldwell population CC2, 2007), with mean values of 0·497 and 0·339 in both years.
Table 3.
Levels of expected heterozygosity (HE) and clonal diversity (R), and inbreeding coefficient (FIS) by population
|
HE |
R |
FIS |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Population | Year 1* | Year 2† | Both | P | Year 1* | Year 2† | Both | Year 1* | Year 2† |
| Straidkilly S1 | 0·583 | 0·540 | 0·566 | 0·106 | 0·773 | 0·965 | 0·580 | 0·497 | 0·575 |
| Straidkilly S2 | 0·591 | 0·546 | 0·582 | 0·128 | 0·708 | 0·805 | 0·478 | 0·419 | 0·417 |
| Straidkilly S3 | 0·672 | NC | 0·672 | NC | 0·875 | NC | 0·636 | 0·367 | NC |
| Straidkilly total | 0·615 | 0·566 | 0·590 | 0·756 | 0·895 | 0·551 | |||
| Castle Caldwell CC2 | 0·309 | 0·374 | 0·336 | 0·121 | 1·000 | 0·750 | 0·571 | 0·813 | 0·686 |
| Castle Caldwell CC3 | 0·596 | 0·417 | 0·584 | 0·020 | 0·778 | 0·571 | 0·471 | 0·530 | 0·125 |
| Castle Caldwell CC5 | 0·545 | 0·525 | 0·525 | 0·142 | 0·750 | 0·571 | 0·333 | 0·579 | 0·335 |
| Castle Caldwell total | 0·527 | 0·497 | 0·511 | 0·833 | 0·667 | 0·489 | |||
| Ely Lodge EL1 | 0·384 | 0·442 | 0·396 | 0·327 | 0·880 | 0·800 | 0·639 | 0·183 | 0·114 |
| Ely Lodge EL2 | 0·455 | NC | 0·455 | NC | 0·933 | NC | 0·737 | 0·509 | NC |
| Ely Lodge EL3 | 0·492 | 0·658 | 0·648 | 0·060 | 1·000 | 0·667 | 0·609 | 0·470 | 0·490 |
| Ely Lodge EL4 | 0·447 | 0·435 | 0·447 | 0·569 | 1·000 | 0·769 | 0·462 | 0·601 | 0·646 |
| Ely Lodge total | 0·496 | 0·600 | 0·543 | 0·898 | 0·765 | 0·598 | |||
| Mean by population | 0·507 | 0·492 | 0·521 | 0·870 | 0·737 | 0·552 | 0·497 | 0·339 | |
Only patches with N ≥ 5 were analysed.
NC, Not calculated due to small sample size.
P, Two-tailed paired t-test probability values for differences in HE between years.
* 2007 for Co. Fermanagh populations, 2009 for Co. Antrim populations.
† 2008 for Co. Fermanagh populations, 2010 for Co. Antrim populations.
The AMOVA analyses revealed no significant genetic differentiation in populations across successive years (data not shown). Significant levels of genetic differentiation between populations within locations were detected in five of the six analyses, with between 7·07 % and 19·59 % of the total observed variation existing between populations within a region (Table 4).
Table 4.
Analysis of molecular variance (AMOVA) for population differentiation by location
| County | Location | Year 1* |
Year 2† |
||
|---|---|---|---|---|---|
| % variation | P | % variation | P | ||
| Co. Antrim | Straidkilly | 7·07 | P < 0·001 | 9·63 | P < 0·001 |
| Co. Fermanagh | Castle Caldwell | 16·77 | P = 0·075 | 18·76 | P < 0·001 |
| Ely Lodge | 19·59 | P < 0·001 | 19·32 | P < 0·001 | |
* 2007 for Co. Fermanagh populations, 2009 for Co. Antrim populations.
† 2008 for Co. Fermanagh populations, 2010 for Co. Antrim populations.
DISCUSSION
Genetic diversity and levels of clonality in peripheral populations of Hypopitys monotropa
Despite occurring in small, highly fragmented populations in Northern Ireland, Hypopitys monotropa exhibited relatively high levels of within-population genetic diversity. The observed mean clonal diversity (R) value of 0·804 was very high compared with the average value for clonal plants (R = 0·17) reported by Ellstrand and Roose (1987). This figure was also at the upper end of the range of more recently published values for understorey herb species [0·08, Uvularia perfoliata (Kudoh et al., 1999); 0·21, Paris quadrifolia (Jacquemyn et al., 2005); 0·33, Convallaria keiskei (Araki et al., 2007); 0·43, Paris quadrifolia (Jacquemyn et al., 2006); 0·47, Mercurialis perennis (Vandepitte et al., 2010); 0·83, Trillium cuneatum (Gonzales et al., 2008); 0·84, Anemone nemorosa (Rusterholz et al., 2009); 0·93, Viola riviniana (Auge et al., 2001)]. These high levels of diversity are in stark contrast to those observed in Orthilia secunda, another member of the Monotropoideae that is restricted to the same two locations in Northern Ireland. A previous study on this species (Beatty et al., 2008) revealed that each population comprised a single clone. Both species currently exist in highly fragmented populations at the edge of their ranges in the same areas in Northern Ireland, but H. monotropa is primarily a temperate species whereas O. secunda generally has a more boreal distribution. It is thus possible that climatic factors have influenced the switch to extensive clonal growth in the latter species. Nevertheless, although H. monotropa exhibited far higher diversity than O. secunda in Northern Ireland, levels of expected heterozygosity were significantly lower than those calculated from populations in the main part of the species' distribution range in Europe (Mann-Whitney U-test P = 0·002; Beatty and Provan, 2011). Such a decrease in genetic variation in range-edge populations has also been reported in other clonal plant species (Lammi et al., 1999; Billingham et al., 2003; Jump et al., 2003; Alberto et al., 2006; Eckstein et al., 2006).
Clones in H. monotropa were small, extending at most over a few tens of centimetres. In most cases, they were pairs of very closely spaced, adjacent MLGs as found in previous studies on other clonal plant species (Harada et al., 1997; Holderegger et al., 1998; Suzuki et al., 2006). In addition, identical MLGs were never found interspersed with other MLGs. Thus, the mode of clonal spread in H. monotropa would appear to conform to the ‘phalanx’ dynamic, where clones are characterized by compact growth forms, rather than the ‘guerrilla’ pattern, where longer rhizomes are intermingled, giving rise to clusters of different ramets (Lovett Doust, 1981; Humphrey and Pyke, 1998). These small clones are again in contrast to those observed previously in O. secunda, where several large monoclonal patches were found, including one comprised of approx. 600 individuals covering an area of approx. 300 m2 (Beatty et al. 2008). Furthermore, populations of H. monotropa in the present study did not appear to be composed of the entirely same clones across both years of study, although the overall genetic difference between years was non-significant.
The observed low levels of clonality suggest that most genets in the populations of H. monotropa studied are fertile and that reproduction is predominantly sexual. The observed levels of inbreeding, however, were high in almost all of the populations studied. A previous study on the reproductive ecology of the genus Monotropa and the related genus Monotropsis identified differences in levels of autogamous seed set in the two colour morphs of H. monotropa that are found in North America (Klooster and Culley, 2009). Both the red and yellow forms (the yellow form being the morph found in Britain and Ireland) were highly self-compatible, but only the yellow form set substantial amounts of autogamous seed after self-pollination. Thus, it is likely that the high levels of inbreeding observed in the populations in the present study are the result of self-pollination, particularly given the small numbers of individuals in most of the patches. Being self-compatible, however, means that H. monotropa does not face the same problems of complete loss of sexual reproduction and/or rapid population extinction that can threaten populations of obligately outcrossing clonal plants. Where mate availability is limited in such species, or where populations are comprised of a small number of large clones, many of which are often related, self-incompatibilty mechanisms and stigma saturation via self-pollination can lead to sexual reproductive failure and subsequent extensive loss of genetic variation (e.g. Willi et al., 2005; Scobie and Wilcock, 2009). Nevertheless, ongoing inbreeding remains a potential threat to the fragmented, peripheral populations investigated in the present study.
Conservation implications
In the present study, the transient nature of H. monotropa was noted at both the population level and at the individual level within populations. Even large populations have been observed to disappear within a few years (Lockton and Walker, 2010), which poses a problem when trying to estimate census numbers for the species, as the actual numbers of individuals will not be truly known if a survey was carried out in any single year. Furthermore, an additional issue when attempting to identify census numbers for H. monotropa is the incidence of clonal growth. As aerial spikes do not necessarily represent separate genets, they may in fact represent multiple ramets of the same genet. Genetic analyses over successive years therefore provide vital information on the dynamics of these threatened populations. Although the present study only considered a 2-year period at each location, no significant differences were observed in genetic diversity or composition of the populations between successive seasons, with the exception of a single decrease in diversity in one population. Given the high levels of inbreeding in Northern Ireland's remaining populations of H. monotropa, however, further genetic monitoring would be advisable to ensure that genetic diversity is maintained. If levels of genetic diversity were to drop to the extent that some form of ‘genetic rescue’ is required, then the genetic distinctness between populations revealed by the AMOVA analyses should be taken into account, both in terms of possibly maximizing genetic diversity, but still considering the potential for outbreeding depression (Frankham, 2010). Furthermore, the small and fragmented nature of remnant populations, with low numbers confirmed by the capture–recapture calculation across successive years, leaves them vulnerable to stochastic extinction events.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We are extremely grateful to Robert and Hannah Northridge for providing the Co. Fermanagh 2007 samples, Robert Beatty for assistance with sampling, Ralph Forbes for helpful discussions, and Neil Reid for GIS advice. We are also grateful to three anonymous referees for comments which greatly improved the manuscript. This research was supported by a PhD studentship awarded to Gemma Beatty by the Department of Agriculture and Rural Development, Northern Ireland.
LITERATURE CITED
- Alberto F, Arnaud-Haond S, Duarte CM, Serrao E. Genetic diversity of a clonal angiosperm near its range limit: the case of Cymodocea nodosa in the Canary Islands. Marine Ecology Progress Series. 2006;309:117–119. [Google Scholar]
- Araki K, Shimatani K, Ohara M. Floral distribution, clonal structure, and their effects on pollination success in a self-incompatible Convallaria keiskei population in northern Japan. Plant Ecology. 2007;189:175–186. [Google Scholar]
- Arnaud-Haond S, Belkhir K. GeneClone: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Molecular Ecology Notes. 2007;7:15–17. [Google Scholar]
- Auge H, Neuffer B, Erlinghagen F, Grupe R, Brandl R. Demographic and random amplified polymorphic DNA analyses reveal high levels of genetic diversity in a clonal violet. Molecular Ecology. 2001;10:1811–1819. doi: 10.1046/j.0962-1083.2001.01311.x. [DOI] [PubMed] [Google Scholar]
- Beatty GE, Provan J. Comparative phylogeography of two related plant species with overlapping ranges in Europe, and the potential effects of climate change on their intraspecific genetic diversity. BMC Evolutionary Biology. 2011 doi: 10.1186/1471-2148-11-29. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty GE, McEvoy PM, Sweeney O, Provan J. Range-edge effects promote clonal growth in peripheral populations of the one-sided wintergreen (Orthilia secunda) Diversity and Distributions. 2008;14:546–555. [Google Scholar]
- Bidartondo MI, Bruns TD. Extreme specificity in epiparasitic Monotropoideae (Ericaceae): widespread phylogenetic and geographic structure. Molecular Ecology. 2001;10:2285–2295. doi: 10.1046/j.1365-294x.2001.01358.x. [DOI] [PubMed] [Google Scholar]
- Billingham MR, Reusch TBH, Alberto F, Serrao EA. Is asexual reproduction more important at geographical limits? A genetic study of the seagrass Zostera marina in the Ria Formosa, Portugal. Marine Ecology Progress Series. 2003;265:77–83. [Google Scholar]
- Booy G, Hendriks RJJ, Smulders MJM, van Groenendael JM, Vosman B. Genetic diversity and the survival of populations. Plant Biology. 2000;2:379–395. [Google Scholar]
- Brown JH. On the relationship between abundance and distribution of species. American Naturalist. 1984;124:255–279. [Google Scholar]
- Caughley D, Grice D, Barker R, Brown B. The edge of range. Journal of Animal Ecology. 1988;57:771–785. [Google Scholar]
- Eckert CG. The loss of sex in clonal plants. Evolutionary Ecology. 2002;15:501–520. [Google Scholar]
- Eckert CG, Samis KE, Lougheed SC. Genetic variation across species' geographical ranges: the central-marginal hypothesis and beyond. Molecular Ecology. 2008;17:1170–1188. doi: 10.1111/j.1365-294X.2007.03659.x. [DOI] [PubMed] [Google Scholar]
- Eckstein RL, O'Neill RA, Danihelka J, Otte SA, Köhler W. Genetic structure among and within peripheral and central populations of three endangered floodplain violets. Molecular Ecology. 2006;15:2367–2379. doi: 10.1111/j.1365-294X.2006.02944.x. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Roose ML. Patterns of genotypic diversity in clonal plant species. American Journal of Botany. 1987;74:123–131. [Google Scholar]
- Excoffier L, Laval LG, Schneider S. Arlequin, Version 3·0: an integrated software package for population genetic data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Challenges and opportunities of genetic approaches to biological conservation. Biological Conservation. 2010;143:1919–1927. [Google Scholar]
- Gonzales E, Hamrick JL, Smouse PE. Comparison of clonal diversity in mountain and piedmont populations of Trillium cuneatum (Melanthiaceae-Trilliaceae), a forest understory species. American Journal of Botany. 2008;95:1254–1261. doi: 10.3732/ajb.2007159. [DOI] [PubMed] [Google Scholar]
- Hampe A, Petit RJ. Conserving biodiversity under climate change: the rear edge matters. Ecology Letters. 2005;8:461–467. doi: 10.1111/j.1461-0248.2005.00739.x. [DOI] [PubMed] [Google Scholar]
- Harada Y, Kawano S, Iwasa Y. Probability of clonal identity: inferring the relative success of sexual versus clonal reproduction from spatial genetic patterns. Journal of Ecology. 1997;85:591–600. [Google Scholar]
- Holderegger R, Stehlik I, Scheller JJ. Estimation of the relative importance of sexual and vegetative reproduction in the clonal woodland herb Anemone nemorosa. Oecologia. 1998;117:105–107. doi: 10.1007/s004420050637. [DOI] [PubMed] [Google Scholar]
- Honnay O, Bossuyt B. Prolonged clonal growth: escape route or route to extinction? Oikos. 2005;108:427–432. [Google Scholar]
- Humphrey LD, Pyke DA. Demographic and growth responses of a guerrilla and a phalanx perennial grass in competitive mixtures. Journal of Ecology. 1998;86:854–865. [Google Scholar]
- Jacquemyn H, Brys R, Honnay O, Hermy M, Roldan-Ruiz I. Local forest environment largely affects below-ground growth, clonal diversity and fine-scale spatial genetic structure in the temperate deciduous forest herb Paris quadrifolia. Molecular Ecology. 2005;14:4479–4488. doi: 10.1111/j.1365-294X.2005.02741.x. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Brys R, Honnay O, Hermy M, Roldan-Ruiz I. Sexual reproduction, clonal diversity and genetic differentiation in patchily distributed populations of the temperate forest herb Paris quadrifolia (Trilliaceae) Oecologia. 2006;147:434–444. doi: 10.1007/s00442-005-0287-x. [DOI] [PubMed] [Google Scholar]
- Jump AS, Woodward FI, Burke T. Cirsium species show disparity in patterns of genetic variation at their range-edge despite similar patterns of reproduction and isolation. New Phytologist. 2003;160:359–370. doi: 10.1046/j.1469-8137.2003.00874.x. [DOI] [PubMed] [Google Scholar]
- Klooster MR, Culley TM. Comparative analysis of the reproductive ecology of Monotropa and Monotropsis: two mycoheterotrophic genera in the Monotropoideae (Ericaceae) American Journal of Botany. 2009;96:1337–1347. doi: 10.3732/ajb.0800319. [DOI] [PubMed] [Google Scholar]
- Klooster MR, Hoenle AW, Culley TM. Characterization of microsatellite loci in the myco-heterotrophic plant Monotropa hypopitys (Ericaceae) and amplification in related taxa. Molecular Ecology Resources. 2009;9:219–221. doi: 10.1111/j.1755-0998.2008.02386.x. [DOI] [PubMed] [Google Scholar]
- Kudoh H, Shibaike H, Takas H, Whigham DF, Kawano S. Genet structure and determinants of clonal structure in a temperate deciduous woodland herb, Uvularia perfoliata. Journal of Ecology. 1999;87:244–257. [Google Scholar]
- Lammi A, Siikamaki P, Mustajarvi K. Genetic diversity, population size, and fitness in central and peripheral populations of a rare plant Lychnis viscaria. Conservation Biology. 1999;13:1069–1078. [Google Scholar]
- Leake JR, McKendrick SL, Bidartondo M, Read DL. Symbiotic germination and development of the myco-heterotroph Monotropa hypopitys in nature and its requirement for locally distributed Tricholoma spp. New Phytologist. 2004;163:405–423. doi: 10.1111/j.1469-8137.2004.01115.x. [DOI] [PubMed] [Google Scholar]
- Lesica P, Allendorf FW. When are peripheral populations valuable for conservation? Conservation Biology. 1995;9:753–760. [Google Scholar]
- Lhullier E, Butaud J-F, Bouvet J-M. Extensive clonality and strong differentiation in the insular Pacific tree Santalum insulare: implications for conservation. Annals of Botany. 2006;98:1061–1072. doi: 10.1093/aob/mcl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockton AJ, Walker KJ. Species account: Monotropa hypopitys. 2010 Botanical Society of the British Isles. http://sppaccounts.bsbi.org.uk/ [Google Scholar]
- Lovett Doust L. Population dynamics and local specialization in a clonal perennial (Ranunculus repens). I. The dynamics of ramets in contrasting habitats. Journal of Ecology. 1981;69:743–755. [Google Scholar]
- Parks JC, Werth CR. A study of spatial features of clones in a population of bracken fern, Pteridium aquilinum (Dennstaedtiaceae) American Journal of Botany. 1993;80:537–544. doi: 10.1002/j.1537-2197.1993.tb13837.x. [DOI] [PubMed] [Google Scholar]
- Provan J, Wilson PJ. Development of microsatellites for the peat moss Sphagnum capillifolium using ISSR cloning. Molecular Ecology Notes. 2007;7:254–256. [Google Scholar]
- Raymond M, Rousset F. Genepop (version 1·2): population genetic software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Rusterholz HP, Kissling M, Baur B. Disturbances by human trampling alter the performance, sexual reproduction and genetic diversity in a clonal woodland herb. Perspectives in Plant Ecology, Evolution and Systematics. 2009;11:17–29. [Google Scholar]
- Scobie AR, Wilcock CC. Limited mate availability decreases reproductiove success of fragmented populations of Linnaea borealis, a rare, clonal self-incompatible plant. Annals of Botany. 2009;103:835–846. doi: 10.1093/aob/mcp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker KM, Babble GR. Patterns of allozymic similarity in ecological central and marginal populations of Hordeum jubatum in Utah. Evolution. 1980;34:110–116. doi: 10.1111/j.1558-5646.1980.tb04793.x. [DOI] [PubMed] [Google Scholar]
- Silvertown J. The evolutionary maintenance of sexual reproduction: evidence from the ecological distribution of asexual reproduction in clonal plants. International Journal of Plant Sciences. 2008;169:157–168. [Google Scholar]
- Smith S, Hughes J, Wardell-Johnson G. High population differentiation and extensive clonality in a rare mallee eucalypt: Eucalyptus curtisii. Conservation Genetics. 2003;4:289–300. [Google Scholar]
- Soulé M. The epistasis cycle: a theory of marginal population. Annual Review of Ecology and Systematics. 1973;4:165–187. [Google Scholar]
- Suzuki J, Herben T, Krahulec F, Storchová H, Hara T. Effects of neighbourhood structure and tussock dynamics on genet demography of Festuca rubra in a mountain meadow. Journal of Ecology. 2006;94:66–76. [Google Scholar]
- Tybjerg H, Vestergaard P. Growth dynamics in the rhizomatous Polygonatum verticillatum. Oikos. 1992;65:395–408. [Google Scholar]
- Vandepitte K, Honnay O, de Meyer T, Jacquemyn H, Roldan-Ruiz I. Patterns of sex ratio variation in the dioecious forest perennial Mercurialis perennis. Plant Ecology. 2010;206:105–114. [Google Scholar]
- Vucetich JA, Waite TA. Spatial patterns of demography and genetic processes across the species' range: null hypotheses for landscape conservation genetics. Conservation Genetics. 2003;4:639–645. [Google Scholar]
- Wallace GD. Studies of the Monotropoideae (Ericaceae): taxonomy and distribution. Wassman Journal of Biology. 1975;33:1–88. [Google Scholar]
- Wallace GD. Studies of the Monotropoideae (Ericaceae). Floral nectarines: anatomy and function in pollination ecology. American Journal of Botany. 1977;64:199–206. [Google Scholar]
- Willi Y, van Buskirk J, Fischer M. A threefold genetic allee effect: population size affects cross-compatibility, inbreeding depression and drift load in the self incompatible Ranunculus reptans. Genetics. 2005;169:2255–2265. doi: 10.1534/genetics.104.034553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.