Abstract
Overall, pediatric high-grade glioma (pHGG) has a poor prognosis, in part due to the lack of understanding of the underlying biology. High-resolution 244 K oligo array comparative genomic hybridization (CGH) was used to analyze DNA from 38 formalin-fixed paraffin-embedded predominantly pretreatment pHGG samples, including 13 diffuse intrinsic pontine gliomas (DIPGs). The patterns of gains and losses were distinct from those seen in HGG arising in adults. In particular, we found 1q gain in up to 27% of our cohort compared with 9% reported in adults. A total of 13% had a balanced genetic profile with no large-scale copy number alterations. Homozygous loss at 8p12 was seen in 6 of 38 (16%) cases of pHGG. This novel deletion, which includes the ADAM3A gene, was confirmed by quantitative real-time PCR (qPCR). Loss of CDKN2A/CDKN2B in 4 of 38 (10%) samples by oligo array CGH was confirmed by fluorescent in situ hybridization on tissue microarrays and was restricted to supratentorial tumors. Only ∼50% of supratentorial tumors were positive for CDKN2B expression by immunohistochemistry (IHC), while ∼75% of infratentorial tumors were positive for CDKN2B expression (P = 0.03). Amplification of the 4q11–13 region was detected in 8% of cases and included PDGFRA and KIT, and subsequent qPCR analysis was consistent with the amplification of PDGFRA. MYCN amplification was seen in 5% of samples being significantly associated with anaplastic astrocytomas (P= 0.03). Overall, DIPG shared similar spectrum of changes to supratentorial HGG with some notable differences, including high-frequency loss of 17p and 14q and lack of CDKN2A/CDKN2B deletion. Informative genetic data providing insight into the underlying biology and potential therapeutic possibilities can be generated from archival tissue and typically small biopsies from DIPG. Our findings highlight the importance of obtaining pretreatment samples.
Keywords: ADAM3A, CDKN2B, diffuse intrinsic pontine glioma, genome wide analysis, MYCN, pediatric high-grade glioma, PDGFRA
High-grade gliomas (HGGs; WHO Grades III and IV) arising in childhood are clinically distinct from those occurring in adults.1 HGGs are the most frequent malignant brain tumor in adults but represent ∼20% of childhood brain tumors.2 Adult HGGs arise predominantly in the cerebral cortex, while in childhood a wide range of anatomical locations might be involved, including the brainstem. Malignant transformation of low-grade gliomas (WHO Grade I and II) is a well-recognized phenomenon in adults, but rarely reported in childhood.3 While the outcome for these highly vascular, invasive, and frequently rapidly advancing tumors is dismal in adults, very young children (children under 5 years) with high-grade astrocytomas appear to have a better outcome.4–6 Prognosis varies depending on the location of the tumor within the brain, the degree of resection, and the treatment possibilities.
Tumors in the brainstem constitute a complex group arising between midbrain and the medullary cervical junction of diverse histological type and grade. The mean age of diagnosis is 7–9 years.7 The majority of brain stem tumors in children are diffuse intrinsic pontine gliomas (DIPGs) characterized by infiltration and expansion of the brainstem and frequently centered on the pons but often extensive.7–9 Characteristic appearances on MRI and the location and infiltrative nature of DIPG allow the possibility of making a diagnosis by imaging without the need for histological confirmation, and surgical biopsies are therefore rarely performed, resulting in a lack of tumor tissue for research.7,9,10 Conventional therapy involves local field radiotherapy, which may result in transient neurological improvement and a transient progression-free survival benefit but provides no improvement in overall survival. The prognosis for these patients regardless of age is appalling and has shown no improvement over time, in part due to the lack of understanding of the underlying molecular pathology of this disease.8 A recognition of the limitations of radiology and re-evaluation of the potential role of biopsy in both helping individual cases and understanding this disease as a whole has occurred in the last 2 years.9,11,12 Indeed, a moral imperative for conducting brainstem biopsies has recently been raised.13
Genetic abnormalities known to occur in adult HGG have been specifically sought in childhood and adolescent HGG (i.e., pediatric HGG [pHGG]) and clear differences in frequency and nature of these reported. For example, amplification of EGFR is the most common genetic abnormality in adult HGG, while in pHGG, although overexpression of epidermal growth factor receptor (EGFR) is common, gene amplification is rare.14 Similarly, loss of p53 is common in adults and children over 3 years, but less so in children under 3 years.15 Deletion of p16 is also seen in 50–70% of adult HGG but is found in <10% of pHGG.2
However, to date there are few unbiased whole genome studies of pretreatment pHGG and in particular of DIPG due largely to the difficulty of obtaining tissue for research, though this is now being addressed.16,17 With recent advances in high throughput genetic techniques on formalin-fixed tissue, we have analyzed and compared the genetic signatures of pHGG arising in the supratentorial region and brainstem (DIPG).
Materials and Methods
Patient Samples
Primary and recurrent pediatric brain tumor samples were identified for entry into this study from the Children's Cancer and Leukaemia Group (CCLG), predominantly from Nottingham Children's Hospitals. Clinical details including diagnostic data were obtained from the CCLG. All cases were centrally reviewed to determine histological diagnosis (J.L., K.R., M.A.-B.) according to the 2007 classification of the World Health Organization (WHO).18 Only cases of WHO Grades III and IV gliomas were included in this study. Areas of viable and representative tumor following review of paraffin blocks were marked by the reviewing pathologist. Data of date of birth, age at diagnosis, imaging (CT/MRI scans or reports), degree of surgical resection, treatment, and current status were obtained from the data center and the case notes. Certain CCLG centers were undertaking biopsies of diffuse brainstem tumors over this time period, providing tissue for the DIPG element of this study.10
The study had Multiple Research Ethics Committee approval, and tumor samples were consented to in accordance with national banking procedures and the Human Tissue Act.
Tissue Microarrays
An extended cohort of pHGG samples was identified from which a tissue microarray (TMA) of 140 formalin-fixed paraffin-embedded (FFPE) samples (69 glioblastomas multiforme [GBMs], 25 anaplastic astrocytomas [AAs], 11 anaplastic oligodendrogliomas [AOs], 31 brainstem tumors, 3 gliomatoses cerebri, and 1 anaplastic pleomorphic xantroastrocytoma) was constructed. All samples were centrally reviewed as above; 3 × 0.6 mm cores were taken from each tumor as marked by the pathologist to provide representative areas within the tumor.
DNA Isolation for Oligo Array Comparative Genomic Hybridization
DNA was isolated from 38 FFPE HGG samples including 13 DIPGs. From each block, 5-µm sections were taken and stained with Harris hematoxylin and eosin for histopathological review. Slides were spotted for areas of high tumor density to aid the removal of cores for DNA isolation. Overall, tumor cells represented ∼70–90% of tissue sections, depending on the degree of infiltration into normal brain. Ten cores or 1-µm scrolls were taken from the blocks, depending on the size and thickness of the sample in the block. DNA was isolated using the RecoverAll Total Nucleic Acid Kit for FFPE (Applied Biosystems). Briefly, tissue was deparaffinized in xylene for 15 minutes at 50°C. Samples were centrifuged to form pellets and washed with ethanol. Pellets were air dried and digestion buffer together with protease were added to each sample. Samples were incubated at 50°C for 40 hours and isolation additive was added together with ethanol. The mixture was applied to filter columns and washed, and DNA was eluted in 60-µL nuclease-free water heated to 95°C.
Oligo Array Comparative Genomic Hybridization
DNA samples were hybridized onto Agilent 244 K oligo comparative genomic hybridization (CGH) arrays (Agilent Technologies) following the manufacturer's protocol. Briefly, 1 µg of sample DNA and 1 µg of control Human Genomic DNA (Promega) were used for each array. DNA was digested with Rsa1/Alu1 restriction enzymes (Promega). Samples were labeled using DNA Labeling Kit PLUS (Agilent Technologies). Tumor DNA and control DNA were differentially labeled with Cy3/Cy5 conjugated deoxyuridine-5' -triphosphate. Dye swap experiments were performed for each sample. Unincorporated nucleotides and dyes were removed using MicroCon YM30 Spin Columns (Millipore). The concentration of labeled DNA was determined using a Nanodrop photospectrometer and appropriate Cy3/Cy5 labeled DNA combined.
DNA was prepared for hybridization using an oligo array CGH (aCGH) hybridization kit (Agilent Technologies) and precipitated with 50 µg Cot-1 DNA, and blocking agent and hybridization buffer were added to each sample. DNA was denatured at 95°C for 3 min followed by preannealing at 37°C for 30 min. A total of 490 µL of hybridization sample mixture was hybridized to the arrays and hybridized for 40 h at 65°C in a hybridization oven with the rotator rack set to rotate at 20 rpm. Hybridized arrays washed in oligo aCGH wash buffer 1 (Agilent Technologies) at room temperature for 5 min, oligo aCGH wash buffer 2 at 37°C for 5 min, acetonitrile (Sigma) at room temperature for 1 min, and stabilization and drying solution (Agilent Technologies) at room temperature for 30 s.
Arrays were scanned using an Agilent DNA Microarray Scanner (Agilent Technologies) at 5 µm resolution, and resulting Tagged Image File images were interpreted by Agilent Feature Extraction software version 9.5.3.1. Data were analyzed using Agilent CGH Analytics version 3.5.14 with the Hg18 genome build of the National Center for Biotechnology Information (NCBI). Dye swaps were combined for each sample and the data centralized. Regions of gain and loss were defined using the z-score algorithm. Genes within aberrant regions were selected as genes of interest if they had previously been implicated in cancer or were involved in cell proliferation, migration, DNA repair, or apoptosis.
Quantitative Real-Time PCR
PCR primers were designed using Primer Blast (NCBI). Quantitative real-time PCR (qPCR) reactions were performed using Stratagene Mastermix (Agilent Technologies) with SYBR green. Twenty-five nanograms of FFPE sample DNA was used for each PCR reaction. A pooled normal DNA was used as a reference. Real-time reactions were performed using an Mx4000p PCR machine (Agilent) and a 40-cycle PCR program with an annealing temperature of 58°C.
Results were analyzed using Mx3000p software (Agilent Technologies). The housekeeping gene PPIA was used as a control to normalize the data, as PPIA copy number was normal across the sample set. Quantification of the target gene copy number in relation to the housekeeping gene was calculated using the Pfaffl equation.19
Fluorescent In situ Hybridization
Fluorescent in situ hybridization (FISH) was performed on FFPE TMA slides (3-µm sections) using the following probes: Vysis 1q25 SpectrumGreen probe and 1p36 SpectrumOrange control probe (Abbott Molecular), Kreatech MYCN (2p24) specific PlatinumBright550 probe plus LAF (2q11) PlatinumBright495 control probe (Stretton Scientific), Kreatech p16 (9p21) PlatinumBright550 probe, and 9q21 PlatinumBright495 control probe (Stretton Scientific). Briefly, slides were deparaffinized in xylene and washed in 100% ethanol, then incubated in formalin for 1 h and washed in warm running water. Slides were placed in steamer with citrate buffer for 1 h at >90°C. Next, tissue was digested in 8 mg/mL pepsin(Dako) in 0.1 M HCl for 30 min. Tissue was fixed using Carnoy's mild fixative for 30 min. A 10-µL probe was applied to tissue, and coverslip was applied. Slides were incubated in hybridizer at 90°C for 12 min, then 37°C overnight. Slides were washed in 2 × standard sodium saline citrate (SSC) for 30 min, and coverslips removed. Slides were washed in 4 M urea for 2 min and 2 × SSC for 1 min. Twenty microliters of 4',6-diamidino-2-phenylindole was applied to each slide and a coverslip was applied and sealed.
Immunohistochemistry
Immunohistochemistry was performed using Dako Envision Detection Kit, as previously described.20 Used in this study were MYCN mouse monoclonal (NCM II 100) antibody (Abcam, Cambridgeshire; 1:100) and CDKN2B/p15INK4b mouse monoclonal antibody (ab4068, Abcam; 1:50).
Statistics
Statistical analysis was perfomed with the Statistical Package for the Social Sciences, version 14. Fisher's exact test was used to explore associations in two-way frequency tables. P-values of ≤0.05 were considered statistically significant.
Results
Large-Scale Copy Number Alterations Reveal Similarities and Differences Based on Anatomical Location
Thirty-eight formalin-fixed HGG samples (28 pretreatment), including 13 DIPGs (3 postmortem and 10 pretreatment), were analyzed by oligo aCGH (Table 1).
Table 1.
Clinical information for 38 HGG samples
| Sample | Sex | Age at Diagnosis (years) | Location | Resection Status | Sample taken pre/posttreatment | Diagnosis | Treatment | Survival (months) |
|---|---|---|---|---|---|---|---|---|
| HGG1 | M | 5.8 | Cerebral | Complete | Pretreatment | GBM | CT + RT | 5 |
| HGG2 | M | 7.3 | Cerebral | Complete | Posttreatment | GBM | RT | 6 |
| HGG3 | M | 4 | Cerebral | Subtotal | Pretreatment | GBM | CT | 3 |
| HGG4 | M | 6 | Cerebral | Complete | Posttreatment | GBM | RT + CT | 12 |
| HGG5 | M | 1 | Cerebral | Biopsy | Pretreatment | GBM | CT | 27 |
| HGG6 | F | 1.2 | Posterior fossa | Subtotal | Pretreatment | GBM | CT | >168 |
| HGG7 | F | 6.7 | Cerebral | Subtotal | Posttreatment | GBM | RT | 24 |
| HGG8 | M | — | Cerebral | Subtotal | Posttreatment | GBM | CT | — |
| HGG9 | M | 8.8 | Cerebral | Complete | Pretreatment | AA | RT | >108 |
| HGG10 | M | — | Cerebral | Subtotal | Pretreatment | AA | CT + RT | — |
| HGG11 | M | 3.1 | — | Posttreatment | AO | — | 21 | |
| HGG12 | M | 18.9 | Frontal lobe | Subtotal | Pretreatment | GBM | CT + RT | 21 |
| HGG13 | M | 12 | Parietal lobe | Subtotal | Pretreatment | GBM | CT + RT | >78 |
| HGG14 | M | — | Frontal | Biopsy | Pretreatment | GBM | CT | 1 |
| HGG15 | F | 5.8 | Chiasm | Subtotal | Pretreatment | GBM | CT | >70 |
| HGG16 | F | 2.4 | Frontal lobe | Subtotal | Pretreatment | AA | CT | >88 |
| HGG17 | M | 16.7 | Parietal lobe | Complete | Pretreatment | GBM | RT | >34 |
| HGG18 | M | 2.7 | Frontal lobe | Subtotal | Pretreatment | AO | CT + RT | >46 |
| HGG19 | F | 7.3 | Posterior Fossa | Biopsy | Pretreatment | AO | — | — |
| HGG20 | M | 0.1 | Cerebral | Subtotal | Pretreatment | GBM | — | 4 |
| HGG21 | M | 12.8 | Thalamus | Subtotal | Pretreatment | GBM | CT + RT | 48 |
| HGG22 | F | 4.5 | Frontal lobe | Complete | Pretreatment | GBM | CT + RT | 20 |
| HGG23 | F | 4.5 | Frontal lobe | Complete | Posttreatment | GBM | CT + RT | 20 |
| HGG24 | M | 2.9 | Frontal lobe | Complete | Posttreatment | GBM | RT | 15 |
| HGG25 | M | 10.1 | Cerebral | Subtotal | Pretreatment | GBM | CT + RT | 11 |
| BSG1 | M | 16.1 | DIPG | Biopsy | Pretreatment | GBM | — | 35 |
| BSG2 | M | 14.1 | DIPG | Biopsy | Pretreatment | GBM | — | — |
| BSG3 | M | 17.3 | DIPG | Biopsy | Pretreatment | LGA | — | 127 |
| BSG4 | F | 7.8 | DIPG | Postmortem | Posttreatment | GBM | CT + RT | 5 |
| BSG5 | M | 8.1 | DIPG | Biopsy | Pretreatment | GBM | RT | 9 |
| BSG6 | F | 5.6 | DIPG | Biopsy | Pretreatment | GBM | CT | 11 |
| BSG7 | M | 7.3 | DIPG | Biopsy | Pretreatment | GBM | CT + RT | 10 |
| BSG8 | M | 12.3 | DIPG | Biopsy | Pretreatment | GBM | RT | 8 |
| BSG9 | M | 12.5 | DIPG | Biopsy | Pretreatment | GBM | — | 1 |
| BSG10 | M | 12.6 | DIPG | Biopsy | Pretreatment | GBM | CT + RT | 16 |
| BSG11 | F | 3.8 | DIPG | Biopsy | Pretreatment | GBM | RT | 15 |
| BSG12 | M | 13.3 | DIPG | Postmortem | Posttreatment | GBM | CT + RT | 7 |
| BSG13 | M | 4.5 | DIPG | Postmortem | Posttreatment | GBM | CT + RT | 5 |
Focal regions of gain or loss in the 38 HGG samples are listed in Tables 2 and 3. There were widespread whole chromosome arm gains and losses across the sample cohort. Whole chromosomal arm gains are seen at 1q, 5p, 5q, 6p, 6q, 7p, 7q, 17p, 20p, and 20q, and whole chromosome arm losses at 1p, 4q, 6p, 6q, 10q, 13q, 14q, 16p, 16q, and 17p.
Table 2.
Gained regions and genes
| Chromosome region | Genomic location | Genes of interest in region | Number of samples with gain |
|---|---|---|---|
| 1q22 | 151370387–155734861 | CLK2 | 1 |
| 1q42 | 223214607–229308654 | WNT3A, ARF1 | 2 |
| 2p24.1 | 15258012–18111407 | MYCN | 2 (1 DIPG) |
| 2q12 | 96692465–101253051 | AFF3, REV1 | 1 |
| 4q11–13 | 54170296–58513820 | PDGFRA, KIT, CLOCK | 3 (2 DIPG) |
| 6p21 | 32796908–38137382 | TAP1, TAP2 | 3 (1 DIPG) |
| 6q26–27 | 162697821–166246965 | QKI, PACRG | 1 (1 DIPG) |
| 10q11.22 | 45411268–47172593 | ANXA8 | 1 |
| 11q13 | 68186386–70325025 | CCND1, FGF3, FGF4 | 13 (3 DIPG) |
| 11q13.4 | 73696165–79188879 | RSF1, PAK1, WNT11 | 1 (1 DIPG) |
| 12q13 | 55348927–58687499 | CDK4 | 3 |
Table 3.
Deleted regions and genes
| Chromosome region | Genomic location | Genes of interest in region | Number of samples with loss |
|---|---|---|---|
| 1p36 | 20897624–4952248 | CDC42, WNT4 | 2 |
| 2p24–22 | 41850167–45751380 | MTA3, SIX2/3, MAP4K3, RAB10, CAD | 1 |
| 3q25 | 163105791–165054849 | NMD3 | 3 |
| 4q13.2 | 68300723–70219426 | UGT2B15, UGT2B17 | 2 (2 DIPG) |
| 6p21 | 32796908–38137382 | TAP1, TAP2 | 1 (1 DIPG) |
| 8p11.23 | 38364631–40491994 | ADAM3A | 6 (1 DIPG) |
| 8p23a | 8140070–11897978 | MSRA, MFHAS1 | 1 |
| 9p22 | 20823666–23049991 | CDKN2A, CDKN2B, MTAP | 4 |
| 11q13 | 68186386–70325025 | CCND1, FGF3, FGF4 | 1 |
| 13q22 | 72858813–74620168 | KLF12, MYCBP2 | 1 |
| 16p12.2 | 21984018–26004132 | ARHGAP17, PRKCB, PALB2 | 1 |
| 17q21.2 | 36655799–37242724 | No genes in region | 1 |
| 20p21.1 | 13541317–16034064 | FLRT3 | 1 |
Gain of the whole of 1q was seen in 21% of all tumors and 23% of DIPGs. Two DIPGs (BSG9 and BSG6) had both loss of 1p and gain of 1q and may reflect the development of an isochromosome 1 in DIPG. 1q FISH was performed on the TMAs, and 41 samples were scorable; 11 of 41 (27%) samples had gain of 1q. Of those 11 samples, 9 were GBMs, including 2 GBM arising in the brainstem, and 2 were AAs.
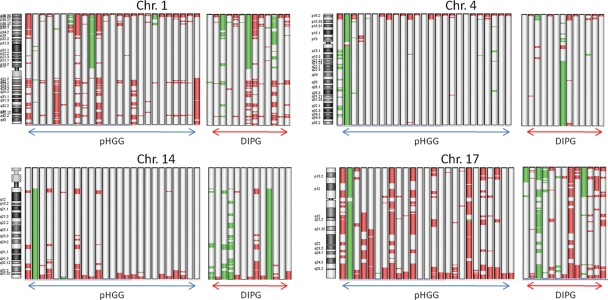
Loss of 10q was found predominantly in supratentorial tumors (20%), but in only 1 of 13 (8%) DIPGs. In contrast, 17p loss was found predominantly in DIPG samples, with 4 of 13 (31%) having 17p loss compared with 1 of 25 (4%) of the remaining HGG cohort. Genomic loss on 14q was most frequently detected in DIPGs, occurring in 4 of the 13 cases (31%), but was lost in just 1 of 25 of the nonbrainstem tumors (4%) (Fig. 1).
Fig. 1.
Ideogram showing all areas of gain and loss on chromosomes 1, 4, 14, and 17 in 25 HGG samples and 13 DIPGs. Red indicates regions of gain, and green indicates regions of loss.
A subgroup of 5 of 38 (13%) tumors displayed no aberrations by aCGH analysis, having a balanced genomic profile. Three of the balanced tumors were primary supratentorial GBMs, 1 was a recurrent supratentorial GBM, and the final balanced tumor was a DIPG. The age at diagnosis of these 5 patients ranged from 1 year to 17.3 years.
Deletion of 8p11 Involving ADAM3A Gene
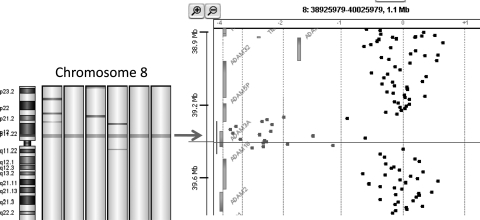
A discrete deletion at 8p11 was seen in 6 of 38 (16%) HGGs samples, including 1 DIPG (BSG7), making this the most commonly deleted gene in our cohort (Fig. 2). The minimal deleted region across the 6 samples was the gene ADAM3A, a member of the ADAM (a disintegrin and metalloproteinase) gene family. The homozygous deletion of ADAM3A was confirmed in 4 samples that had DNA available for validation by qPCR.
Fig. 2.
Heatmap showing loss of 8p12 in 6 of the samples (green indicates regions of loss, red indicates regions of gain), and gene view showing specific loss of ADAM3A in 1 sample (probes in regions of loss are highlighted in green).
Loss of CDKN2A and CDKN2B
Four samples, HGG1, HGG4, HGG8, and HGG17, showed deletions of CDKN2A and CDKN2B. Samples HGG1, HGG4, and HGG17 showed homozygous deletions by qPCR, while HGG8 showed a heterozygous deletion of CDKN2A and CDKN2B. Although 10% (4/38) of samples demonstrated loss of CDKN2A and CDKN2B, interestingly this did not include any of the 13 DIPGs.
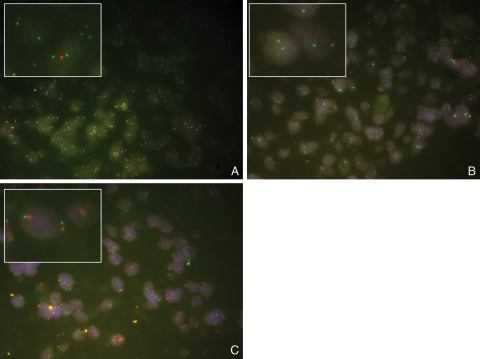
All TMAs were subjected to p16 FISH, and there were 61 scorable samples. Twenty-one samples were infratentorial tumors, including 5 DIPGs; 28 were supratentorial tumors, 3 were spinal tumors, and 9 tumors were of unknown location. Out of the 61 samples, 3 had heterozygous loss of p16 and 5 had homozygous loss of p16 (Fig. 3). Interestingly, all the tumors with loss of p16 were supratentorial GBMs, occurring in the cerebral lobes. None of the 21 infratentorial samples on the TMAs showed loss of p16, the same result seen in the aCGH data. Unfortunately, sample numbers were too small for this to be statistically significant (P = 0.17).
Fig. 3.
p16 FISH on pHGG TMAs: (A) sample with heterozygous loss of p16; (B) sample with homozygous loss of p16; (C) sample with normal copy of p16.
CDKN2B IHC was also evaluated, and 93 samples on the TMA were scorable; 38 samples were negative for CDKN2B expression, 35 samples showed low-level expression of CDKN2B, and 20 samples showed high-level expression of CDKN2B. All 4 samples found to have loss of CDKN2B by aCGH were also negative for the expression of CDKN2B by IHC. Of the 2 samples with heterozygous loss of CDKN2B by aCGH, 1 was negative for CDKN2B expression and the other showed low-level expression of CDKN2B.
Of the 55 samples positive for CDKN2B expression, 46 had location data available: 16 of 46 were infratentorial and 30 of 46 were supratentorial. Of the 38 samples negative for CDKN2B expression, 5 were infratentorial tumors and 33 were supratentorial tumors. Therefore, around half the supratentorial tumors were negative (52.3%), while only a quarter of infratentorial tumors were negative (23.8%). The difference between CDKN2B expression in supratentorial and infratentorial tumors is statistically significant (P = 0.03).
Amplification of 4q11–13 PDGFRA and KIT
Three samples showed amplification at 4q11–13 by aCGH, with 2 of the 3 samples being DIPG. The amplified region includes the genes PDGFRA and KIT. Quantitative real-time PCR gave a derived copy number of PDGFRA of 28 copies in BSG9 and 14 copies in HGG21, confirming amplification (>6 copies). Sample BSG8 gave a derived copy number of 5.4 copies for PDGFRA, which indicates a high-level gain rather than amplification. No gain or amplification of EGFR was seen in this study.
Amplification of MYCN at 2p24 Is Associated with Anaplastic Astrocytoma
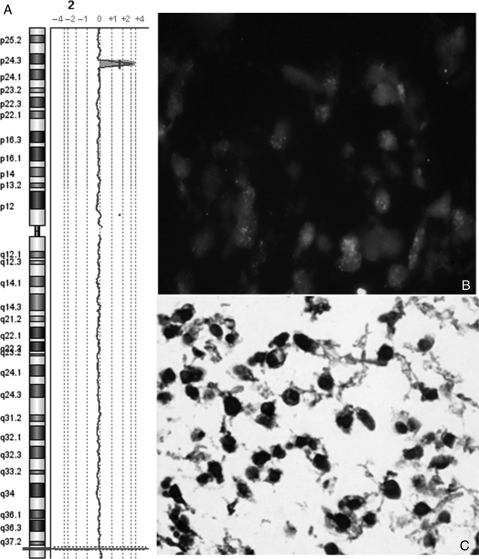
Samples BSG9 and HGG10 showed a high-level amplification at 2p24 that includes MYCN, which is commonly amplified in neuroblastoma and has been seen amplified in other tumor types. Results of qPCR gave a derived copy number of MYCN of 72 copies and 16 copies for samples BSG9 and HGG10, respectively.
FISH was performed on HGG TMAs for the MYCN gene at 2p24.1 locus. Fifty-eight samples on the TMAs were scorable for MYCN FISH: 29 GBMs, 12 DIPGs, 11 AAs, 5 AOs, and 1 gliomatosis cerebri. Of those 58 samples, 4 had amplification of MYCN (7%), including BSG9 (Fig. 4) and HGG10 previously identified by aCGH. Of the 4 samples shown to have MYCN amplification, 3 were AAs and 1 was DIPG. Fisher's exact test was used to show that MYCN amplification is significantly associated with AAs in primary tumors (P = 0.03).
Fig. 4.
(A) Oligo aCGH profile showing amplification of MYCN in sample BSG9. (B) FISH showing high-level amplification of MYCN in sample BSG9. (C) Immunohistochemistry showing high nuclear expression of MYCN in sample BSG9.
MYCN IHC was performed on the same TMAs, with 81 samples scorable: 14 were positive and 67 were negative for MYCN expression. This included the 4 samples that had previously shown amplification of MYCN, and hence overexpression was as predicted. Five of the 14 samples had previously shown normal copy number of MYCN by aCGH or FISH, indicating a different mechanism of overexpression in these samples. No MYCN copy number data were available for the remaining 5 samples with overexpression of MYCN, and so the mechanism of overexpression was undefined.
Amplification of 11q13
Amplification at 11q13 was seen in sample BSG4. This region contains the gene cyclin D1, a well-characterized cell cycle regulator, and FGF3, a member of the fibroblast growth factor family. This amplification was confirmed by qPCR, which gave a derived copy number of 10 copies of cyclin D1 and 9 copies of FGF3 in this sample.
Low-level gain of this region was seen in a number of other HGG samples. For cyclin D1, qPCR failed to validate this gain, although qPCR for FGF3 did validate gain in 2 samples with DNA available for validation. High-frequency gain of this region has not been identified in single nucleotide polymorphism (SNP) array studies of pHGG. Unfortunately, DNA was unavailable for further validation of gain of this region.
Discussion
This unbiased genome-wide study of predominantly pretreated childhood and adolescent pHGG provides further biological insight into this devastating tumor.
Homozygous loss at 8p12 was seen in 16% of pHGG and has not been previously found in adult HGG or pHGG in high-resolution studies using the Affymetrix SNP platforms.16,17 This is likely due to the lack of probes for this particular gene on the Affymetrix 500 and 250 K platforms. The minimal deleted region is of the gene ADAM3A. The ADAM family of genes are widely expressed and have many potential functions relating to cell-to-cell communication and adhesion. Little is known about ADAM3A.21 However, several members of the ADAM gene family have been implicated in cancer. Notable examples include overexpression of ADAM28 in lung and breast carcinomas22 and loss of expression of ADAM23 in breast tumors correlating with increasing grade.23,24 ADAM17 has been shown to be involved in EGFR regulation, and overexpression of ADAM17 in astrocytes promotes an increase in cell proliferation and invasion.25 This novel homozygous deletion of ADAM3A therefore merits further study.
Deletion of CDKN2A/CDKN2B in 19% of pHGG has been recently reported.16 In this study, deletion of CDKN2A/CDKN2B was seen in 10% of samples by aCGH and 13% of samples by FISH. Interestingly, none of the DIPGs or infratentorial tumors showed deletion at 9p21 by aCGH or FISH, and all deletions were seen in supratentorial GBMs. Zarghooni et al.26 also noted loss of 9p in supratentorial tumors but did not define specific loss of CDKN2A/CDKN2B. Loss of CDKN2A/CDKN2B has previously been reported in another supratentorial high-grade malignancy.27 The differential loss of CDKN2A/CDKN2B depending on tumor location may point to a cell of origin effect as seen in ependymoma.28
MYCN is a proto-oncogene and part of the MYC transcription factor family.29 It is highly amplified in aggressive neuroblastoma (a sympathetic nervoussystem–derived tumor) and has been reported in primitive neuroectodermal tumors of the central nervous system (CNS PNETs), medulloblastoma, and pHGG.16,30,31 Overall MYCN amplification was detected in 5% of both brainstem and supratentorial HGG. Correlation of MYCN amplification with histological grade revealed that MYCN amplification is significantly associated with AAs. The relevance and clinical significance of this finding need further study in a larger cohort.
Loss of 17p was seen in 4 of 13 (31%) DIPGs and just 1 of 25 (4%) nonbrainstem tumors. Region 17p is the site of the well-characterized tumor suppressor gene p53, which has been widely implicated in cancer.32 Mutation of p53 has been reported in adult HGG, but less so in pHGG.16 The high frequency of loss of 17p in DIPG suggests that the loss of the 17p arm is the mechanism for the loss of p53 in this subset of tumors. Zarghooni et al.26 also reported 17p loss associated with pediatric DIPG compared with nonbrainstem HGGs. Combining data from this study and that of Zarghooni et al. reveals 11 of 24 (46%) DIPGs had loss of 17p, while only 3 of 36 (8%) nonbrainstem tumors had loss of 17p, which is statistically significant (P = 0.02). This site-related difference in the inactivation of the TP53 tumor suppressor gene deserves further study.
Loss of 14q was is seen in 4 of 13 (31%) DIPGs, but only 1 supratentorial pHGG (4%); however, we were not able to identify a focal region of loss. Zarghooni et al.26 also identified 14q loss as being more common in DIPG.
We report an increased frequency gain of 1q in pHGG compared with adult tumors.21 In this study 21% of pHGG demonstrated gain of 1q by aCGH, while only 9% of adult HGG have been reported to have 1q gain.16,33 A recent study by Bax et al.34 found gain of 1q in 12 of 63 (19%) pHGG samples. Combining data from this study with those of Bax et al. reveals 20 of 101 (20%) pHGGs have gain of 1q compared with 17 of 189 (9%) adult tumors. This is statistically significant (P = 0.03). Gain of 1q is commonly seen in pediatric brain tumors, including CNS PNETs, ependymoma, and HGG, in addition to other pediatric malignancies.16,35 We have recently shown a significant association between 1q gain and decreased survival, as found in some other pediatric malignancies.16,35 Nearly 25% of DIPGs have gain of the whole long arm of chromosome 1, which is comparable to the level of gain seen in other pHGG. In the recent study by Zarghooni et al.,24 PARP-1, which is located at 1q41–42, was highlighted as gained in 3 of 11 (27%) DIPGs but in only 1 of the 2 pretreatment cases. PARP-1 is involved in repairing DNA damage, and overexpression of PARP-1 could be responsible for resistance to chemotherapy and radiotherapy, making it an attractive potential target for therapy. While focal gains were identified on 1q in this cohort, this did not include the PARP-1 locus at 1q41–42. Further assessment of the frequency of PARP-1 involvement in DIPG is needed.
Loss of 10q is seen in 6 of 38 (16%) of all pHGG samples and only 1 of 13 DIPGs in this study, which is notably lower than the 35% loss of 10q previously reported in pHGG.16 Loss of 10q is the most common whole-arm aberration in adult GBM (∼80%), which includes loss of PTEN (10q23).36 The difference between the frequency of 10q loss in pHGG in this study (6/38) compared with adult HGG (152/189) is statistically significant (P = 0.0001).33 PTEN is a tumor suppressor gene that negatively regulates the Akt signaling pathway, and PTEN loss or mutation is seen in a number of different cancers.1 The lack of 10q loss in this DIPG cohort may indicate a different mechanism of Akt signaling pathway activation or that different pathways are involved in these pediatric tumors. However, a recent study by Zarghooni et al.26 reported PTEN loss in 27% of both DIPG and nonbrainstem pHGG. An increased sample cohort is needed to determine whether 10q loss is a predominant feature of supratentorial tumors.
Expression of platelet-derived growth factor receptor-α (PDGFRA) correlates with malignant grade in pHGG, with high expression being associated with malignant histology (WHO Grades II and IV).37 We have recently reported amplification of PDGFRA in 12% of Grade III and 17% of Grade IV pHGG tumors in a large recent series based on high-resolution copy number and expression profiling.16 Similar levels of focal amplification are reported in this series based on aCGH data and IHC. In this same collaborative study, amplification of PDGFRA was found at a significantly higher frequency (50%) in irradiation-induced childhood and adolescent HGG.16 This later finding might explain the relatively high frequency of PDGFRA expression reported by Zarghooni et al., as 82% of their cohort was analyzed posttreatment.26 Consistent with this possibility is that amplification of PDGFRA in adult tumors is associated with a secondary GBM,38 although the time course for the latter is likely longer. Further studies of the role of PDGFRA in childhood and adolescent HGG are needed to understand the influence of treatment on PDGFRA amplification and to determine whether this tyrosine kinase receptor is a conditional target for therapy in the subset of pHGG with PDGFRA amplification.
Conversely, EGFR amplification, the most common focal amplification in adult HGG,1,36,39 does not appear to be a feature of pHGG; indeed, no amplification of EGFR was seen in the 38 pHGG samples in our cohort.
The findings in this study have shown that pediatric and adult HGG represent 2 different genetic entities that are distinguished by differences in copy number changes. Overall there are marked differences in the degree of genomic imbalance between adult and childhood/adolescent HGG. Four focal regions of amplification were seen across this data set compared with 27 different sites of amplification identified in a dataset of 97 adult HGG samples by metaphase CGH.36 Moreover, a subgroup of pHGG tumors (13%) have balanced genomes by aCGH analysis. This has also been seen in other pediatric brain tumors, including ependymoma and CNS PNETs.40,41 In our series of pHGG, this was not defined by young age at diagnosis as for ependymoma.40 This important difference in the number of genomic imbalances and regions of amplification in adults and children may reflect the development of the tumor in a narrower time frame/developmental window. It might also be that other mechanisms are important in the pathogenesis of these tumors and suggests that relatively fewer mutations may be required to initiate the disease if these arise within a specific developmental period. We are currently undertaking genome-wide methylation analysis to determine whether epigenetic mechanisms may play a role in these cases.
Zarghooni et al.26 identified a number of genes involved in DNA repair pathways as being lost in DIPG. Loss of some of these DNA repair pathway genes was seen in this study as part of whole-arm deletions, such as 17p (RPA1 and MYH1), 14q (MNAT1 and RAD51L1), and 1p (MSH4). However, losses of other DNA repair pathway genes identified by Zarghooni were not seen in this study (GTF2H3, LIG4, XRCC4, XRCC5, MLH1, BRCA1, BRCA2, RAD50, DUT, and PMS1).26 Importantly their cohort included just 2 pretreatment samples compared with 10 pretreatment samples in this study, suggesting that treatment with DNA-damaging agents such as radiotherapy and temozolomide may be responsible for the loss of DNA repair pathway genes and possibly other genetic aberrations. We have previously shown that diagnostic biopsy of diffuse brain stem glioma is relatively safe10 and can now demonstrate that pretreatment tumor tissue can generate high-quality biological information on which to base future strategies.
This study adds to the body of knowledge regarding this poor prognosis tumor, providing further insight into underlying biology, and highlights potential therapeutic possibilities. Overall, pediatric DIPGs share many genetic changes with supratentorial pHGG. However, the frequency of aberrations differs in some instances, notably the low occurrence of 10q loss, the lack of any CDKN2A/CDKN2B loss in DIPG, and the high frequency of 14q and 17p loss in DIPG relative to their supratentorial counterpart. Larger studies need to be done on pretreatment DIPG based on tumor biopsies at diagnosis to comprehensively map the genetics of these tumors.
We also report the novel homozygous deletion of ADAM3A, one of the ADAM family of genes that are thought to play a role in cancer, confirm and extend the database of genetic aberrations that have been previously reported in pHGG, such as amplification of PDGFRA, MYCN, and cyclin D1, and raise the association between MYCN amplification and AA.
Supplementary Material
Supplementary material is available at Neuro-Oncology Journal online.
Funding
Financial support was provided by the Samantha Dickson Brain Tumor Trust and Air and Ground.
Acknowledgments
This study was a Children's Cancer and Leukaemia Group (CCLG) biological study. The authors acknowledge Dr Lisa Storer for sample collection, Lee Ridley for TMA construction, and the Molecular Biology Service team at the University of Warwick for their time and the use of equipment.
Conflict of interest statement. None declared.
References
- 1.Maher EA, Furnari FB, Bachoo RM, et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. doi:10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 2.Broniscer A, Gajjar A. Supratentorial high-grade astrocytoma and diffuse brainstem glioma: two challenges for the pediatric oncologist. Oncologist. 2004;9:197–206. doi: 10.1634/theoncologist.9-2-197. doi:10.1634/theoncologist.9-2-197. [DOI] [PubMed] [Google Scholar]
- 3.Broniscer A, Baker SJ, West AN, et al. Clinical and molecular characteristics of malignant transformation of low-grade glioma in children. J Clin Oncol. 2007;25:682–689. doi: 10.1200/JCO.2006.06.8213. doi:10.1200/JCO.2006.06.8213. [DOI] [PubMed] [Google Scholar]
- 4.Duffner PK, Krischer JP, Burger PC, et al. Treatment of infants with malignant gliomas: the Pediatric Oncology Group experience. J Neurooncol. 1996;28:245–256. doi: 10.1007/BF00250203. [DOI] [PubMed] [Google Scholar]
- 5.Grundy RG, Wilne SH, Robinson KJ, et al. Primary postoperative chemotherapy without radiotherapy for treatment of brain tumours other than ependymoma in children under 3 years: results of the first UKCCSG/SIOP CNS 9204 trial. Eur J Cancer. 2010;46:120–133. doi: 10.1016/j.ejca.2009.09.013. doi:10.1016/j.ejca.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 6.Dufour C, Grill J, Lellouch-Tubiana A, et al. High-grade glioma in children under 5 years of age: a chemotherapy only approach with the BBSFOP protocol. Eur J Cancer. 2006;42:2939–2945. doi: 10.1016/j.ejca.2006.06.021. doi:10.1016/j.ejca.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 7.Jallo GI, Biser-Rohrbaugh A, Freed D. Brainstem gliomas. Childs Nerv Syst. 2004;20:143–153. doi: 10.1007/s00381-003-0870-6. doi:10.1007/s00381-003-0870-6. [DOI] [PubMed] [Google Scholar]
- 8.Hargrave D, Bartels U, Bouffet E. Diffuse brainstem glioma in children: critical review of clinical trials. Lancet Oncol. 2006;7:241–248. doi: 10.1016/S1470-2045(06)70615-5. doi:10.1016/S1470-2045(06)70615-5. [DOI] [PubMed] [Google Scholar]
- 9.Leach PA, Estlin EJ, Coope DJ, Thorne JA, Kamaly-Asl ID. Diffuse brainstem gliomas in children: should we or shouldn't we biopsy? Br J Neurosurg. 2008;22:619–624. doi: 10.1080/02688690802366198. doi:10.1080/02688690802366198. [DOI] [PubMed] [Google Scholar]
- 10.Cartmill M, Punt J. Diffuse brain stem glioma. A review of stereotactic biopsies. Childs Nerv Syst. 1999;15:235–237. doi: 10.1007/s003810050379. discussion 238. [DOI] [PubMed] [Google Scholar]
- 11.Rachinger W, Grau S, Holtmannspotter M, Herms J, Tonn JC, Kreth FW. Serial stereotactic biopsy of brainstem lesions in adults improves diagnostic accuracy compared with MRI only. J Neurol Neurosurg Psychiatry. 2009;80:1134–1139. doi: 10.1136/jnnp.2009.174250. doi:10.1136/jnnp.2009.174250. [DOI] [PubMed] [Google Scholar]
- 12.Frazier JL, Lee J, Thomale UW, Noggle JC, Cohen KJ, Jallo GI. Treatment of diffuse intrinsic brainstem gliomas: failed approaches and future strategies. J Neurosurg Pediatr. 2009;3:259–269. doi: 10.3171/2008.11.PEDS08281. doi:10.3171/2008.11.PEDS08281. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson R, Harris J. Moral and legal reasons for altruism in the case of brainstem biopsy in diffuse glioma. Br J Neurosurg. 2008;22:617–618. doi: 10.1080/02688690802482896. doi:10.1080/02688690802482896. [DOI] [PubMed] [Google Scholar]
- 14.Rickert CH, Strater R, Kaatsch P, et al. Pediatric high-grade astrocytomas show chromosomal imbalances distinct from adult cases. Am J Pathol. 2001;158:1525–1532. doi: 10.1016/S0002-9440(10)64103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollack IF, Finkelstein SD, Burnham J, et al. Age and TP53 mutation frequency in childhood malignant gliomas: results in a multi-institutional cohort. Cancer Res. 2001;61:7404–7407. [PubMed] [Google Scholar]
- 16.Paugh BS, Qu C, Jones C, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu HQ, Jacob K, Fatet S, et al. Genome-wide profiling using single-nucleotide polymorphism arrays identifies novel chromosomal imbalances in pediatric glioblastomas. Neuro Oncol. 2010;12:153–163. doi: 10.1093/neuonc/nop001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis DN, Ohgaki H, Wiestlet OD, Cavenee WK. WHO Classification of Tumours of the Central Nervous System. 4th ed. Lyon: IARC; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. doi:10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ridley L, Rahman R, Brundler MA, et al. Multifactorial analysis of predictors of outcome in pediatric intracranial ependymoma. Neuro Oncol. 2008;10:675–689. doi: 10.1215/15228517-2008-036. doi:10.1215/15228517-2008-036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLaughlin EA, Frayne J, Bloomerg G, Hall L. Do fertilin beta and cyritestin play a major role in mammalian sperm—oolemma interactions? A critical re-evaluation of the use of peptide mimics in identifying specific oocyte recognition protiens. Mol Hum Reprod. 2001;7:313–317. doi: 10.1093/molehr/7.4.313. doi:10.1093/molehr/7.4.313. [DOI] [PubMed] [Google Scholar]
- 22.Mochizuki S, Okada Y. ADAM28 as a target for human cancers. Curr Pharm Des. 2009;15:2349–2358. doi: 10.2174/138161209788682424. doi:10.2174/138161209788682424. [DOI] [PubMed] [Google Scholar]
- 23.Seniski GG, Camargo AA, Ierardi DF, et al. ADAM33 gene silencing by promoter hypermethylation as a molecular marker in breast invasive lobular carcinoma. BMC Cancer. 2009;9:80. doi: 10.1186/1471-2407-9-80. doi:10.1186/1471-2407-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa FF, Verbisck NV, Salim AC, et al. Epigenetic silencing of the adhesion molecule ADAM23 is highly frequent in breast tumors. Oncogene. 2004;23:1481–1488. doi: 10.1038/sj.onc.1207263. doi:10.1038/sj.onc.1207263. [DOI] [PubMed] [Google Scholar]
- 25.Katakowski M, Jiang F, Zheng X, Gutierrez JA, Szalad A, Chopp M. Tumorigenicity of cortical astrocyte cell line induced by the protease ADAM17. Cancer Sci. 2009;100:1597–1604. doi: 10.1111/j.1349-7006.2009.01221.x. doi:10.1111/j.1349-7006.2009.01221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarghooni M, Bartels U, Lee E, et al. Whole-Genome Profiling of Pediatric Diffuse Intrinsic Pontine Gliomas Highlights Platelet-Derived Growth Factor Receptor {alpha{ and Poly (ADP-ribose) Polymerase As Potential Therapeutic Targets. J Clin Oncol. 2010;28:1337–1344. doi: 10.1200/JCO.2009.25.5463. [DOI] [PubMed] [Google Scholar]
- 27.Pfister S, Remke M, Toedt G, et al. Supratentorial primitive neuroectodermal tumors of the central nervous system frequently harbor deletions of the CDKN2A locus and other genomic aberrations distinct from medulloblastomas. Genes Chromosomes Cancer. 2007;46:839–851. doi: 10.1002/gcc.20471. doi:10.1002/gcc.20471. [DOI] [PubMed] [Google Scholar]
- 28.Taylor MD, Poppleton H, Fuller C, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. doi:10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Tonini GP, Boni L, Pession A, et al. MYCN oncogene amplification in neuroblastoma is associated with worse prognosis, except in stage 4s: the Italian experience with 295 children. J Clin Oncol. 1997;15:85–93. doi: 10.1200/JCO.1997.15.1.85. [DOI] [PubMed] [Google Scholar]
- 30.Fruhwald MC, O'Dorisio MS, Rush LJ, et al. Gene amplification in PNETs/medulloblastomas: mapping of a novel amplified gene within the MYCN amplicon. J Med Genet. 2000;37:501–509. doi: 10.1136/jmg.37.7.501. doi:10.1136/jmg.37.7.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfister S, Remke M, Benner A, et al. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol. 2009;27:1627–1636. doi: 10.1200/JCO.2008.17.9432. doi:10.1200/JCO.2008.17.9432. [DOI] [PubMed] [Google Scholar]
- 32.King R. Cancer Biology. 2nd ed. Harlow, UK: Pearson Education Limited; 2000. pp. 71–95. [Google Scholar]
- 33.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bax DA, Mackay A, Little SE, et al. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin Cancer Res. 2010;16:3368–3377. doi: 10.1158/1078-0432.CCR-10-0438. doi:10.1158/1078-0432.CCR-10-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendrzyk F, Korshunov A, Benner A, et al. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res. 2006;12:2070–2079. doi: 10.1158/1078-0432.CCR-05-2363. doi:10.1158/1078-0432.CCR-05-2363. [DOI] [PubMed] [Google Scholar]
- 36.Mohapatra G, Bollen AW, Kim DH, et al. Genetic analysis of glioblastoma multiforme provides evidence for subgroups within the grade. Genes Chromosomes Cancer. 1998;21:195–206. doi:10.1002/(SICI)1098-2264(199803)21:3<195::AID-GCC3>3.0.CO;2-V. [PubMed] [Google Scholar]
- 37.Thorarinsdottir HK, Santi M, McCarter R, et al. Protein expression of platelet-derived growth factor receptor correlates with malignant histology and PTEN with survival in childhood gliomas. Clin Cancer Res. 2008;14:3386–3394. doi: 10.1158/1078-0432.CCR-07-1616. doi:10.1158/1078-0432.CCR-07-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toepoel M, Joosten PH, Knobbe CB, et al. Haplotype-specific expression of the human PDGFRA gene correlates with the risk of glioblastomas. Int J Cancer. 2008;123:322–329. doi: 10.1002/ijc.23432. doi:10.1002/ijc.23432. [DOI] [PubMed] [Google Scholar]
- 39.Nishizaki T, Ozaki S, Harada K, et al. Investigation of genetic alterations associated with the grade of astrocytic tumor by comparative genomic hybridization. Genes Chromosomes Cancer. 1998;21:340–346. doi: 10.1002/(sici)1098-2264(199804)21:4<340::aid-gcc8>3.0.co;2-z. doi:10.1002/(SICI)1098-2264(199804)21:4<340::AID-GCC8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 40.Dyer S, Prebble E, Davison V, et al. Genomic imbalances in pediatric intracranial ependymomas define clinically relevant groups. Am J Pathol. 2002;161:2133–2141. doi: 10.1016/S0002-9440(10)64491-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natrajan R, Williams RD, Hing SN, et al. Array CGH profiling of favourable histology Wilms tumours reveals novel gains and losses associated with relapse. J Pathol. 2006;210:49–58. doi: 10.1002/path.2021. doi:10.1002/path.2021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.