Abstract
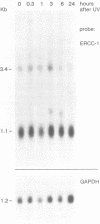
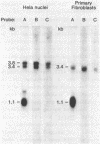
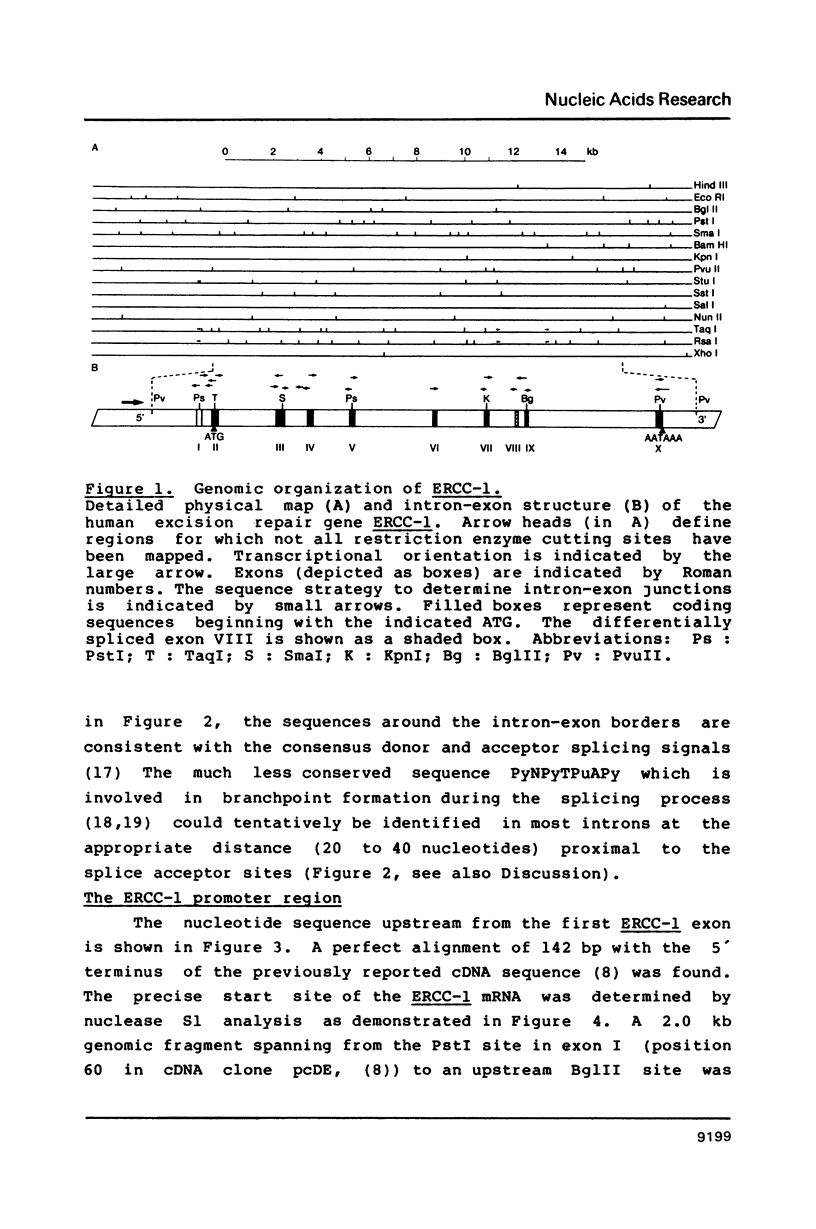
In this report the genomic characterization of the human excision repair gene ERCC-1 is presented. The gene consists of 10 exons spread over approximately 15 kb. By means of a transfection assay the ERCC-1 promoter was confined to a region of +/- 170 bp upstream of the transcriptional start site. Classical promoter elements like CAAT, TATA and GC-boxes are absent from this region. Furthermore, ERCC-1 transcription is not UV-inducible. A possible explanation is provided for the previously reported alternative splicing of exon VIII. Analysis of ERCC-1 cDNA clones revealed the occurrence of differential polyadenylation which gives ERCC-1 transcripts of 3.4 and 3.8 kb in addition to the major 1.1 kb mRNA. Apparent evolutionary conservation of differential polyadenylation of ERCC-1 transcripts suggests a possible role for this mode of RNA processing in the ERCC-1 repair function.
Full text
PDF

















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Benham F. J., Hodgkinson S., Davies K. E. A glyceraldehyde-3-phosphate dehydrogenase pseudogene on the short arm of the human X chromosomes defines a multigene family. EMBO J. 1984 Nov;3(11):2635–2640. doi: 10.1002/j.1460-2075.1984.tb02186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S. M. Are U4 small nuclear ribonucleoproteins involved in polyadenylation? Nature. 1984 May 10;309(5964):179–182. doi: 10.1038/309179a0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Brawerman G. Determinants of messenger RNA stability. Cell. 1987 Jan 16;48(1):5–6. doi: 10.1016/0092-8674(87)90346-1. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Burke J. F. High-sensitivity S1 mapping with single-stranded [32P]DNA probes synthesized from bacteriophage M13mp templates. Gene. 1984 Oct;30(1-3):63–68. doi: 10.1016/0378-1119(84)90105-7. [DOI] [PubMed] [Google Scholar]
- Chen M. J., Shimada T., Moulton A. D., Cline A., Humphries R. K., Maizel J., Nienhuis A. W. The functional human dihydrofolate reductase gene. J Biol Chem. 1984 Mar 25;259(6):3933–3943. [PubMed] [Google Scholar]
- De Weerd-Kastelein E. A., Keijzer W., Bootsma D. Genetic heterogeneity of xeroderma pigmentosum demonstrated by somatic cell hybridization. Nat New Biol. 1972 Jul 19;238(81):80–83. doi: 10.1038/newbio238080a0. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Johnson M. S., Husain I., Van Houten B., Thomas D. C., Sancar A. Domainal evolution of a prokaryotic DNA repair protein and its relationship to active-transport proteins. Nature. 1986 Oct 2;323(6087):451–453. doi: 10.1038/323451a0. [DOI] [PubMed] [Google Scholar]
- Fischer E., Keijzer W., Thielmann H. W., Popanda O., Bohnert E., Edler L., Jung E. G., Bootsma D. A ninth complementation group in xeroderma pigmentosum, XP I. Mutat Res. 1985 May;145(3):217–225. doi: 10.1016/0167-8817(85)90030-6. [DOI] [PubMed] [Google Scholar]
- Friedberg E. C. The molecular biology of nucleotide excision repair of DNA: recent progress. J Cell Sci Suppl. 1987;6:1–23. doi: 10.1242/jcs.1984.supplement_6.1. [DOI] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3' end formation. Cell. 1987 May 8;49(3):399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- Grosveld G. C., Koster A., Flavell R. A. A transcription map for the rabbit beta-globin gene. Cell. 1981 Feb;23(2):573–584. doi: 10.1016/0092-8674(81)90153-7. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Wellauer P. K., Cribbs D. L., Schibler U. Termination of transcription in the mouse alpha-amylase gene Amy-2a occurs at multiple sites downstream of the polyadenylation site. Cell. 1984 Oct;38(3):737–744. doi: 10.1016/0092-8674(84)90269-1. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J. H., van Duin M., Westerveld A., Yasui A., Bootsma D. Identification of DNA repair genes in the human genome. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):91–101. doi: 10.1101/sqb.1986.051.01.012. [DOI] [PubMed] [Google Scholar]
- Ishii S., Imamoto F., Yamanashi Y., Toyoshima K., Yamamoto T. Characterization of the promoter region of the human c-erbB-2 protooncogene. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4374–4378. doi: 10.1073/pnas.84.13.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S., Xu Y. H., Stratton R. H., Roe B. A., Merlino G. T., Pastan I. Characterization and sequence of the promoter region of the human epidermal growth factor receptor gene. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4920–4924. doi: 10.1073/pnas.82.15.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W. The RNA lariat: a new ring to the splicing of mRNA precursors. Cell. 1984 Dec;39(3 Pt 2):423–425. doi: 10.1016/0092-8674(84)90449-5. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Moores J. C., David D., Respess J. G., Jolly D. J., Friedmann T. The organization of the human HPRT gene. Nucleic Acids Res. 1986 Apr 11;14(7):3103–3118. doi: 10.1093/nar/14.7.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Bifunctional messenger RNAs in eukaryotes. Cell. 1986 Nov 21;47(4):481–483. doi: 10.1016/0092-8674(86)90609-4. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- LeMeur M. A., Galliot B., Gerlinger P. Termination of the ovalbumin gene transcription. EMBO J. 1984 Dec 1;3(12):2779–2786. doi: 10.1002/j.1460-2075.1984.tb02209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDevitt M. A., Hart R. P., Wong W. W., Nevins J. R. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 1986 Nov;5(11):2907–2913. doi: 10.1002/j.1460-2075.1986.tb04586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijlink F., Curran T., Miller A. D., Verma I. M. Removal of a 67-base-pair sequence in the noncoding region of protooncogene fos converts it to a transforming gene. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4987–4991. doi: 10.1073/pnas.82.15.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama H., Berg P. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Mol Cell Biol. 1983 Feb;3(2):280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Propst F., Rosenberg M. P., Iyer A., Kaul K., Vande Woude G. F. c-mos proto-oncogene RNA transcripts in mouse tissues: structural features, developmental regulation, and localization in specific cell types. Mol Cell Biol. 1987 May;7(5):1629–1637. doi: 10.1128/mcb.7.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Robinson G. W., Nicolet C. M., Kalainov D., Friedberg E. C. A yeast excision-repair gene is inducible by DNA damaging agents. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1842–1846. doi: 10.1073/pnas.83.6.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin J. S., Prideaux V. R., Willard H. F., Dulhanty A. M., Whitmore G. F., Bernstein A. Molecular cloning and chromosomal localization of DNA sequences associated with a human DNA repair gene. Mol Cell Biol. 1985 Feb;5(2):398–405. doi: 10.1128/mcb.5.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Thompson L. H., Busch D. B., Brookman K., Mooney C. L., Glaser D. A. Genetic diversity of UV-sensitive DNA repair mutants of Chinese hamster ovary cells. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3734–3737. doi: 10.1073/pnas.78.6.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5' element and c-fos 3' sequences. Cell. 1985 Oct;42(3):889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- Westerveld A., Hoeijmakers J. H., van Duin M., de Wit J., Odijk H., Pastink A., Wood R. D., Bootsma D. Molecular cloning of a human DNA repair gene. Nature. 1984 Aug 2;310(5976):425–429. doi: 10.1038/310425a0. [DOI] [PubMed] [Google Scholar]
- Wood R. D., Burki H. J. Repair capability and the cellular age response for killing and mutation induction after UV. Mutat Res. 1982 Aug;95(2-3):505–514. doi: 10.1016/0027-5107(82)90281-0. [DOI] [PubMed] [Google Scholar]
- van Duin M., de Wit J., Odijk H., Westerveld A., Yasui A., Koken M. H., Hoeijmakers J. H., Bootsma D. Molecular characterization of the human excision repair gene ERCC-1: cDNA cloning and amino acid homology with the yeast DNA repair gene RAD10. Cell. 1986 Mar 28;44(6):913–923. doi: 10.1016/0092-8674(86)90014-0. [DOI] [PubMed] [Google Scholar]