Abstract
CD1 molecules are specialized in presenting lipids to T lymphocytes, but identification and isolation of CD1-restricted lipidspecific T cells has been hampered by the lack of reliable and sensitive techniques. We here report the construction of CD1d–glycolipid tetramers from fully denatured human CD1d molecules by using the technique of oxidative refolding chromatography. We demonstrate that chaperone- and foldase-assisted refolding of denatured CD1d molecules and β2-microglobulin in the presence of synthetic lipids is a rapid method for the generation of functional and specific CD1d tetramers, which unlike previously published protocols ensures isolation of CD1d tetramers loaded with a single lipid species. The use of human CD1d–α-galactosylceramide tetramers for ex vivo staining of peripheral blood lymphocytes and intrahepatic T cells from patients with viral liver cirrhosis allowed for the first time simultaneous analysis of frequency and specificity of natural killer T cells in human clinical samples. Application of this protocol to other members of the CD1 family will provide powerful tools to investigate lipid-specific T cell immune responses in health and in disease.
CD1 molecules are nonpolymorphic transmembrane proteins that associate with β2-microglobulin (β2m) and present lipid and glycolipid antigens to T lymphocytes (1). Based on amino acid sequence homology, the five members of the human CD1 family have been assigned to either group 1, which comprises CD1a, -b, -c, and -e molecules, or group 2, which consists of the CD1d molecule (2–6).
Group 1 CD1 molecules are not expressed in mice and rats, whereas CD1d molecules are highly conserved in all mammalian species studied thus far (1). In humans, group 1 CD1 molecules have been shown to present bacterial and mycobacterial lipids to T cells expressing a wide range of T cell receptors (TCR) (1, 7). In contrast, very little is known about the natural ligand(s) of CD1d recognized by T cells; to date, the only human T cell subset restricted by CD1d molecules has been shown to be specific for α-anomeric galactosyl- and glucosylceramides (8–10), glycolipids that are not found in mammals or bacteria. Because this subset of T cells shares some markers with natural killer (NK) cells [i.e., expression of CD161 in humans (11, 12) and NK1.1 in mice (13, 14)], they have been named NKT cells. Although natural ligand(s) recognized by NKT cells are not known, it is likely that they recognize a single ligand, because NKT cells express a highly conserved TCR, which in humans corresponds to the gene products Vα 24JaQ and Vβ11 and in mice corresponds to Vα 14Ja281, which pairs with diverse β chains (8–10, 15). The capacity to secrete large amounts of IL-4 and IFN-γ after TCR engagement (16–18) and their presence across species suggest that NKT cells may play an important role in the immune response. Recent studies suggest that NKT cells are involved in type 1 diabetes mellitus (19) and viral hepatitis (20) and have shown that NKT cells play an important role in tumor immunity (21–23).
The ability to characterize the specificity of CD1d-restricted NKT cells in ex vivo assays is of paramount importance in understanding their physiological relevance and harnessing their effector functions for clinical purposes. Development of ex vivo assays for establishing the specificity of CD1d-restricted cells would aid the identification of the natural ligands of these cells and hence contribute to the understanding of the physiological role of CD1d-restricted T cells in health and disease.
The use of soluble tetrameric class I HLA–peptide complexes (“tetramers”) for the identification of antigen-specific CTL has shown that these reagents allow rapid and accurate analysis of human cytotoxic T lymphocyte (CTL) responses (24–26). The tetramer technology has been applied recently to the monitoring of CD1d-restricted T lymphocytes. Murine and human CD1d–β2m monomers purified from Drosophila expression systems have been used successfully to generate murine CD1d tetramers and human CD1d dimers, which were capable of staining NKT cells ex vivo (27, 28) after exogenous loading with α-galactosylceramide (GalCer). However, because binding of lipid ligands to CD1 monomers is known to be very stable (27), it is likely that a proportion of insect cell-derived CD1d monomers are loaded with ligands acquired during their intracellular refolding and secretion from insect cells. Indeed, analysis of the crystal structure of Drosophila-derived mouse CD1d molecules has revealed the presence of an electron density in the binding groove of CD1d molecules, which is consistent with the presence of high-affinity ligand(s) occupying the groove of CD1d molecules (29). The presence of “contaminant” hydrophobic ligands in CD1d monomers secreted by insect cells may affect the efficiency of CD1d loading with exogenous ligands negatively and compromise their ability to identify low-frequency NKT cells in ex vivo assays. This reasoning led us to pursue in vitro-refolding approaches by using recombinant denatured CD1d protein purified from bacterial inclusion bodies, which ensure generation of CD1d tetramers loaded with a single lipid species.
Here we describe the successful use of the technique of in vitro oxidative refolding chromatography (30, 31) to engineer human CD1d–β2m–glycolipid tetramers from fully denatured and reduced recombinant CD1d proteins. This technique combines the benefits of the GroEL minichaperone, which prevents hydrophobic interactions of early folding intermediates with disulphide and prolyl isomerases, which catalyze native disulphide bond formation and proline isomerization, respectively.
Materials and Methods
Generation of Tetramers.
Purified CD1d heavy chain and β2m were synthesized by using a prokaryotic expression system (pET, Novagen). Both recombinant proteins CD1d and β2m were expressed in the Escherichia coli strain BL21 and purified from inclusion bodies as described (25). Oxidative refolding chromatography was applied for in vitro refolding of CD1d–β2m complexes in the presence of either α-GalCer (Kirin Brewery, Gunma, Japan), α-Mannosylceramide (Kirin Brewery), β-GalCer (Fluka) or ganglioside GM1 (G-7641 Sigma–Aldrich) lipid ligands, following a protocol previously described (30–32). Soluble protein fractions were concentrated to a volume of 3 ml and biotinylated directly as described (25). Biotinylated monomeric CD1d–β2m–glycolipid complexes were separated from soluble protein aggregates and free biotin by ÅKTA FPLC size-exclusion chromatography by using a Superdex 75 h 10/30 gel filtration column (Amersham Pharmacia) and conjugated to Streptavidin-PE (Sigma–Aldrich).
Cell Line.
α-GalCer-specific NKT lines were generated from peripheral blood mononuclear cells (PBMC) of healthy donors as described (15). On day 20, CD1d–β2m–α-GalCer tetramer/CD4 double-positive cells were sorted, by using VANTAGE FACS sorter, and in vitro expanded in the presence of rIL-2 and α-GalCer.
Patient Samples.
Heparinized whole blood was obtained from seven patients (patients 1–7) with active hepatitis C virus (HCV) infection during routine follow up visits and also from four healthy donors. Human liver specimens were obtained from four patients with end-stage liver failure caused by chronic inflammatory liver disease of various etiologies: patients 8–10, HCV; patient 11, HCV plus Hepatitis B virus (HBV). Informed consent was obtained from all individuals.
Mice.
β2m−/− mice were deficient for murine MHC class I molecules [β2m(−/−) D(b−/−)] and transgenic for a chimeric HLA-A*0201/D(b) molecule covalently bound to the human β2m [HHD(+/+)] (33).
Cell Preparation and Tetramer Staining.
Intrahepatic mononuclear cells were separated from the patient liver tissue specimens by cutting tissue specimens into small pieces using a scalpel, followed by incubation at 37°C for 4 h in complete medium containing 0.5 mg/ml type IV collagenase (Sigma–Aldrich) and 0.02 mg/ml DNase (Roche Molecular Biochemicals). The digested tissue was passed through a 100-μm metal-mesh filter, washed twice, layered over Ficoll/Hypaque gradients, and centrifuged at 400 × g at room temperature for 20 min. PBMC, NKT lines, and intrahepatic lymphocytes were incubated with CD1d tetramers for 20 min at 37°C. After one wash with warm PBS/1% FCS, samples were stained with monoclonal antibodies on ice. Propidium iodide was added immediately before FACS analysis to gate out dead cells.
Results
Refolding of CD1d with Glycolipid Ligands.
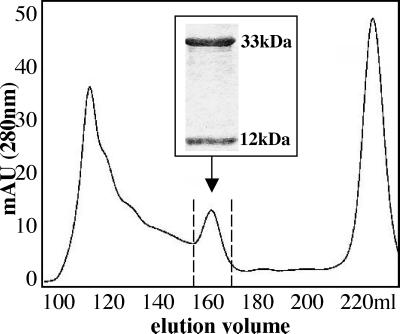
Analysis of CD1d, β2m, and α-GalCer containing refolding mix by FPLC size-exclusion chromatography revealed a UV-light absorption profile at 280 nm similar to the profiles obtained after refolding of HLA class I/β2m/peptide. Monomeric CD1d–β2m–α-GalCer complexes eluted as a defined peak at the expected size of 45 kDa (Fig. 1). The same protocol was used successfully to refold denatured CD1d and β2m in the presence of β-GalCer, α-Mannosylceramide, and ganglioside GM1 (data not shown). SDS/PAGE analysis of material eluted between 155 and 170 ml demonstrates the purity of monomeric CD1d–β2m complexes. Tetrameric human CD1d–β2m–α-GalCer complexes then were tested for their ability to bind to Vα24/Vβ11-positive cells.
Figure 1.
Purification of refolded CD1d–β2m monomers loaded with α-GalCer. FPLC and UV-light absorption profile at 280 nm wave length of a refolding mix containing CD1d and β2m in the presence of α-GalCer. The arrow indicates the CD1d–β2m monomeric complexes. (Inset) SDS/PAGE analysis of proteins eluted between 155 and 170 ml, which demonstrates the presence of two proteins of 33 and 12 kDa, corresponding to unglycosylated CD1d and β2m, respectively.
Specific Staining of Human NKT Cells by CD1d–α-GalCer Tetramers.
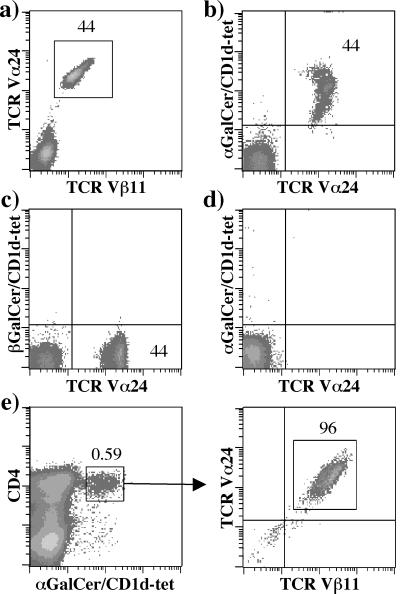
To test the specificity of the CD1d–α-GalCer tetramers, we generated T cell lines enriched in TCR Vα24/Vβ11 double-positive cells by in vitro stimulation of PBMC from healthy donors with α-GalCer. Enzyme-linked immunospot analysis demonstrated their ability to secrete IFN-γ and IL-4 after addition of α-GalCer but not β-GalCer and α-Mannosylceramide, hence confirming specificity of NKT cell lines for α-GalCer (data not shown). Fig. 2 shows staining of a representative PBMC culture stimulated with α-GalCer, in which 44% of T cells coexpressed Vα24 and Vβ11 TCR chains (Fig. 2a). Staining of the same cell culture with CD1d–α-GalCer tetramer, anti-Vα24 antibody, and anti-CD3 antibody revealed an identical frequency of CD1d–α-GalCer tetramer+/Vα24+ T cells (Fig. 2b), consistent with the ability of CD1d–α-GalCer tetramers to stain Vα24+/Vβ11+ T cells. Further experiments confirmed the specificity of binding of the CD1d/α-GalCer tetramers (Fig. 2 c–e). Vα24+/Vβ11+ cells were not stained by CD1d tetramers loaded with β-GalCer (Fig. 2c), and preincubation of the same cells with unlabeled anti-Vα24 and -Vβ11 antibodies impaired binding of CD1d–α-GalCer tetramer (Fig. 2d), hence demonstrating that CD1d–α-GalCer tetramers bind to the Vα24+/Vβ11+ TCR. HLA A02*01 (A2) -restricted T cell clones with specificity for the influenza A matrix protein epitope 58–66 (34) were not stained by CD1d–α-GalCer tetramers, although they were stained efficiently by an A2–matrix 58–66 tetramer (data not shown). Specificity of the CD1d–α-GalCer tetramers was confirmed further by demonstrating the ability of CD1d–α-GalCer tetramer to enrich cultures for the presence of Vα24+/Vβ11+ T cells (Fig. 2e).
Figure 2.
Specificity of CD1d–α-GalCer tetramers. (a–d) Vα24+/Vβ11+ T cells were expanded after stimulation of PBMC with α-GalCer and were stained with CD1d tetramers and antibodies shown in the figure. Cells in d were incubated first with unlabeled anti-Vα24 and anti-Vβ11 antibodies and subsequently stained with FITC-conjugated anti-Vα24 antibody and CD1d–α-GalCer tetramers. All density plots were gated on the CD3+ propidium iodide− cells. (e) CD4+ CD1d–α-GalCer tetramer+ cells were sorted from an in vitro-expanded α-GalCer-stimulated T cell line containing 0.59% positive cells. Enrichment for NKT cells was confirmed by using anti-Vα24 and -Vβ11 antibodies after two rounds of stimulation.
Together these results demonstrate that human CD1d–α-GalCer tetramers generated from fully denatured and reduced CD1d molecules by oxidative refolding chromatography can be used specifically to detect human CD1d–α-GalCer-specific T cells.
Staining of Murine NKT Cells by Human CD1d–α-GalCer Tetramers.
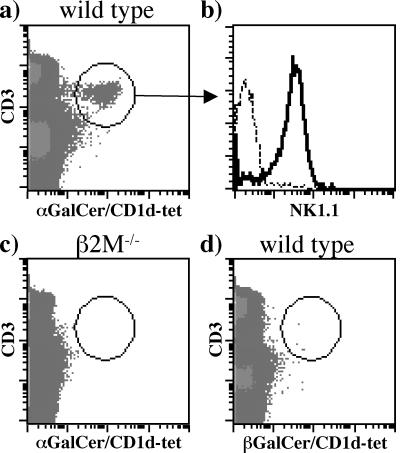
Murine NKT cells have been shown to kill human CD1d-expressing target cells pulsed with α-GalCer efficiently (11, 35, 36). Moreover, insect cell-derived human CD1d dimers have been reported to stain mouse α-GalCer-specific NKT cells (28). We tested the ability of CD1d–α-GalCer tetramers to stain mouse NKT cells. Human CD1d tetramers were capable of staining NKT cells in the spleen (Fig. 3), thymus, and liver (data not shown) of wild-type C57BL/6 mice but not in β2m-knockout mice (Fig. 3c). Lack of staining of NKT cells in wild-type C57BL/6 mice with β-GalCer tetramers confirmed the specificity of murine NKT cells for α-GalCer (Fig. 3d).
Figure 3.
Staining of mouse NK1.1 cells by human CD1d–α-GalCer tetramers. (a and c) Splenocytes from wild-type C57BL/6 mice and β2m−/− mice were stained with human CD1d–α-GalCer tetramers and antibodies specific to CD3 and NK1.1. (b) Cells in histogram plot were gated on the CD3+ CD1d–α-GalCer+ tetramer+ cell population shown in a. Dashed line shows staining with a control antibody, and the continuous line shows staining with anti-NK1.1 (d) As a negative control, splenocytes from wild-type C57BL/6 mice were stained with CD1d–β-GalCer tetramers and anti-CD3 antibody.
Ex Vivo Tetramer Staining of Circulating and Intrahepatic NKT Cells in Patients with Viral Hepatitis and Liver Cirrhosis.
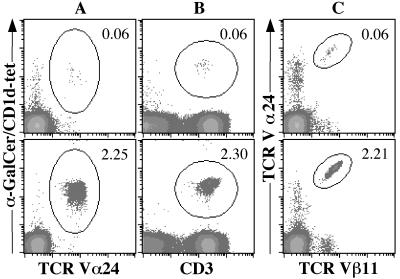
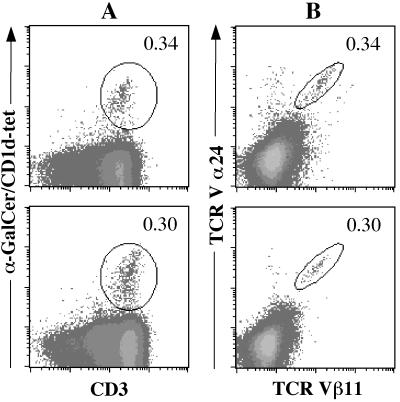
Ex vivo staining of PBMC from viral hepatitis patients with CD1d–α-GalCer tetramers showed frequencies ranging from 0.06 to 2.3% (Fig. 4 and Table 1). Similar frequencies were obtained whether anti-Vα24 or -CD3 antibodies were added together with CD1d–α-GalCer tetramers (Fig. 4). The frequency of NKT cells, as defined by the percentage of Vα24+/Vβ11+ cells, was very similar to the frequency of CD1d–α-GalCer tetramer+CD3+ cells, hence defining in an ex vivo assay the antigen specificity and restriction element of circulating Vα24+/Vβ11+ cells.
Figure 4.
Detection of CD1d–α-GalCer-specific T cells in peripheral blood from patients with HCV. Aliquots from each blood sample were stained with CD1d–α-GalCer tetramer and anti-Vα24 antibody (A), CD1d–α-GalCer tetramer and anti-CD3 antibody (B), or anti-Vα24 and -Vβ11 antibodies (C). Percentages of double-positive cells (circled regions) are indicated on each density plot. Top row corresponds to patient 1, and the bottom row corresponds to patient 5.
Table 1.
Frequency of NKT cells in PBMC from HCV-infected patients
| Patient | CD3+ CD1d/αGalCer-tet+ | Vα24+ CD1d/αGalCer-tet+ | Vα24+ Vβ11+ |
|---|---|---|---|
| P1 | 0.06 | 0.06 | 0.06 |
| P2 | 0.18 | 0.20 | 0.22 |
| P3 | 0.24 | 0.24 | 0.26 |
| P4 | 0.06 | 0.08 | 0.08 |
| P5 | 2.25 | 2.30 | 2.21 |
| P6 | 0.60 | 0.64 | 0.66 |
| P7 | 0.11 | 0.10 | 0.09 |
| HD1 | 0.08 | 0.07 | 0.08 |
| HD2 | 0.09 | 0.08 | 0.09 |
| HD3 | ND | ND | 0.50 |
Percentages of double-positive cells are shown. P1–7, HCV patients; HD1–3, healthy donors; ND, not determined.
Recent studies have shown that lymphocytes infiltrating liver parenchyma in response to viral hepatitis are enriched in Vα24+ T cells (20). These results suggest that NKT cells may play a role in the immune response against hepatitis virus. Consistent with this hypothesis, it has been shown that injection of α-GalCer into HBV-transgenic mice abolished virus replication (37). To assess whether CD1d–α-GalCer tetramers can be used to monitor the ex vivo frequency and phenotype of Vα24/Vβ11 NKT cells in liver samples, staining of intrahepatic T lymphocytes was carried out from four patients with viral hepatitis-induced cirrhosis (Fig. 5 and Table 2). Intrahepatic T lymphocytes from patients with viral hepatitis-induced cirrhosis had detectable Vα24/Vβ11-positive T cells with frequencies ranging from 0.03 up to 0.34%. Fig. 5 shows staining of intrahepatic T lymphocytes from two patients (P9 and P11) with the highest frequency of Vα24+/Vβ11+-positive T cells. Aliquots of the same intrahepatic T lymphocyte samples were stained with CD1d–α-GalCer tetramers and anti-CD3 antibody (Fig. 5). These results showed frequencies of double-positive cells (i.e., CD1d–α-GalCer+–CD3+) matching the frequencies determined by Vα24- and Vβ11-specific antibodies (Fig. 5 A and B). Further experiments were carried out to characterize the Va24+/Vβ11+ population better and to confirm the specificity of binding of CD1d–α-GalCer tetramers to intrahepatic T lymphocytes. Analysis of samples from patients 9 and 11 confirmed the presence of a distinct CD1d–α-GalCer tetramer+/Vα24+ population, which was not stained by either CD1d tetramers loaded with ganglioside GM1 tetramers or HLA–A23 tetramers (data not shown). Phenotypic analysis of the intrahepatic Vα24/Vβ11+ cells was consistent with an activated phenotype as defined by expression of CD45R0 and CD69 molecules (data not shown).
Figure 5.
Staining of intrahepatic lymphocytes in patients with HCV and HBV infection. Samples from patients (P9, top row; P11, bottom row) were stained with CD1d–α-GalCer tetramers and anti-CD3 antibody (A) or anti-Vα24 and -Vβ11 antibodies (B). Percentages of CD1d–α-GalCer tetramer+/CD3+ and Vα24+/Vβ11+ cells are shown.
Table 2.
Frequency of intrahepatic NKT cells in patients with end-stage viral-induced liver cirrhosis
| Patient | CD3+ CD1d/αGalCer-tet+ | Vα24+ CD1d/αGalCer-tet+ | Vα24+ Vβ11+ |
|---|---|---|---|
| P8 | 0.03 | ND | 0.03 |
| P9 | 0.30 | 0.29 | 0.30 |
| P10 | 0.04 | ND | 0.06 |
| P11 | 0.34 | 0.35 | 0.34 |
Percentages of double-positive cells are shown. Patients' diagnoses are described in Materials and Methods. ND, not determined.
Discussion
In this report we have shown that oxidative refolding chromatography is an effective and rapid protocol for the construction of CD1d-tetrameric complexes loaded with a single lipid species. We demonstrated that human CD1d–α-GalCer tetramers are highly sensitive and specific staining reagents that can be used for ex vivo monitoring of NKT cells in humans and mice. Refolding of denatured CD1d molecules ensures that a high proportion of CD1d–β2m complexes is loaded with a single lipid species, hence minimizing the presence of monomeric CD1d molecules loaded with endogenous lipids derived from CD1d-secreting cells.
Standard refolding techniques, used for the refolding of HLA class I molecules, and detergent-assisted refolding methods were unsuccessful for the refolding of CD1d–β2m–glycolipid complexes. It is possible that these difficulties were partly caused by the high hydrophobicity of CD1d molecules and its ligand(s) and possibly by the presence of an unpaired cysteine at position 12 in the α1 domain of CD1 molecules. In Fig. 1 we showed that oxidative refolding chromatography of denatured CD1d molecules allowed these problems to be overcome and yielded monomeric CD1d–β2m–α-GalCer in amounts sufficient for further purification and tetramer generation. We extended the application of this refolding protocol to other glycolipids and were capable of generating CD1d tetramers loaded with β-GalCer, α-Mannosylceramide, and GM1. It remains to be established whether assembly of CD1d molecules with β2m requires the presence of lipid ligands or whether, similar to CD1a and CD1b molecules (32), assembly of CD1d–β2m complexes is ligand independent.
The specificity of tetramer binding to Vα24/Vβ11-positive T cells was confirmed by several experiments. First, we showed that preincubation of NKT cells with unlabeled anti-Vα24 and -Vβ11 antibodies prevented their staining by CD1d–α-GalCer tetramers (Fig. 2d). Second, CD1d-tetrameric complexes loaded with either β-GalCer (Fig. 2c) or GM1 (data not shown) failed to stain Vα24+/Vβ11+ T cells. Furthermore, no cells were stained by CD1d–α-GalCer tetramers in β2m-knockout mice, which lacked surface expression of folded CD1d molecules (Fig. 3c). These results demonstrate the specificity of binding of CD1d–α-GalCer tetramers to NKT cells and show that glycosylation of CD1d molecules is not required to stabilize the interaction between E. coli-derived human CD1d–α-GalCer tetramers and NKTs TCR in both humans and mice.
By using CD1d tetramers we were able to characterize in ex vivo assays CD1d-restricted α-GalCer-specific T cell responses in liver and peripheral blood of patients with viral hepatitis. Staining of human samples with CD1d–α-GalCer tetramers rather than Vα24/Vβ11 antibodies defined both frequency and specificity of NKT cells, hence establishing restriction and specificity of Vα24+/Vβ11+ cells. The frequency of Vα24+/Vβ11+ cells closely matched the frequency of CD1d–α-GalCer tetramer+/CD3+ T cells in all patients' samples (Figs. 4 and 5; Tables 1 and 2) and in vitro-expanded NKT lines (Fig. 2). Moreover, similar frequencies were obtained by double staining patients' samples and NKT lines with CD1d–α-GalCer tetramers and Vα24-specific antibodies (Figs. 2, 4, and 5; Tables 1 and 2).
Frequency of NKT cells in patients' PBMC was in the same range as observed in healthy controls, with the only exception of patient 5 (Fig. 4; Table 1), in whom we found 2.3% of NKT cells. The significance of this striking elevation of NKT cells in patient 5's PBMC is unclear. However, it is unlikely that the enhanced frequency of NKT cells in patient 5 may be accounted for by HCV infection, because other HCV-infected patients had a lower frequency of NKT cells (Table 1). Tetramer staining of liver samples confirmed the specificity of intrahepatic Vα24/Vβ11-positive cells for CD1d–α-GalCer complexes (Fig. 5) as frequency of Vα24/Vβ11-positive cells closely matched the frequency of CD1d–α-GalCer tetramer-positive/CD3-positive T cells.
In conclusion, we have described a protocol to generate CD1d tetramers from fully denatured CD1d molecules and confirmed their specificity of binding to murine and human NKT cells. The role of NKT cells in vivo remains ill defined, and to date, the identity of NKT cells' ligand(s) is not known. Optimization of protocols to generate CD1d tetramers loaded rapidly with different lipids will aid the identification of NKT cells' natural ligand(s). We anticipate that the use of oxidative refolding chromatography to generate CD1 tetramers will be a powerful tool for studying in depth CD1-restricted T cell immune responses in infectious, autoimmune, and neoplastic disorders.
Acknowledgments
We thank Rod Dunbar, Mariolina Salio, and Michael Palmowski for helpful discussion. We also thank Dr. Hans Zweerink (Merck) for provision of the influenza matrix peptide T cell clone and Dr. Sergey Beresten for help with the FPLC column. We are grateful to Mie Nieda (Japanese Red Cross Central Blood Center, Tokyo) for providing protocols to establish NKT cell lines. This work was supported by the Cancer Research Campaign, the Medical Research Council, the Cancer Research Institute of U.S.A. and the National Health Institutes. S.G. was supported by The National Swiss Science Foundation, Novartis Foundation, and Hoffmann LaRoche Research Foundation.
Abbreviations
- β2m
β2-microglobulin
- TCR
T cell receptor
- NK
natural killer
- CTL
cytotoxic T lymphocyte
- GalCer
galactosylceramide
- PBMC
peripheral blood mononuclear cells
- HCV
hepatitis C virus
- HBV
hepatitis B virus
Footnotes
See commentary on page 2950.
References
- 1.Porcelli S A, Modlin R L. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- 2.Bilsland C A, Milstein C. Eur J Immunol. 1991;21:71–78. doi: 10.1002/eji.1830210112. [DOI] [PubMed] [Google Scholar]
- 3.Calabi F, Jarvis J M, Martin L, Milstein C. Eur J Immunol. 1989;19:285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 4.Albertson D G, Fishpool R, Sherrington P, Nacheva E, Milstein C. EMBO J. 1988;7:2801–2805. doi: 10.1002/j.1460-2075.1988.tb03135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin L H, Calabi F, Milstein C. Proc Natl Acad Sci USA. 1986;83:9154–9158. doi: 10.1073/pnas.83.23.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin L H, Calabi F, Lefebvre F A, Bilsland C A, Milstein C. Proc Natl Acad Sci USA. 1987;84:9189–9193. doi: 10.1073/pnas.84.24.9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park S H, Bendelac A. Nature (London) 2000;406:788–792. doi: 10.1038/35021233. [DOI] [PubMed] [Google Scholar]
- 8.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, et al. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 9.Brossay L, Chioda M, Burdin N, Koezuka Y, Casorati G, Dellabona P, Kronenberg M. J Exp Med. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Exley M, Garcia J, Balk S P, Porcelli S. J Exp Med. 1997;186:109–120. doi: 10.1084/jem.186.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nieda M, Nicol A, Koezuka Y, Kikuchi A, Takahashi T, Nakamura H, Furukawa H, Yabe T, Ishikawa Y, Tadokoro K, et al. Hum Immunol. 1999;60:10–19. doi: 10.1016/s0198-8859(98)00100-1. [DOI] [PubMed] [Google Scholar]
- 12.Exley M, Porcelli S, Furman M, Garcia J, Balk S. J Exp Med. 1998;188:867–876. doi: 10.1084/jem.188.5.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lantz O, Bendelac A. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bendelac A. Curr Opin Immunol. 1995;7:367–374. doi: 10.1016/0952-7915(95)80112-x. [DOI] [PubMed] [Google Scholar]
- 15.Brossay L, Naidenko O, Burdin N, Matsuda J, Sakai T, Kronenberg M. J Immunol. 1998;161:5124–5128. [PubMed] [Google Scholar]
- 16.Burdin N, Brossay L, Kronenberg M. Eur J Immunol. 1999;29:2014–2025. doi: 10.1002/(SICI)1521-4141(199906)29:06<2014::AID-IMMU2014>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 17.Carnaud C, Lee D, Donnars O, Park S H, Beavis A, Koezuka Y, Bendelac A. J Immunol. 1999;163:4647–4650. [PubMed] [Google Scholar]
- 18.Singh N, Hong S, Scherer D C, Serizawa I, Burdin N, Kronenberg M, Koezuka M, Van Kaer L. J Immunol. 1999;163:2373–2377. [PubMed] [Google Scholar]
- 19.Wilson S B, Kent S C, Patton K T, Orban T, Jackson R A, Exley M, Porcelli S, Schatz D A, Atkinson M A, Balk S P, et al. Nature (London) 1998;391:177–181. doi: 10.1038/34419. [DOI] [PubMed] [Google Scholar]
- 20.Nuti S, Rosa D, Valiante N M, Saletti G, Caratozzolo M, Dellabona P, Barnaba V, Abrignani S. Eur J Immunol. 1998;28:3448–3455. doi: 10.1002/(SICI)1521-4141(199811)28:11<3448::AID-IMMU3448>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Terabe M, Matsui S, Noben-Trauth N, Chen H, Watson C, Donaldson D, Carbone D, Paul W, Berzofsky J. Nat Immun. 2000;1:515–520. doi: 10.1038/82771. [DOI] [PubMed] [Google Scholar]
- 22.Moodycliffe A, Nghiem D, Clydesdale G, Ullrich S. Nat Immun. 2000;1:521–525. doi: 10.1038/82782. [DOI] [PubMed] [Google Scholar]
- 23.Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 24.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 25.Dunbar P R, Ogg G S, Chen J, Rust N, van der Bruggen P, Cerundolo V. Curr Biol. 1998;8:413–416. doi: 10.1016/s0960-9822(98)70161-7. [DOI] [PubMed] [Google Scholar]
- 26.Ogg G S, McMichael A J. Curr Opin Immunol. 1998;10:393–396. doi: 10.1016/s0952-7915(98)80110-6. [DOI] [PubMed] [Google Scholar]
- 27.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda J L, Naidenko O V, Gapin L, Nakayama T, Taniguchi M, Wang C R, Koezuka Y, Kronenberg M. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng Z-H, Castano A R, Segelke B W, Stura E A, Peterson P A, Wilson I A. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 30.Altamirano M M, Golbik R, Zahn R, Buckle A M, Fersht A R. Proc Natl Acad Sci USA. 1997;94:3576–3578. doi: 10.1073/pnas.94.8.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altamirano M M, Garcia C, Possani L D, Fersht A R. Nat Biotechnol. 1999;17:187–191. doi: 10.1038/6192. [DOI] [PubMed] [Google Scholar]
- 32.Altamirano M M, Woolfson A, Donda A, Shamshiev A, Briseño-Roa L, Foster N W, Veprintsev D B, De Libero G, Fersht A R, Milstein C. Proc Natl Acad Sci USA. 2001;98:3288–3293. doi: 10.1073/pnas.041596598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pascolo S, Bervas N, Ure J, Smith A, Lemonnier F, Perarnau B. J Exp Med. 1997;185:2043–2050. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bednarek M A, Sauma S Y, Gammon M C, Porter G, Tamhankar S, Williamson S R, Zweerink H J. J Immunol. 1991;147:4047–4053. [PubMed] [Google Scholar]
- 35.Ishihara S, Nieda M, Kitayama J, Osada T, Yabe T, Kikuchi A, Koezuka Y, Porcelli S A, Tadokoro K, Nagawa H, et al. J Immunol. 2000;165:1659–1664. doi: 10.4049/jimmunol.165.3.1659. [DOI] [PubMed] [Google Scholar]
- 36.Nicol A, Nieda M, Koezuka Y, Porcelli S, Suzuki K, Tadokoro K, Durrant S, Juji T. Immunology. 2000;99:229–234. doi: 10.1046/j.1365-2567.2000.00952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kakimi K, Guidotti L, Koezuka Y, Chisari F. J Exp Med. 2000;192:921–930. doi: 10.1084/jem.192.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]