Abstract
Several small molecules that inhibit the PI3 kinase (PI3K)-Akt signaling pathway are in clinical development. Although many of these molecules have been effective in preclinical models, it remains unclear whether this strategy alone will be sufficient to interrupt the molecular events initiated and maintained by signaling along the pathways because of the activation of other pathways that compensate for the inhibition of the targeted kinase. In this study, we performed a synthetic lethality screen to identify genes or pathways whose inactivation, in combination with the PI3K inhibitors PX-866 and NVPBEZ-235, might result in a lethal phenotype in glioblastoma multiforme (GBM) cells. We screened GBM cells (U87, U251, and T98G) with a large-scale, short hairpin RNA library (GeneNet), which contains 43 800 small interfering RNA sequences targeting 8500 well-characterized human genes. To decrease off-target effects, we selected overlapping genes among the 3 cell lines that synergized with PX-866 to induce cell death. To facilitate the identification of potential targets, we used a GSE4290 dataset and The Cancer Genome Atlas GBM dataset, identifying 15 target genes overexpressed in GBM tissues. We further analyzed the selected genes using Ingenuity Pathway Analysis software and showed that the 15 genes were closely related to cancer-promoting pathways, and a highly interconnected network of aberrations along the MYC, P38MAPK, and ERK signaling pathways were identified. Our findings suggest that inhibition of these pathways might increase tumor sensitivity to PX-866 and therefore represent a potential clinical therapeutic strategy.
Keywords: glioma, shRNA library, synthetic lethality
GBM is the most common primary brain tumor. Recent progress in understanding the molecular events that occur during GBM development and progression has increased interest in developing signal pathway–specific small-molecule kinase inhibitors for treatment of cancer in general and GBM in particular.1–3 Among these inhibitors are erlotinib and gifitinib, which target the epidermal growth factor receptor;4,5 rapamycin and RAD001, which target mammalian target of rapamycin (mTOR);6,7 and PX-866 and NVPBEZ-235, which target phosphatidylinositol-3 kinases (PI3 kinases or PI3Ks).8,9 Although all these agents have shown good preclinical inhibitory activity, it remains unclear whether inhibition by these molecules alone will be sufficient to interrupt the molecular events initiated and maintained by signaling along the pathway because of activation of other compensatory or collateral pathways compensating for the inhibition of the targeted kinase.9 It is therefore important to identify combinations of specific molecular agents that together can shut down these compensatory and collateral pathways and enhance the efficacy of inhibiting the PI3K pathway.
Large-scale RNA interference screening may be useful for identifying novel targets for such drugs;10 functional genetic screening has already been found to be highly effective in the unbiased identification of novel genes involved in biological processes.11,12 In the present study, we performed synthetic lethality screening to identify genes or pathways whose inactivation, in combination with targeting of the PI3 kinase pathway by small-molecule inhibitors, would result in a lethal phenotype in GBM cells.13 We used the GeneNet human short hairpin RNA (shRNA) library, which contains 43 800 small interfering RNA (siRNA) sequences targeting 8500 well-characterized human genes listed in the National Center for Biotechnology Information RefSeq database.14 The siRNA template sequences were designed to hybridize to oligonucleotide probes on the GeneChip Human Genome Focus Array (Affymetrix) to facilitate the identification of siRNA inserts. The shRNA library was used in combination with a new PI3K inhibitor, PX-866,15,16 and a genome-wide screening was performed using microarrays to identify targets or pathways whose inactivation sensitized cells to PX-866–induced cell death. We describe here a functional genetic screen designed to identify synergistic targets that would complement the effects of a PI3K inhibitor and help provide the rationale for combining the targeted therapies in GBM patients.
Materials and Methods
Cell Lines and Reagents
To identify novel synergistic targets in human GBM, we used glioma cells with different genetic backgrounds: U87 (bearing wild-type p53), U251 (bearing mutant p53), and T98G (bearing wild-type phosphatase and tensin homolog [PTEN]). All cell lines were maintained as monolayer cultures in Dulbecco's modified Eagle's medium/F12 medium supplemented with 10% fetal bovine serum and penicillin/streptomycin (all from Life Technologies).17
PX-866 is a biologically stable synthetic viridin related to wortmannin11–13 that inhibits PI3K by binding covalently to Lys-802 of the catalytic site of p110α (more potently than wortmannin) (14) and Lys-883 of p110γ (16); PX-866 also inhibits p110δ. PX-866 was obtained from G. Powis, who is a consultant to Oncothyreon, which is developing PX-866. A second PI3K inhibitor, NVP-BEZ235 (Novartis), was also tested to validate the screening process.18
shRNA Library Screening Assay
We used the GeneNet Human 8.5K shRNA Library (System Biosciences) to screen target genes whose inactivation in combination with PI3K inhibition by the lead drug, PX-866, would lead to a lethal phenotype in the U87, T98G, and U251 GBM cell lines. The library was cloned into a feline immunodeficiency virus (FIV)–based pFIV-H1-cop green fluorescent protein (GFP) shRNA expression vector.19 The cells were infected with the shRNA expression vector using the manufacturer's protocol (System Biosciences). In brief, 1 × 106/10 cm2 of each glioma cell line was infected with 2 multiplicities of infection (MOI) of the lentiviral shRNA library that target 8500 genes with 4 or 5 siRNAs per gene. After 48 h, one plate was treated with 5-µM PX-866 for an additional 72 h and the other plate was left untreated. Viable cells from both plates were harvested to extract total RNA.
Inserted siRNAs were amplified by means of PCR using shRNA library–specific biotinylated primers for subsequent hybridization to an Affymetrix Genome Focus Array (Affymetrix) in the MD Anderson Cancer Center microarray core facility. Each microarray consisted of 8793 probe sets, representing 8137 distinct UniGene clusters (UniGene Build 186, August 2005). Scan outputs of hybridization signals were modeled and normalized as summarized gene expression values using DNA-Chip Analyzer software (Affymetrix). Expression measurements of each array were obtained using the GeneNet software package (System Biosciences). Expression data were normalized using the R statistical computing platform and packages from Bioconductor bioinformatics software project (version 2.3.1). Expression profiling of cells treated with PX-866 alone performed with Affymetrix GeneChip arrays was used as a control to test if there were significantly overlapped genes with that of genes produced by PX-866 treatment with the shRNA pools. Since cell viability is our functional assay for shRNA library–transduced cells followed by PX-866 treatment, the shRNA signal absent in the PX-866–treated array but not in the control-treated array will be taken as evidence for synthetic lethality.
To identify differentially expressed genes, we performed the unequal variance t-test on each probe set. Because of the multiple comparisons, we expected many low P values due to chance alone. Given our null hypothesis that no genes would provide useful information about predicting response to combination treatment, we expected an overabundance of small P values. We captured this situation by modeling the distribution of the P value as a beta-uniform mixture. This analysis has been used to estimate false discovery rates that accompany particular P values derived from the t-test.20
To identify the potential targets, we used a GSE4290 dataset21 and The Cancer Genome Atlas (TCGA) dataset22 as validation datasets that facilitated our identification and selection of potential targets. In addition, we used Ingenuity Pathway Analysis (IPA; Ingenuity Systems) software to identify relevant pathways or networks (P for all <0.05).23
Lentiviral-Expressing shRNA
Retroviral-expressing shRNA specific to the RACK1 available from Open Biosystems was used to inactivate gene expression in glioma cell lines. We generated at least 3 shRNA vectors for each target in a lentiviral vector with the H1 promoter driving the expression of shRNA. This vector also contains a puromycin selectable marker. Lentiviral production was done as described (39). Briefly, the packaging cell line 293T (8 × 105) was plated on poly-L-lysine-coated 100-mm tissue culture plates (Corning) and transfected the following day. FuGene6 (Invitrogen) cotransfected with 1 μg of the transducing RACK1 shRNA, 1 μg of the packaging vector pCMV, and 1 μg of the vesicular stomatitis virus envelope vector, according to the manufacturer's instructions. The medium was changed the next day, and cells were cultured for another 24 h. Conditioned medium was then collected and cleared of debris by filtration through a 0.45-μm filter and stored at −70°C. This collection was repeated daily for 3 more days, and media from the 3 days were pooled and added to U87 and U251 cells at a density of 1 × 105 cells for 3 days. Following shRNA transduction, clones resistant to puromycin were isolated and assayed for gene expression using western blotting analysis. We tested 3 shRNAs for RACK1 and selected cell lines expressing at least <70% of the target gene expression for further validation experiments.
Cell Proliferation Assay
Cells were seeded in 96-well plates (1000 cells/well) and incubated at 37°C for 24 h before addition of serial dilutions of PX-866. Growth inhibition was determined using the sulforhodamine B (SRB) viability assay. The half maximal inhibitory concentration value was calculated as the mean drug concentration required to inhibit cell proliferation by 50% compared with vehicle controls.
Statistical Analyses
We used the 2-sample t-test to identify genes that were differentially expressed between 2 classes of treated and untreated shRNA library–infected cells. In brief, the P value for the differential expression of each probe (gene) between groups of treated and untreated shRNA library–infected cells was computed on the basis of the test statistic. The false discovery rate was then used to identify genes that would predict response to combination treatment, as described previously.20 All in vitro assays were performed in triplicate.
Results
shRNA Library Screen for Genes Sensitizing to PX-866
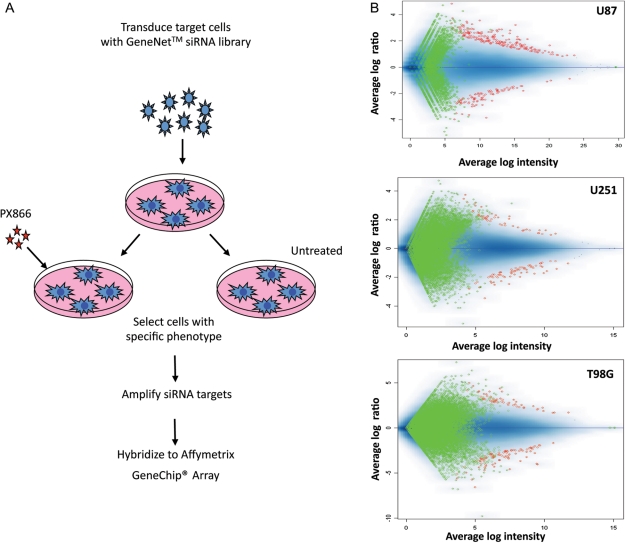
To screen shRNA libraries for target genes whose inactivation led to a lethal phenotype when combined with PX-866 treatment in cancer cells, we used U87, U251, and T98G cell lines. Detailed experimental procedures are outlined in Figure 1A. Using a lentiviral vector carrying a GFP reporter gene, we established that a MOI of 2 resulted in 95–100% transduction efficiency in U87 cells. Next, one plate was left untreated and the other plate was treated with 5 µM of PX-866 for 72 h. In cells infected with the library alone, GFP was uniformly distributed. By contrast, PX-866 treatment resulted in significant phenotypic variability. Our previous study showed that treatment with PX-866 alone induced a marked increase in the number of U87 cells in the G1 phase after 48 h.16 These initial assays showed that the integrated systems in the U87 cell line behaved as expected, showing >35% cell death. Therefore, this system was used to screen the shRNA library, targeting 8137 genes. Instead of sequencing individual cell lines, we used a high-throughput approach. The array dataset of each cell line consisted of 6 Affymetrix GeneChip Human Genome Focus Arrays. Data were normalized to analyze, and calculating a t-score generated a hit list. The scatter plot of the analysis, shown in Figure 1B, verified that each GeneNet shRNA library was efficiently transduced and expressed in the 3 cell lines without a significant loss of representation by amplifying siRNA inserts from pseudoviral RNA that had been isolated from the cell's total RNA.
Fig. 1.
(A) Outline of functional screening with GeneNet shRNA libraries showing the general steps required to identify genes that modulate a specific phenotype: transduction of the library into target cells, selection of cells with the desired phenotype, and identification of phenotype-inducing siRNAs and corresponding target genes by hybridizing amplified shRNA cassettes with a GeneChip Array. (B) Representative results of an analysis of shRNA inserts in U87, U251, and T98G GBM cells infected with the Human 8.5K shRNA Library in pFIV-H1-Puro vector. Cells were infected separately and harvested 72 h after PX-866 treatment. shRNA inserts were amplified and hybridized as described in Materials and Methods. Green and red reflect the threshold and identified target genes, respectively. Mean log intensity of gene expression is represented along the x-axis. The y-axis shows the mean log ratio. The ratio for most probes was scattered around 1.0 (no change), with a range between 0.5 and 2.0.
To identify shRNAs that modulate the lethal phenotype, we measured the viability of shRNA library–transduced cells after PX-866 treatment and compared it with the viability of untreated cells separately for each cell line; the absence of an shRNA signal in the drug-treated array but not the untreated array was considered evidence of synthetic lethality. On the basis of the experimental design, genes whose shRNA expression in untreated cells was 2 times higher than that in PX-866–treated cells were selected for further analysis. To avoid off-target effects, as shown in Table 1, we selected 30 overlapping genes among 3 cell lines identified by the screen and showed that knockdown of these genes together with PX-866 treatment could lead to a lethal phenotype regardless of the genetic background of the cell lines.
Table 1.
Overlapping genes among three cell lines using PX-866
| ACC no | Gene symbol | Fold ratio | Description |
|---|---|---|---|
| NM016270 | KLF2 | −5.8 | Kruppel-like factor 2 lung |
| U11282 | FUT7 | −5.0 | fucosyltransferase 7 alpha 13 fucosyltransferase |
| NM012418 | FSCN2 | −4.9 | fascin Strongylocentrotus purpuratus homolog 2 |
| NM006098* | RACK1 | −4.8 | receptor for activated C-kinase 1 |
| BC005241 | MRE11A | −4.7 | meiotic recombination S. cerevisiae 11 homolog A |
| U48297 | PTP4A2 | −4.6 | protein tyrosine phosphatase type IVA member 2 |
| NM003299 | HSP90B1 | −4.0 | heat shock protein 90kDa beta Grp94 member 1 |
| NM015995 | KLF13 | −3.9 | Kruppel-like factor 13 |
| BC001360 | RHOA | −3.7 | Ras homolog gene family member A |
| L19184 | PRDX1 | −3.6 | peroxiredoxin 1 |
| U06935 | TEF | −3.6 | cystatin B stefin B |
| U17714 | ST13 | −3.5 | suppression of tumorigenicity 13 colon carcinoma Hsp70 interacting protein |
| NM004369 | COL6A3 | −3.5 | collagen type VI alpha 3 |
| U43328 | HAPLN1 | −3.3 | hyaluronan and proteoglycan link protein 1 |
| NM002896 | RBM4 | −3.3 | RNA binding motif protein 4 |
| AL039831 | JAK1 | −3.1 | janus kinase 1 a protein tyrosine kinase |
| NM020365 | EIF2B3 | −3.1 | eukaryotic translation initiation factor 2B subunit 3 gamma 58kD |
| BC000733 | EIF3S4 | −3.0 | eukaryotic translation initiation factor 3 subunit G |
| NM001438 | ESRRG | −3.0 | estrogen-related receptor gamma |
| NM019076 | UGT1A6 | −3.0 | UDP glycosyltransferase 1 family polypeptide A6 |
| NM006078 | CACNG2 | −2.8 | calcium channel voltage-dependent gamma subunit 2 |
| NM003668 | MAPKAPK5 | −2.7 | mitogen-activated protein kinase-activated protein kinase 5 |
| NM001408 | CELSR2 | −2.6 | cadherin EGF LAG seven-pass G-type receptor 2 |
| NM005271 | GLUD1 | −2.6 | glutamate dehydrogenase 1 |
| AI761759 | CANX | −2.6 | calnexin |
| AF283777 | CD72 | −2.5 | CD72 molecule |
| NM006494 | CDC42BPA | −2.5 | CDC42 binding protein kinase alpha DMPK-like |
| AF028333 | GDF11 | −2.5 | growth differentiation factor-11 |
| NM022443 | MLF1 | −2.2 | myeloid leukemia factor 1 |
| AF196175 | TRPV1 | −2.2 | transient receptor potential cation channel subfamily V member 1 |
Primary Target Validation Using Public Databases
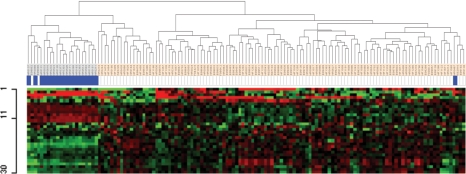
After identifying genes that produced a lethal phenotype in the 3 GBM cell lines, we performed the first round of validation of these genes using public GBM databases. To gain insight into the expression of the identified target genes in GBM tissues, we assessed an independent public GBM gene dataset (the GSE4290 dataset).24 We performed hierarchical cluster analysis of the genes highly expressed in all 3 cell lines using a 2-sample t-test (P = .01). Hierarchical cluster analysis of the 30 overlapping genes revealed overexpression patterns of 15 genes in specimens from 116 GBM patients represented in the GSE4290 dataset, as compared with 21 nontumor tissues (Fig. 2). Therefore, we selected the 15 genes as a subset of our target genes shown in bold type in Table 1. Of the 116 GBM patients, 67 were considered short-term survivors (<2 y) and 16 were considered long-term survivors (≥2 y) (Supplementary Fig. S1). Kaplan–Meier plots and log-rank survival analyses showed that the median overall survival time of all the patients with overexpression of the 15 genes was shorter (40.2 wks) than that in a group with lower expression of the genes (59.7 wks), indicating that overexpression of the 15 target genes was associated with activation of cancer-related pathways. To confirm that overexpression of these 15 genes constituted an unfavorable phenotype in GBM, we assessed data from the TCGA project (Supplementary Fig. S2). In this dataset (which contains data for 200 short-term survivors and 54 long-term survivors), the correlations among the expression levels of the 15 genes were highly consistent with those for the GSE4290 dataset. In addition, data in the GSE4290 dataset allowed us to determine that these genes were also overexpressed in stage II and stage III glioma (data not shown), suggesting that the 15 genes are involved in glioma progression.
Fig. 2.
Hierarchical clustering of the GSE4290 dataset of 116 GBM samples from patients with GBM and 21 nontumor tissues from the same patients based on the 30 genes we found overexpressed in all 3 of our cell lines. Genes with an expression ratio that differed by a factor of at least 2 were selected for hierarchical analysis (8015 gene features). Thirty probe sets (22 genes) were generated using a two-sample t-test (P = .01) between 2 populations. Fifteen genes were overexpressed in the GBM samples, and 7 genes were downregulated, with 8 genes not showing in this analysis. The 30 probe sets are presented here in matrix format showing supervised hierarchical clustering, where rows represent individual genes and columns represent each tissue. Each cell in the matrix thus represents the expression level of a particular gene in an individual tissue. Red and green cells reflect high and low expression levels, respectively. Each blue bar represents a nontumor tissue.
Secondary Target Validation Using Another PI3K Inhibitor
For secondary validation, we performed a similar shRNA library screening with the U87 cell line using a dual PI3K/mTOR inhibitor, NVP-BEZ235. We have shown previously that PX-866 and NVP-BEZ235 at micromolar and nanomolar concentrations can effectively suppress U87, U251, LN229, and LN18 cell growth.16,18 We also observed that the 2 agents caused the same pattern of dose-dependent growth inhibition and G1 cell cycle arrest in glioma cells. Therefore, the same shRNA library was used to test the effect of NVP-BEZ235 in U87 cells. The NVP-BEZ235 screen also identified the 15 genes set similar to the PX-866 screen, with very consistent expression levels of the inhibited targets in the comparison analysis (Table 2). These findings further confirm the reproducibility of the synthetic lethality screen and suggest that the 15 genes identified may be potential novel targets that have synergistic effects with PI3K inhibition in GBM.
Table 2.
Fifteen genes regulated by PX-866 and NVP-BEZ235 in U87 cells
| ACC no | Gene symbol | PX-866 | NVP-BEZ235 |
|---|---|---|---|
| NM006098 | RACK1 | −9.6 | −2.0 |
| BC005241 | MRE11A | −3.6 | −2.0 |
| NM015995 | KLF13 | −3.4 | −5.6 |
| BC001360 | RHOA | −2.7 | −1.1 |
| L19184 | PRDX1 | −5.8 | −3.3 |
| NM004369 | COL6A3 | −2.2 | −2.3 |
| NM002896 | RBM4 | −2.4 | −4.8 |
| BC000733 | EIF3S4 | −3.8 | −2.2 |
| NM003668 | MAPKAPK5 | −5.3 | −1.8 |
| AI761759 | CANX | −3.4 | −2.0 |
| AF283777 | CD72 | −2.1 | −1.6 |
| NM006494 | CDC42BPA | −2.5 | −1.6 |
| AF028333 | GDF11 | −2.0 | −1.3 |
| NM022443 | MLF1 | −4.3 | −2.1 |
| AF196175 | TRPV1 | −3.1 | −1.8 |
IPA
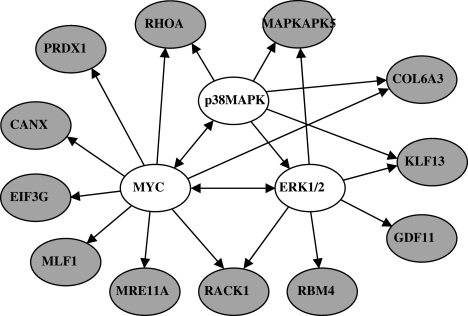
We further analyzed the signal pathway relationship of the 15 genes using IPA software to gain integrated biological insight into the underlying molecular pathways that are differentially expressed in functional units of the targets. We focused on pathways that are associated with cell death and growth arrest because of our selection schema for cell viability loss based on our findings regarding the effects of shRNA inhibition of these targets on cell viability in the context of PI3K inhibition. A protein–protein interaction network analysis was performed using existing knowledge about GBM-related networks, in particular the PI3K and Akt signaling pathways. The protein–protein interaction analysis identified several interacting genes among the 15 target genes identified from the public databases; these included genes encoding the G-protein coupled receptor (RACK1); kinases (CDC42BPA and MAPKAPK5); a growth factor (GDF11); and transcription regulators (KLF13 and EIF3S4). We also mapped clusters of targets to functional networks and cellular pathways in order of the strength of correlation, as determined by the P value. Figure 3A shows the results of the gene ontology functional analysis of various sets of the 15 target genes; these genes are known to be involved in cancer-initiated cellular processes and functional disorders in many cancers. Figure 3B shows the interactions among these genes in various cancer-promoting pathways, in particular cancer-specific signaling, cell-cycle regulation, cellular growth and proliferation, and cytokine-specific signaling (Fig. 3A and B). When inspecting the two analyses, we found that the 15 gene sets have the functional relevance of cancer-related features, consistent with the results of the glioma-related databases. This analysis also identified a highly interconnected network of aberrations, including 12 genes that interact with MYC, P38MAPK, and ERK1/2 (Fig. 4), which suggests that inhibition of these pathways might increase cellular sensitivity to either PX-866 or other PI3K inhibitors.
Fig. 3.
(A) Gene ontology analysis using IPA shows stratification of 15 genes in cancer-initiating functions. (B) IPA shows that cancer-promoting pathways were major pathways for genes identified in the synthetic lethality screening. Each function (or pathway) shows a statistically significant change, and the number in parentheses represents the number of genes in each category. The same gene could be represented multiple times in each category.
Fig. 4.
Network mapping of 12 gene sets found to have significant changes in pathways by IPA. Each gene connects the top 3 signaling pathways, that is, the MYC, ERK, and p38MAPK pathways. Red indicates a target gene; the uncolored ovals show the names of the 3 main interacting genes. Arrows indicate gene products that act on other gene products.
Validation of Target
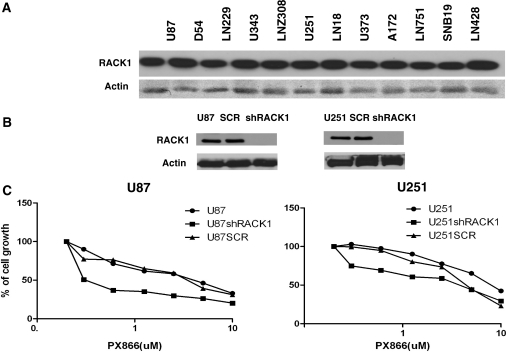
We validated one of the targets, RACK1, in this study. First, endogenous expression of RACK1 was checked in a battery of glioma cells using anti-RACK1 antibody in Western blot analysis, and Figure 5A shows that glioma cells express high levels of RACK1. We selected U87 and U251 glioma cells that express high levels of RACK1 and generated U87 and U251 RACK1 knockdown cell lines. RACK1 knockdown cell lines were generated using a recombinant shRNA retroviral vector with 4 different shRNA species against RACK1 using Open Biosystems retroviral shRNA against RACK1 (Fig. 5B). In an attempt to validate RACK1 as a synergistic target, we tested the sensitivity of RACK1 knockdown cells with the PI3K inhibitor PX-866. The cell viability assay showed that RACK1 knockdown cells showed enhanced sensitivity to PX-866, confirming that inactivation of RACK1 sensitizes glioma cells to PX-866–induced cell death, thereby showing a strong synergism of inactivating PI3 kinase and RACK1 (Fig. 5C).
Fig. 5.
(A) Western blot showing RACK1 expression in glioma cells. (B) Stable knockdown of RACK1 in glioma cells. U87 and U251 cells were treated with a shRNA retroviral vector with different shRNAs against RACK1. RACK1 knockdown was observed by decreased expression of RACK1. (C) Cell viability assay to determine the synergistic effect of knocking down the PI3 kinase pathway (PX-866) and RACK1 expression. Inactivation of RACK1 differentially sensitizes glioma cells to PX-866–induced cell death.
Discussion
Several small molecules that inhibit the PI3K-Akt signaling pathway are in clinical development. In preclinical models bearing PTEN/PIK3CA mutations, however, single-agent Akt and dual PI3K/mTOR inhibitors only delay tumor growth and do not shrink existing tumors.25,26 Even when PI3K/Akt inhibition is effective in preclinical cancer models, apoptosis is not induced.27 Thus, single-agent PI3K pathway inhibitors often do not yield the expected responses in cancers that are supposed to be sensitive to these inhibitors.28 One potential reason for the limited efficacy of PI3K inhibition alone is the existence of compensatory or redundant pathways in tumor cells. For example, inhibition of mTORC1 leads to activation of the ERK and PI3K signaling pathways, showing increased Akt activity and downstream signals.29 Taken together, the evidence from the present and prior studies suggests that combining multiple pathway inhibitors might be more effective.
In this study, we developed a comprehensive method for identifying molecular targets whose inactivation could enhance the therapeutic efficacy of PI3K inhibitors, suggesting that combinations of PI3K inhibitors and agents targeting those molecules can act synergistically to improve therapeutic efficacy, thus, affirming that testing for synthetic lethality is useful for identifying new targets that would complement the effects of drugs of interest.30,31 Here, we used a retrovirus-based library that can target approximately one-third of human genes and a platform whereby we could systematically assess the loss-of-function phenotypes.32 Identifying the genes that are essential for cell survival may facilitate the discovery of compensatory signaling pathways that impair the effectiveness of PI3K pathway inhibitors clinically.33 Using a synthetic lethality screen, we confirmed the functional relevance of 15 cancer-related genes through the use of the glioma-related databases and by analyzing molecular pathways, showing that most, but not all, of the target genes can lead to activation of the ERK signaling pathway. Remarkably, recent studies showed that for many cancers, combined PI3K-Akt and Raf-MEK-ERK inhibition was required to effectively shut off PI3K/mTOR signaling and promote apoptosis through Bcl-2 family proteins.34
The genome-scale screening method used in our study has been reported to be powerful, because it can reveal interactions not only between gene products that have direct contact but also between those whose signaling occurs along the same or parallel pathways.35 An advantage of a large-scale shRNA library is that it is genome-wide and is robust for most if not all relevant targets. The key practical issues when performing RNA interference screening in mammalian cells are assay development, library selection, target validation, robust statistical algorithms, and functional validation experiments.36 We used retroviral vectors under optimized conditions so that cells are infected by only a single virion during retroviral transduction.37 Monitoring phenotypic changes simplified the subsequent identification of functional clones from the library and allowed us to rapidly screen millions of cells. This approach can be readily adapted to other cell types and phenotypic changes. Some siRNAs have “off-target” effects, and these effects are often the result of the RNAs being partially homologous to other transcripts. A sequence identity of as few as 11 or 12 nucleotides between an siRNA and an mRNA may be sufficient for interference to occur,38 indicating that cross-reactivity is a substantial problem. However, the shRNA library used in our study was designed to avoid off-target effects by minimizing the similarities between shRNAs and other transcripts.
We performed a target-specific screening with another PI3K/mTOR inhibitor, NVP-BEZ235, which confirmed the functional validity of the high-throughput screening, suggesting that our robust screening strategies facilitated the selection of high-confidence target genes that could be rationally combined with PI3K pathway inhibitors. In addition, the target validation was done using the shRNA approach to inactivate the individual target, and inactivation of the target gene showed an additive effect with PX-866 in growth inhibition.
The success of this large-scale screening approach is illustrated by our bioinformatics and molecular discovery that the target genes might be a significant contributor to the molecular mechanisms that underlie and regulate selective sensitivity of GBM cells to PI3K inhibitors. Our data strongly established the effectiveness of searching for synthetic lethality using an shRNA library with a specific molecular inhibitor to perform genome-wide screening of potential targets or pathways whose inactivation would synergize the antitumor effects of the drug. We expect that our results will not only advance knowledge of the interdependency among complex signaling pathways and their response to particular targeted therapies, but also increase the number of combination therapies for GBM.
Supplementary Material
Funding
This study was supported by grants from the National Cancer Institute (CA 123304 to W.K.A.Y., P30 CA 016672 to MD Anderson, and P30 CA 127001 to W.K.A.Y.).
Acknowledgments
We thank Ann M. Sutton and Kathryn B. Carnes (Department of Scientific Publications, The University of Texas MD Anderson Cancer Center) for editing the manuscript.
Conflict of interest statement. None declared.
References
- 1.Chang SM, Lamborn KR, Kuhn JG, et al. Neurooncology clinical trial design for targeted therapies: lessons learned from the North American Brain Tumor Consortium. Neuro Oncol. 2008;10(4):631–642. doi: 10.1215/15228517-2008-021. doi:10.1215/15228517-2008-021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Groot JF, Gilbert MR. New molecular targets in malignant gliomas. Curr Opin Neurol. 2007;20(6):712–718. doi: 10.1097/WCO.0b013e3282f15650. doi:10.1097/WCO.0b013e3282f15650. [DOI] [PubMed] [Google Scholar]
- 3.Omuro AM. Exploring multi-targeting strategies for the treatment of gliomas. Curr Opin Investig Drugs. 2008;9(12):1287–1295. [PubMed] [Google Scholar]
- 4.Cappuzzo F. Erlotinib in gliomas: should selection be based on EGFR and Akt analyses? J Natl Cancer Inst. 2005;97(12):868–869. doi: 10.1093/jnci/dji169. doi:10.1093/jnci/dji169. [DOI] [PubMed] [Google Scholar]
- 5.Reardon DA, Egorin MJ, Quinn JA, et al. Phase II study of imatinib mesylate plus hydroxyurea in adults with recurrent glioblastoma multiforme. J Clin Oncol. 2005;23(36):9359–9368. doi: 10.1200/JCO.2005.03.2185. doi:10.1200/JCO.2005.03.2185. [DOI] [PubMed] [Google Scholar]
- 6.Fasolo A, Sessa C. mTOR inhibitors in the treatment of cancer. Expert Opin Investig Drugs. 2008;17(11):1717–1734. doi: 10.1517/13543784.17.11.1717. doi:10.1517/13543784.17.11.1717. [DOI] [PubMed] [Google Scholar]
- 7.Yang G, Sun Q, Teng Y, Li F, Weng T, Yang X. PTEN deficiency causes dyschondroplasia in mice by enhanced hypoxia-inducible factor 1alpha signaling and endoplasmic reticulum stress. Development. 2008;135(21):3587–3597. doi: 10.1242/dev.028118. doi:10.1242/dev.028118. [DOI] [PubMed] [Google Scholar]
- 8.Opel D, Westhoff MA, Bender A, Braun V, Debatin KM, Fulda S. Phosphatidylinositol 3-kinase inhibition broadly sensitizes glioblastoma cells to death receptor- and drug-induced apoptosis. Cancer Res. 2008;68(15):6271–6280. doi: 10.1158/0008-5472.CAN-07-6769. doi:10.1158/0008-5472.CAN-07-6769. [DOI] [PubMed] [Google Scholar]
- 9.Goudar RK, Shi Q, Hjelmeland MD, et al. Combination therapy of inhibitors of epidermal growth factor receptor/vascular endothelial growth factor receptor 2 (AEE788) and the mammalian target of rapamycin (RAD001) offers improved glioblastoma tumor growth inhibition. Mol Cancer Ther. 2005;4(1):101–112. [PubMed] [Google Scholar]
- 10.Iorns E, Lord CJ, Turner N, Ashworth A. Utilizing RNA interference to enhance cancer drug discovery. Nat Rev Drug Discov. 2007;6(7):556–568. doi: 10.1038/nrd2355. doi:10.1038/nrd2355. [DOI] [PubMed] [Google Scholar]
- 11.Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell. 2003;12(3):627–637. doi: 10.1016/s1097-2765(03)00348-4. doi:10.1016/S1097-2765(03)00348-4. [DOI] [PubMed] [Google Scholar]
- 12.Mukherji M, Bell R, Supekova L, et al. Genome-wide functional analysis of human cell-cycle regulators. Proc Natl Acad Sci USA. 2006;103(40):14819–14824. doi: 10.1073/pnas.0604320103. doi:10.1073/pnas.0604320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5(9):689–698. doi: 10.1038/nrc1691. doi:10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 14.Information. NCfB. NCBI RefSeq database. 2005. www.ncbi.nlm.nih.gov/refseq/
- 15.Ihle NT, Williams R, Chow S, et al. Molecular pharmacology and antitumor activity of PX-866, a novel inhibitor of phosphoinositide-3-kinase signaling. Mol Cancer Ther. 2004;3(7):763–772. [PubMed] [Google Scholar]
- 16.Koul D, Shen R, Kim YW, et al. Cellular and in vivo activity of a novel PI3K inhibitor, PX-866, against human glioblastoma. Neuro Oncol. 2010;12(6):559–569. doi: 10.1093/neuonc/nop058. doi:10.1093/neuonc/nop058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerrato JA, Khan T, Koul D, et al. Differential activation of the Fas/CD95 pathway by Ad-p53 in human gliomas. Int J Oncol. 2004;24(2):409–417. [PubMed] [Google Scholar]
- 18.Liu TJ, Koul D, LaFortune T, et al. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol Cancer Ther. 2009;8(8):2204–2210. doi: 10.1158/1535-7163.MCT-09-0160. doi:10.1158/1535-7163.MCT-09-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curran MA, Nolan GP. Nonprimate lentiviral vectors. Curr Top Microbiol Immunol. 2002;261:75–105. doi: 10.1007/978-3-642-56114-6_4. [DOI] [PubMed] [Google Scholar]
- 20.Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19(10):1236–1242. doi: 10.1093/bioinformatics/btg148. doi:10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- 21.Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102(16):5814–5819. doi: 10.1073/pnas.0402870102. doi:10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holleman A, Cheok MH, den Boer ML, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351(6):533–542. doi: 10.1056/NEJMoa033513. doi:10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 24.Li A, Walling J, Ahn S, et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009;69(5):2091–2099. doi: 10.1158/0008-5472.CAN-08-2100. doi:10.1158/0008-5472.CAN-08-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serra V, Markman B, Scaltriti M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68(19):8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. doi:10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 26.She QB, Chandarlapaty S, Ye Q, et al. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS One. 2008;3(8):3065. doi: 10.1371/journal.pone.0003065. doi:10.1371/journal.pone.0003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan QW, Cheng CK, Nicolaides TP, et al. A dual phosphoinositide-3-kinase alpha/mTOR inhibitor cooperates with blockade of epidermal growth factor receptor in PTEN-mutant glioma. Cancer Res. 2007;67(17):7960–7965. doi: 10.1158/0008-5472.CAN-07-2154. doi:10.1158/0008-5472.CAN-07-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simi L, Pratesi N, Vignoli M, et al. High-resolution melting analysis for rapid detection of KRAS, BRAF, and PIK3CA gene mutations in colorectal cancer. Am J Clin Pathol. 2008;130(2):247–253. doi: 10.1309/LWDY1AXHXUULNVHQ. doi:10.1309/LWDY1AXHXUULNVHQ. [DOI] [PubMed] [Google Scholar]
- 29.Carracedo A, Ma L, Teruya-Feldstein J, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118(9):3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tucker CL, Fields S. Lethal combinations. Nat Genet. 2003;35(3):204–205. doi: 10.1038/ng1103-204. doi:10.1038/ng1103-204. [DOI] [PubMed] [Google Scholar]
- 31.Iglehart JD, Silver DP. Synthetic lethality–a new direction in cancer-drug development. N Engl J Med. 2009;361(2):189–191. doi: 10.1056/NEJMe0903044. doi:10.1056/NEJMe0903044. [DOI] [PubMed] [Google Scholar]
- 32.Berns K, Hijmans EM, Mullenders J, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. doi:10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 33.Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14(10):2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. doi:10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 34.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–1356. doi: 10.1038/nm.1890. doi:10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simons AH, Dafni N, Dotan I, Oron Y, Canaani D. Genetic synthetic lethality screen at the single gene level in cultured human cells. Nucleic Acids Res. 2001;29(20):100. doi: 10.1093/nar/29.20.e100. doi:10.1093/nar/29.20.e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moffat J, Sabatini DM. Building mammalian signalling pathways with RNAi screens. Nat Rev Mol Cell Biol. 2006;7(3):177–187. doi: 10.1038/nrm1860. doi:10.1038/nrm1860. [DOI] [PubMed] [Google Scholar]
- 37.Kustikova OS, Wahlers A, Kuhlcke K, et al. Dose finding with retroviral vectors: correlation of retroviral vector copy numbers in single cells with gene transfer efficiency in a cell population. Blood. 2003;102(12):3934–3937. doi: 10.1182/blood-2003-05-1424. doi:10.1182/blood-2003-05-1424. [DOI] [PubMed] [Google Scholar]
- 38.Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–637. doi: 10.1038/nbt831. doi:10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.