Abstract
The biosynthetic gene clusters for the glycopeptide antitumor antibiotics bleomycin (BLM), tallysomycin (TLM), and zorbamycin (ZBM) have been recently cloned and characterized from Streptomyces verticillus ATCC15003, Streptoalloteichus hindustanus E465-94 ATCC31158, and Streptomyces flavoviridis ATCC21892, respectively. The striking similarities and differences among the biosynthetic gene clusters for the three structurally related glycopeptide antitumor antibiotics prompted us to compare and contrast their respective biosynthetic pathways and to investigate various enzymatic elements. The presence of different numbers of isolated nonribosomal peptide synthetase (NRPS) domains in all three clusters does not result in major structural differences of the respective compounds. The seemingly identical domain organization of the NRPS modules responsible for heterocycle formation, on the other hand, is contrasted by the biosynthesis of two different structural entities, bithiazole and thiazolinyl-thiazole, for BLM/TLM and ZBM, respectively. Variations in sugar biosynthesis apparently dictate the glycosylation patterns distinct for each of the BLM, TLM, and ZBM glycopeptide scaffolds. These observations demonstrate nature’s ingenuity and flexibility in achieving structural differences and similarities via various mechanisms and will surely inspire combinatorial biosynthesis efforts to expand on natural product structural diversity.
Natural products are a vital source of current clinical drugs. The Actinomycetales have clearly been the richest microbial source of bioactive compounds.1 Consequently, the biosynthetic machineries responsible for the construction of these diverse and complex compounds have been intensely studied.
The exponential growth in cloning and characterization of natural product biosynthetic machinery in the past two decades, in particular gene clusters encoding the biosynthesis of polyketides and nonribosomal peptides, members of two of the largest families of natural products, has presented several new opportunities to produce natural products and generate natural product analogues. Central to these discoveries is the observation that genes responsible for natural product biosynthesis are often clustered in the microbial genome and that variations of a few common biosynthetic machineries can account for the vast structural diversity found in natural products. These findings have inspired the exploration of an emerging technology, referred to as combinatorial biosynthesis,2–7 as a promising methodology to prepare complex natural products and their analogues biosynthetically. Specific structural alterations in the presence of abundant functional groups can often be achieved by precise rational manipulation of the biosynthetic machinery.
In a laboratory setting, a minimum of four requirements must be met before combinatorial biosynthesis can be successfully used to generate structural diversity of natural products: (i) availability of the gene clusters encoding the production of a particular natural product or family of natural products, (ii) genetic and biochemical characterizations of the biosynthetic machinery for the targeted natural products to a degree that the combinatorial biosynthesis principles can be rationally applied to engineer the novel analogues, (iii) expedient genetic systems for in vivo manipulation of genes governing the production of the target molecules in their native producers or heterologous hosts, and (iv) production of the natural products or their engineered analogues to levels that are appropriate for detection, isolation, and structural and biological characterization.5
In nature, however, the processes of evolution have generated and horizontally transferred variations of a few common biosynthetic machineries in a manner similar to combinatorial biosynthesis, but without all the knowledge and requirements mentioned above. Thus, amongst the vast variety of natural products produced by Actinomycetes it is frequently seen that a set of compounds exhibits the same structural core differing only in its “decorations”. The producing organisms of these compounds, however, are not necessarily closely related. The 9-membered enediynes maduropeptin, neocarzinostatin, and C-1027, for example, are produced by organisms as different as Actinomadura madurae, Streptomyces carzinostaticus, and Streptomyces globisporus, respectively,8–10 and the native producers of the glycopeptide derived antibiotics tallysomycin (TLM), bleomycin (BLM), and zorbamycin (ZBM), are represented by Streptoalloteichus hindustanus E465-94 ATCC31158, Streptomyces verticillus ATCC15003, and Streptomyces flavoviridis ATCC21892, respectively,11–13 while the native producers of the aminocoumarin antibiotics novobiocin, clorobiocin, and coumermycin A1 are all members of the Streptomyces genus (Streptomyces caeruleus, Streptomyces roseochromogenes, and Streptomyces rishiriensis, respectively).14–16 The biosynthetic gene clusters for all of these compounds have previously been cloned and their analysis revealed significant similarities among the clusters of each family.8–16 These observations lead one to ask three questions: (i) how close/similar can related biosynthetic gene clusters be, yet still make a different compound; (ii) how distant/different can biosynthetic gene clusters be, yet still make the same structural entity; and (iii) what evidence can be found as to whether nature evolved these clusters by “adopting” combinatorial biosynthetic strategies to generate structural diversity?
In this report we focus on the comparative analysis of the biosynthetic gene clusters for the three structurally related glycopeptide antitumor antibiotics BLM, ZBM, and TLM, thereby shedding insight into how the above questions might best be answered.
Similar and still different – formation of a bithiazole vs. a thiazolinyl-thiazole moiety
One characteristic difference between the structures of BLM/TLM and ZBM is the presence of a bithiazole unit in BLM and TLM and a thiazolinyl-thiazole moiety in ZBM (Figure 1). Thiazolinyl moieties in nonribosomal peptides are typically formed via cyclization of cysteine by cyclization (Cy) domains17 and may subsequently be oxidized to thiazole rings by an oxidation (Ox) domain.17–20 The BLM,12 TLM,11 and ZBM13 biosynthetic gene clusters, however, do not show any differences regarding their domain organization in the respective nonribosomal peptide synthetase (NRPS) modules: (i) only one of the two adenylation (A) domains in NRPS-1 and -0 modules is functional and the single A domain is predicted to load cysteine to the peptidyl carrier protein (PCP) of both NRPS-1 and -0 modules; (ii) the two Cy domains presumably are responsible for the cyclization of two cysteine moieties; and (iii) only one Ox domain can be found in NRPS-1 and -0 modules (Figure 2A). On the structural level, this should account for the presence of a thiazolinyl-thiazole unit in all three molecules. However, this is true only for ZBM, whereas BLM and TLM contain bithiazole moieties, the formation of which typically requires the presence of a second Ox domain. No such additional Ox domain was identified within the BLM/TLM biosynthetic machinery. Although all three gene clusters look alike, they form two chemically distinct units.
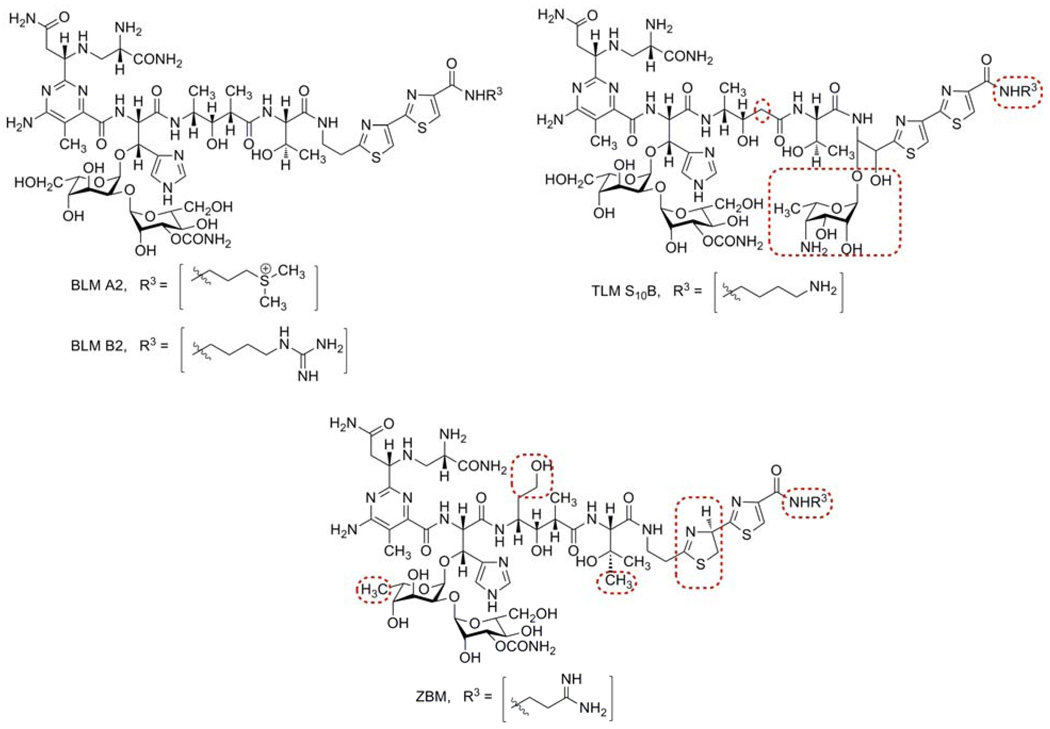
Figure 1.
Structures of selected members of the bleomycin (BLM) family of antitumor antibiotics: BLM A2 and B2, tallysomycin (TLM) S10B, and zorbamycin (ZBM). Structural differences between BLMs and other members of this family are highlighted by boxes.
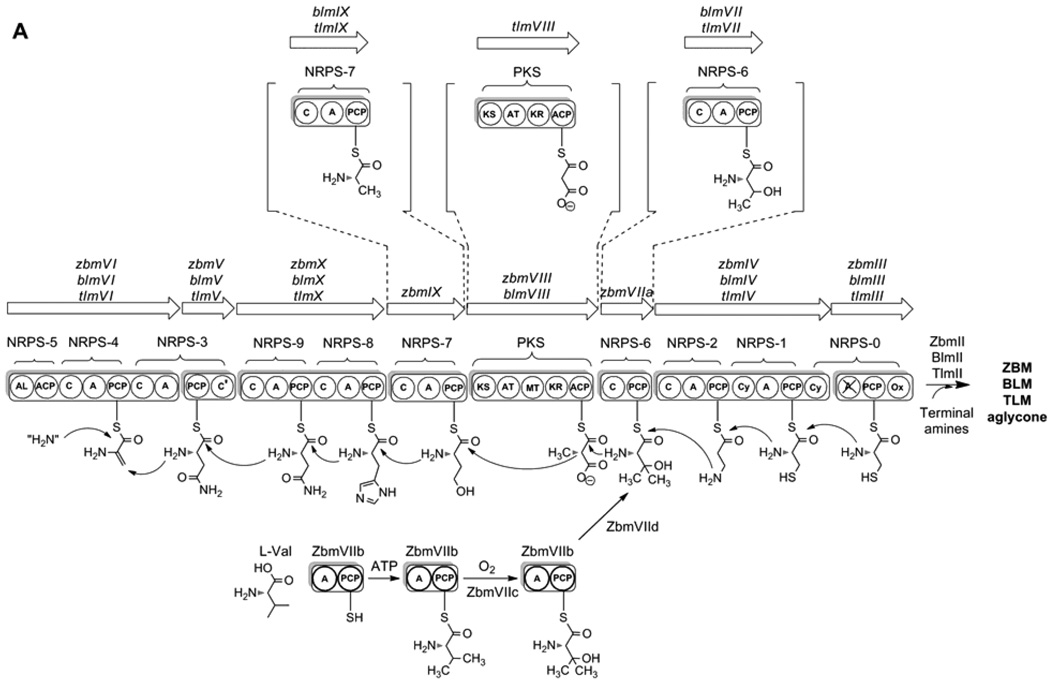
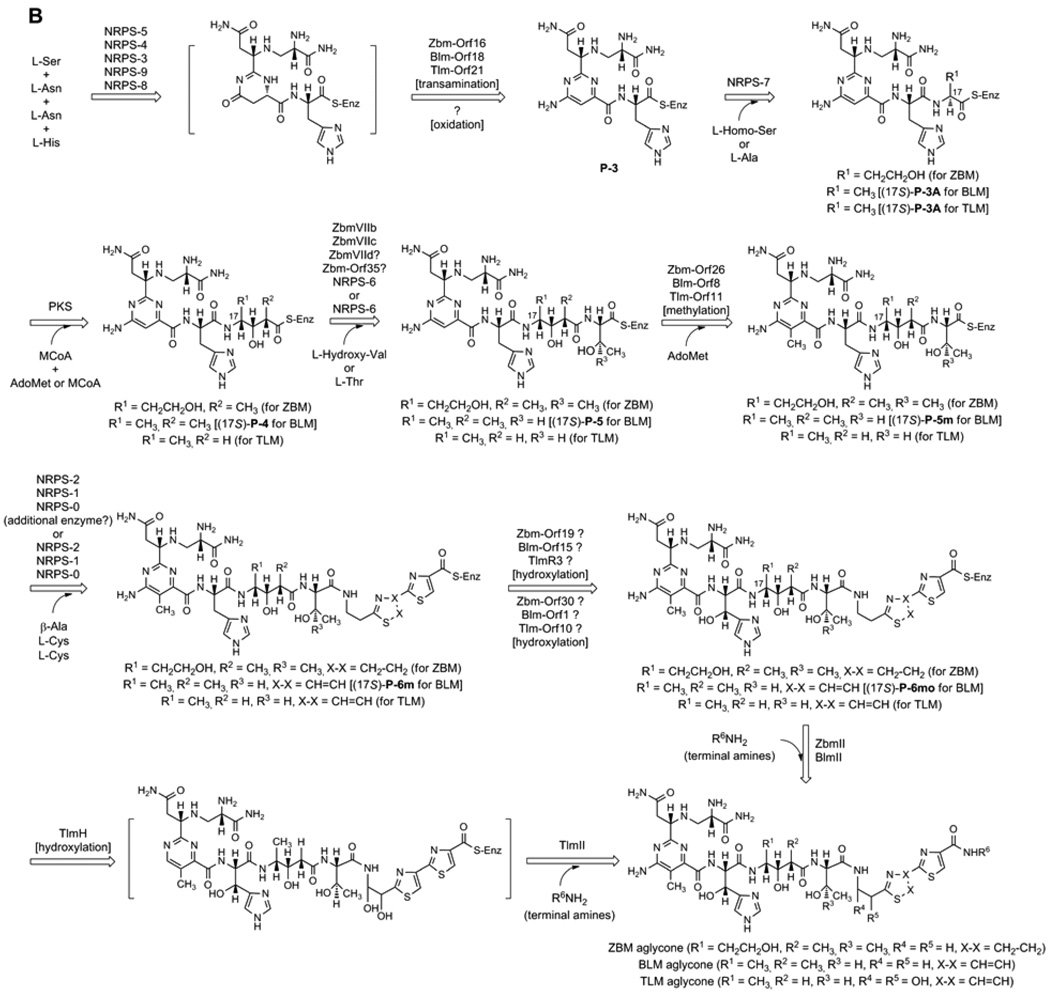
Figure 2.
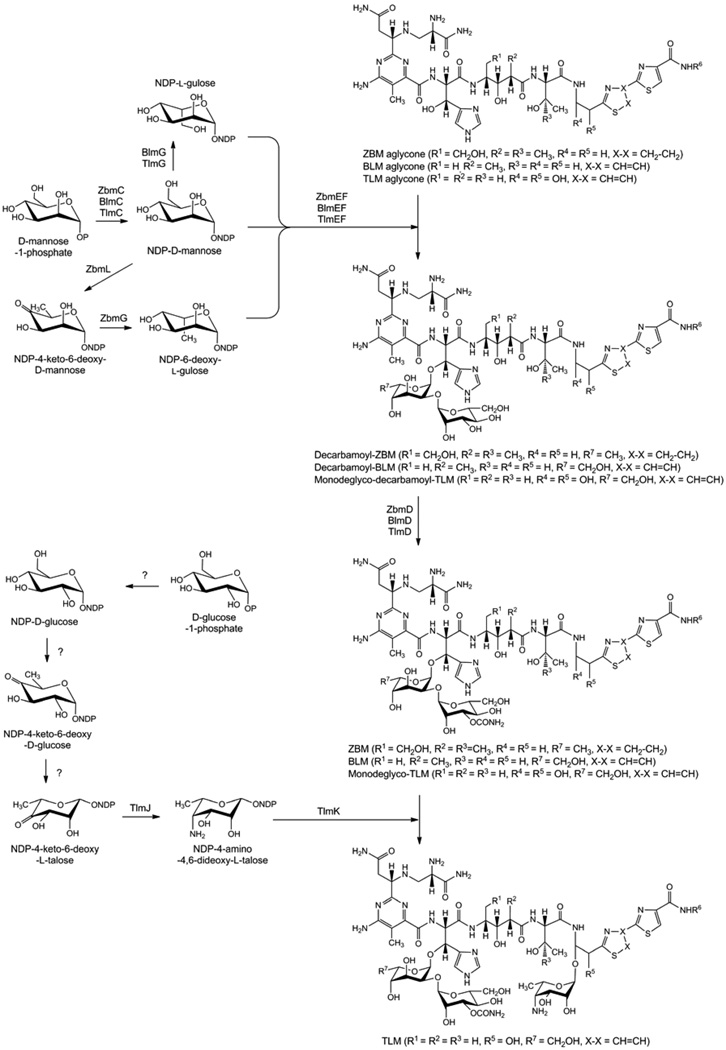
(A) A linear model for the BLM, TLM, and ZBM hybrid NRPS-PKS templated assembly of the BLM, TLM, and ZBM aglycones from nine amino acids and one acetate. Abbreviations for NRPS and PKS domains are: A, adenylation; ACP, acyl carrier protein; AL, acyl CoA ligase; AT, acyltransferase; C and C’, condensation; Cy, cyclization; KR, ketoreductase; KS, ketosynthase; MT, methyltransferase; Ox, oxidation; PCP, peptidyl carrier protein. (B) Proposed pathway for BLM, TLM, and ZBM aglycone biosynthesis. [?] indicates a step whose enzyme activity could not be identified within the sequenced BLM, TLM, and ZBM clusters. While all intermediates for TLM and ZBM biosynthesis are hypothetical, the analogous compounds, except the ones in brackets, have been identified in BLM biosynthesis from S. verticillus fermentation as the corresponding free acids.12
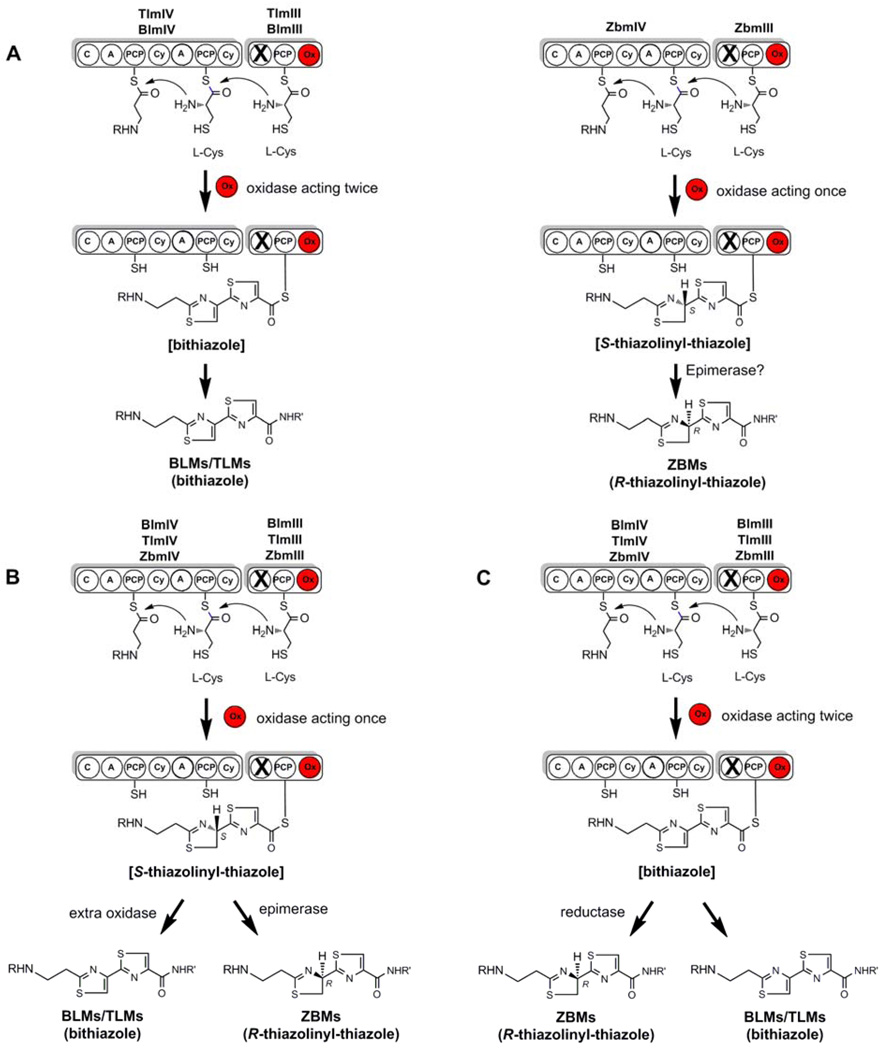
Three scenarios can be envisioned for the formation of two different structural features by the these clusters (Figure 3). First, a non-obvious Ox domain difference between the BLM/TLM and ZBM NRPS modules may have evolved, turning the BLM/TLM Ox domain into a twice-acting domain, while the ZBM Ox domain remained a single-acting domain. This scenario would either require the unlikely activation of one D- and one L-cysteine by the A domain of ZbmIV (NRPS-1) or the presence of an epimerase gene in the ZBM cluster to account for the formation of the R-thiazolinyl-thiazole in ZBM (Figure 3A). Evidence for neither can be found in the ZBM biosynthetic gene cluster and no significant differences on the amino acid level were discovered between the BLM/TLM and ZBM Ox domain. On the contrary, biochemical characterization of the BLM NRPS-1 and -0 modules has confirmed that the single A domain in the NRPS-1 module loads L-cysteine to both PCPs.18 Alternatively, the single Ox domain may, as expected, form just one thiazole moiety in all three biosynthetic pathways, and the Ox domain activity for the BLM NRPS-0 has been experimentally confirmed.19,20 This scenario would require (i) the presence of an extra oxidase in BLM/TLM biosynthesis to subsequently convert the remaining thiazoline moiety into a thiazole ring and (ii) either the activation of one D- and one L-cysteine by the A domain of ZbmIV (NRPS-1) or the presence of an epimerase gene in the ZBM cluster to account for the formation of the R-thiazolinyl-thiazole in ZBM (see above) (Figure 3B). No such additional oxidase has been identified yet in either the BLM or the TLM biosynthetic gene cluster. Finally, the single Ox domain could represent a twice-acting Ox domain and be responsible for the formation of a bithiazole ring in all three molecules. This would require the presence of a reductase exclusively in the ZBM cluster in order to subsequently reduce one thiazole back to a thiazoline ring (Figure 3C). One candidate for such a reduction in ZBM biosynthesis is Zbm-Orf2 with similarity to a putative dehydrogenase. As previously reported, replacement of the zbm-orf2 gene by the apramycin resistance gene aac(3)IV resulted in the complete abolishment of ZBM production.13 However, no accumulation of the expected fully oxidized bithiazole-ZBM intermediate was observed. Despite the fact that zbm-orf1, zbm-orf2, and zbm-orf3 appear to be translationally coupled, ZBM production was successfully restored to ~50% of wild-type level by the introduction of both a complementation construct containing zbm-orfs1–3 and a complementation construct harbouring exclusively zbm-orf2 (Supporting Information). This result indicates that zbm-orf2 is essential for ZBM production, however, it does not provide conclusive data as to whether Zbm-Orf2 indeed represents the expected thiazole reductase, supporting the proposed pathway depicted in Figure 3C. While unlikely, our current studies also cannot rule out the presence of promiscuous epimerases, reductases, or oxidases residing outside the sequenced BLM, TLM and ZBM gene clusters that could be recruited for their biosynthesis.
Figure 3.
Schematic representation of the three potential scenarios for bithiazole vs. thiazolinyl-thiazole formation in the BLM/TLM and ZBM biosynthetic pathways, respectively. (A) Non-visible difference between twice-acting Ox domain in BlmIII/TlmIII and single-acting Ox domain in ZbmIII with additional epimerase accounting for change in configuration. (B) Single-acting Ox domain in BlmIII/TlmIII and ZbmIII with an additional oxidase in BLM/TLM biosynthesis and an additional epimerase in ZBM biosynthesis. (C) Twice-acting Ox domain in BlmIII/TlmIII and ZbmIII with additional reductase in ZBM biosynthesis.
Different and still similar – freestanding condensation (C) domains
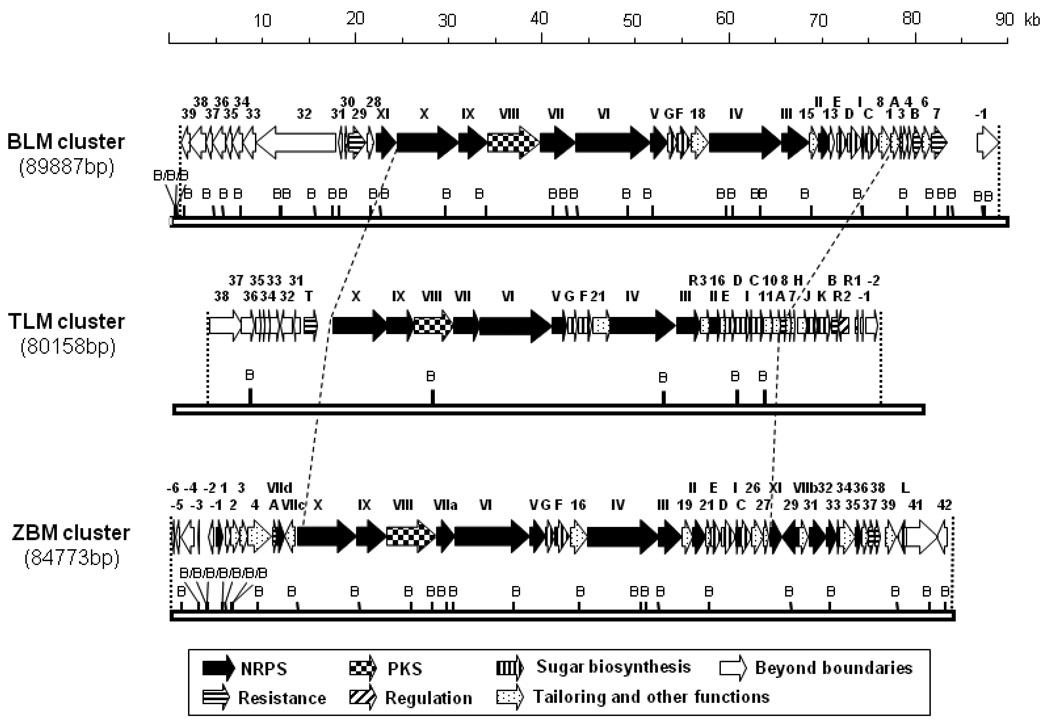
In addition to the expected genes encoding the NRPS-polyketide synthase (PKS) enzymatic machinery accountable for the formation of the BLM, TLM, and ZBM hybrid peptide-polyketide backbones, several genes encoding freestanding C domains (blmII, blmXI, tlmII, zbmII, zbmXI, zbm-orf31) were identified in the BLM, TLM, and ZBM biosynthetic gene clusters (Figure 4). For some of these genes (blmII, tlmII, zbmII) direct counterparts were found in the related clusters, while others were present in only one (zbm-orf31) or two (blmXI, zbmXI) of the three clusters. What are the functions of these six freestanding C domains in BLM, TLM, and ZBM biosynthesis?
Figure 4.
Comparison of the organization of the BLM, TLM, and ZBM biosynthetic gene clusters. Proposed functions for individual ORFs have been reported previously.11–13
BlmII and its counterparts TlmII and ZbmII were hypothesized to play a role in amide bond formation between the respective aglycone and terminal amines, especially since all three clusters lack a TE domain typically responsible for aglycone release from the NRPS machinery (Figure 2). The conserved motif (HHXXXDG) typically found in intact C domains21 is altered to HXXXXDX in BlmII (HTLLLDT), TlmII (HQMLLDA), and ZbmII (HFLVADL) (Table 1). This may account for the amine substrates in the proposed pathways differing somewhat from the typical amino acid substrate of classical C domains (Figure 2B). To support this proposal, zbmII was inactivated by in-frame deletion. The resulting mutant strain, however, did not accumulate the ZBM aglycone, but instead showed complete abolishment of ZBM production (Supporting Information). This indicates that ZbmII is indeed required for ZBM biosynthesis, which, in a functional analogy, is most likely also true for its homologs BlmII and TlmII. However, whether these proteins truly catalyze the attachment of the terminal amine still remains obscure.
Table 1.
Conserved C and Cy domain motifs identified in the BLM, TLM, and ZBM biosynthetic gene clusters.
| Conserved C domain motif |
H | H | Xa | X | X | D | G | ||
|---|---|---|---|---|---|---|---|---|---|
| BlmII | H | T | L | L | L | D | T | ||
| TlmII | H | Q | M | L | L | D | A | ||
| ZbmII | H | F | L | V | A | D | L | ||
| BlmIV (NRPS-2) | H | H | A | V | T | D | G | ||
| TlmIV (NRPS-2) | H | H | I | A | I | D | G | ||
| ZbmIV (NRPS-2) | H | H | A | V | T | D | G | ||
| BlmV (NRPS-3, C’) | H | H | L | V | L | D | G | ||
| TlmV (NRPS-3, C’) | H | H | L | I | L | D | G | ||
| ZbmV (NRPS-3, C’) | H | H | L | I | L | D | G | ||
| BlmVI (NRPS-3) | S | S | F | A | L | D | G | ||
| TlmVI (NRPS-3) | S | S | F | G | L | D | G | ||
| ZbmVI (NRPS-3) | S | S | F | G | L | D | G | ||
| BlmVI (NRPS-4) | H | H | L | V | A | D | F | ||
| TlmVI (NRPS-4) | H | H | L | L | A | D | F | ||
| ZbmVI (NRPS-4) | H | H | L | V | A | D | Y | ||
| BlmVII (NRPS-6) | H | H | I | A | S | D | G | ||
| TlmVII (NRPS-6) | H | H | I | A | S | D | G | ||
| ZbmVII (NRPS-6) | H | H | I | A | G | D | G | ||
| BlmIX (NRPS-7) | H | H | I | V | F | D | G | ||
| TlmIX (NRPS-7) | H | H | I | V | F | D | G | ||
| ZbmIX (NRPS-7) | H | H | I | V | F | D | G | ||
| BlmX (NRPS-8) | H | H | E | I | V | D | G | ||
| TlmX (NRPS-8) | H | H | E | I | V | D | G | ||
| ZbmX (NRPS-8) | H | H | E | I | V | D | G | ||
| BlmX (NRPS-9) | H | A | L | V | A | D | R | ||
| TlmX (NRPS-9) | H | A | L | V | G | D | R | ||
| ZbmX (NRPS-9) | S | V | L | A | A | D | R | ||
| BlmXI | P | H | I | T | A | D | L | ||
| ZbmXI | H | H | V | A | V | D | L | ||
| Zbm-Orf31 | H | H | C | I | V | D | L | ||
| Conserved Cy-domain motif | D | Xa | X | X | X | D | X | X | S |
| BlmIV (NRPS-0) | D | L | L | I | A | D | A | H | S |
| TlmIV (NRPS-0) | D | L | L | I | A | D | A | H | S |
| ZbmIV (NRPS-0) | D | L | L | I | A | D | A | H | S |
| BlmIV (NRPS-1) | D | A | L | I | C | D | A | H | S |
| TlmIV (NRPS-1) | D | A | L | I | C | D | A | Y | S |
| ZbmIV (NRPS-1) | D | S | L | V | C | D | A | H | S |
X indicates a variable amino acid within the determined code.
In contrast, the C domain proteins encoded by blmXI and zbmXI were thought to be dispensable for BLM and ZBM biosynthesis, respectively. No biosynthetic function could be envisioned for BlmXI and ZbmXI in their respective biosynthetic pathways. Moreover, the TLM cluster lacks a gene encoding a counterpart for these proteins. Surprisingly, inactivation of blmXI by gene replacement abolished BLM production, and a ZBM non-producing phenotype was also obtained from in-frame deletion of zbmXI (Supporting Information). Wild-type level ZBM production was restored by the introduction of a zbmXI complementation construct. The ZBM-producing phenotype, however, could not be restored by the introduction of the corresponding cross-complementation construct containing blmXI from the BLM biosynthetic gene cluster. Although both BlmXI and ZbmXI were shown to be essential for BLM and ZBM biosynthesis, respectively, BlmXI was apparently not similar enough to ZbmXI to cross-complement for ZbmXI functionality. The precise function of BlmXI and ZbmXI within the BLM and ZBM biosynthetic pathways could not be deduced from these data, and the question of why the TLM cluster lacks an equivalent protein although it is required for BLM and ZBM biosynthesis, remains open. It may be speculated that BlmXI and ZbmXI are needed to complement an inactive C domain within the regular NRPS machinery of the respective pathway in trans, while the TLM counterpart of this NRPS embedded C domain is intact, making in trans complementation superfluous.
To identify such potential differences, the conserved motifs of NRPS embedded and freestanding C and Cy domains of all three clusters were compared (Table 1). The C domains of NRPS-2, NRPS-3 (C’), NRPS-6, NRPS-7, and NRPS-8 of all three clusters appear to contain the intact HHXXXDG motif indicating that they are fully functional as regular C domains catalyzing amino acid condensation.21 The NRPS-0 and NRPS-1 conserved motifs of all three clusters deviate from the HHXXXDG motif, and instead exhibit a DXXXXDXXS motif characteristic for Cy domains responsible for the cyclization of cysteine.22 The NRPS-3 C domain motif is replaced by an SSXXXDG motif in all three clusters indicating that it might not be functional as a regular C domain motif but instead catalyzes a conjugated addition, as reflected by the proposed BLM, TLM, and ZBM biosynthetic pathways (Figure 2A) and in agreement with earlier reports.12 The conserved C domain motif of NRPS-4 deviates from the classical HHXXXDG by possessing an F, F, and Y instead of a G at the last position in the respective BLM, TLM, and ZBM enzymes (Table 1). The NRPS-4 C domain is located in the starter module and may either be non-functional or involved in dehydroalanine formation or aminolysis reactions.12 The NRPS-9 C domains appear to be inactive for regular transpeptidation since they exhibit HALVADR, HALVGDR, and SVLAADR motifs instead of the HHXXXDG motif in the BLM, TLM, and ZBM clusters, respectively. This may account for a different reaction, the cyclization required for pyrimidine ring formation, being catalyzed by these domains, which is in contrast to previous reports assuming that the NRPS-9 represents a regular C domain and the NRPS-3 (C’) domain coordinates the cyclization reaction.12 As discussed above, BlmII, TlmII, and ZbmII are thought to be involved in attachment of the different terminal amines to the BLM, TLM, and ZBM aglycones, respectively. The BlmXI (PHITADL) and ZbmXI (HHVAVDL) C domain motifs both differ from the classical HHXXXDG motif and would therefore be predicted to be inactive or at least dispensible for biosynthesis of BLM and ZBM, respectively. However, because both were found to be required for biosynthesis of their respective molecules and because none of the C domains discussed above was found to obviously be active in the TLM, but inactive in the BLM and ZBM enzymatic machinery, the function of BlmXI and ZbmXI remains obscure.
In-frame deletion of zbm-orf31 encoding the only C domain present exclusively in the ZBM cluster reduced but failed to abolish ZBM biosynthesis (Supporting Information). These data are inconclusive and will need further investigation. The conserved C domain motif of Zbm-Orf31 (HHCIVDL) only slightly deviates from the classical motif and should therefore still be functional (Table 1).
Combinatorial biosynthesis in the lab and in nature
Variations in sugar biosynthesis apparently dictate the glycosylation patterns distinct for each of the BLM, TLM, and ZBM glycopeptide scaffolds (Figure 5), and a des-talose TLM analogue has been previously generated by manipulating the TLM biosynthetic machinery.23,24 In order to generate novel ZBM derivatives by combinatorial biosynthesis, redirection of ZBM disaccharide biosynthesis may represent an option.
Figure 5.
Proposed pathways for BLM, TLM, and ZBM sugar biosynthesis and attachment to the respective aglycones.11–13
One difference between ZBM and BLM/TLM biosynthesis is the incorporation of NDP-6-deoxy-l-gulose instead of NDP-l-gulose into the disaccharide moieties of the respective molecules. Only one additional enzymatic step, the dehydration of NDP-d-mannose to NDP-4-keto-6-deoxy-d-mannose, is expected to distinguish between the pathways for NDP-6-deoxy-l-gulose formation in ZBM and NDP-l-gulose formation in BLM and TLM biosynthesis (Figure 5). Analysis of the ZBM biosynthetic gene cluster suggested that this reaction in ZBM biosynthesis would be catalyzed by a GDP-mannose-4,6-dehydratase encoded by zbmL (Figure 4). Replacement of zbmL by an apramycin resistance cassette, however, completely abolished ZBM production and did not result in the expected accumulation of ZBM aglycone.13 Introduction of the integrative zbmL complementation construct, pBS9019, into the ΔzbmL mutant strain SB9003 restored ZBM production to ~70% of previous production levels.13
The ΔzbmL mutant strain SB9003 is thought to accumulate NDP-d-mannose instead of the NDP-4-keto-6-deoxy-d-mannose produced by its parent strain. The ZbmG catalyzed epimerization step in ZBM biosynthesis is expected to require NDP-4-keto-6-deoxy-d-mannose as substrate (Figure 4 and Figure 5), and the transfer of any non-native mono- or disaccharide to the ZBM aglycone by the glycosyltransferases ZbmE or ZbmF or both may be significantly impaired. In contrast, the epimerase BlmG/TlmG is predicted to convert NDP-d-mannose into NDP-l-gulose in BLM/TLM biosynthesis, and the corresponding glycosyltransferases BlmE/TlmE and BlmF/TlmF are expected to be capable of transferring the resulting mono- or disaccharide to a very similar aglycone. Therefore, cross-complementation of the ΔzbmL mutant strain with constructs containing combinations of the epimerase gene blmG and either one or both of the predicted glycosyltransferase genes, blmE and blmF, from the BLM biosynthetic gene cluster may represent a viable option for the generation of a new ZBM analogue carrying the BLM disaccharide.
How does nature “adopt” combinatorial biosynthesis strategies to create natural product structural diversity? We can speculate by comparing and contrasting biosynthetic machineries that make similar but distinct natural products. In addition to sugar biosynthesis discussed above, comparison of the BLM, TLM, and ZBM hybrid peptide-polyketide backbones revealed that two amino acids incorporated into the ZBM backbone differ from the BLM and TLM scaffolds – the two amino acids flanking the polyketide unit of the backbone are l-alanine and l-threonine in both, BLM and TLM, but l-homoserine and l-OH-valine in ZBM. All of the other amino acids incorporated into the peptide backbone of BLM, TLM, and ZBM are identical in the three molecules (Figure 1). The A domains of the respective NRPS modules show the expected amino acid substrate specificities (Table 2). In the case of l-homoserine, which is suggested to be incorporated by ZbmIX, nature appears to have evolved a different substrate specificity of the A domain thereby accounting for structural diversity. The A domains of BlmIX and TlmIX are predicted to activate l-alanine according to their signature motifs. In contrast, the ZbmIX A domain signature motif shows some degree of similarity to a d-lysergic acid and an l-homoserine activating A domain and therefore is clearly different from its corresponding A domains in the BLM and TLM clusters (Table 2). Whether the ZbmIX A domain is indeed responsible for l-homoserine incorporation remains to be confirmed experimentally.
Table 2.
Predictions of substrate specificity of BLM, TLM, and ZBM NRPSs based on the specificity-conferring codes of A domains (shown in bold).32–34
| Domain | 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | 517 | Similarity (%)a |
|---|---|---|---|---|---|---|---|---|---|---|---|
| L-Cys(2) | D | L | Y | N | L | S | L | I | W | K | |
| BlmIII (NRPS-0) | P | L | Y | H | L | G | L | P | W | R | 60 |
| TlmIII (NRPS-0) | G | F | Y | H | L | G | L | L | W | R | 60 |
| ZbmIII (NRPS-0) | E | R | Y | S | A | S | L | I | W | R | 70 |
| BlmIV (NRPS-1) | D | L | Y | N | L | S | L | I | W | K | 100 |
| TlmIV (NRPS-1) | D | L | Y | N | M | S | L | I | W | K | 100 |
| ZbmIV (NRPS-1) | D | L | Y | N | L | S | L | I | W | K | 100 |
| β-Ala | V | D | Xb | V | I | S | Xb | G | D | K | |
| BlmIV (NRPS-2) | V | D | W | V | I | S | L | A | D | K | 80 |
| TlmIV (NRPS-2) | V | D | W | V | V | S | L | A | D | K | 80 |
| ZbmIV (NRPS-2) | V | D | A | L | V | S | L | A | D | K | 80 |
| L-Asn | D | L | T | K | L | G | E | V | G | K | |
| BlmVI (NRPS-3) | D | L | T | K | V | G | E | V | G | K | 100 |
| TlmVI (NRPS-3) | D | L | T | K | V | G | E | V | G | K | 100 |
| ZbmVI (NRPS-3) | D | L | T | K | V | G | E | V | G | K | 100 |
| BlmX (NRPS-9) | D | L | T | K | V | G | E | V | G | K | 100 |
| TlmX (NRPS-9) | D | L | T | K | V | G | E | V | G | K | 100 |
| ZbmX (NRPS-9) | D | F | T | K | V | G | E | V | G | K | 90 |
| L-Ser | D | V | W | H | L | S | L | I | D | K | |
| BlmVI (NRPS-4) | D | V | W | H | V | S | L | V | D | K | 100 |
| TlmVI (NRPS-4) | D | V | W | H | V | S | L | V | D | K | 100 |
| ZbmVI (NRPS-4) | D | V | W | H | L | S | L | I | D | K | 100 |
| L-Thr | D | F | W | N | I | G | M | V | H | K | |
| BlmVII (NRPS-6) | D | F | W | S | V | G | M | I | H | K | 90 |
| TlmVII (NRPS-6) | D | F | W | G | V | G | M | V | H | K | 90 |
| ZbmVIIa (NRPS-6) | A-domain missing | - | |||||||||
| L-Val (1) | D | A | F | W | I | G | G | T | F | K | |
| ZbmVIIb (NRPS-6, A-T) | D | A | F | W | L | G | G | T | F | K | 100 |
| L-Ala | D | L | F | N | N | A | L | T | Y | K | |
| BlmIX (NRPS-7) | D | L | F | N | N | A | L | T | Y | K | 100 |
| TlmIX (NRPS-7) | D | L | F | N | N | A | L | T | Y | K | 100 |
| D-Lyserg | D | V | F | S | V | G | L | Y | M | K | |
| ZbmIX (NRPS-7) | D | V | F | S | N | G | L | T | H | K | 70 |
| ps2 (Q8J0L6, D-Lyserg) | D | V | F | S | V | G | L | Y | M | K | 100 |
| FUSS A (AAT28740, L-Homoser) | D | M | T | F | S | A | G | I | I | K | 60 |
| L-His | D | S | Xb | L | Xb | A | E | V | Xb | K | |
| BlmX (NRPS-8) | D | S | A | L | I | A | E | V | W | K | 70 |
| TlmX (NRPS-8) | D | S | A | L | V | A | E | V | W | K | 70 |
| ZbmX (NRPS-8) | D | S | V | L | T | A | E | V | W | K | 70 |
Similarity is calculated by AlignX in the Vector NTI Advance™ 10 program from Invitrogen.
X indicates a variable amino acid within the determined code. In similarity calculations, X is not recognized as arbitrary amino acid, hence similarity values appear to be lower than calculated manually (100% for ZbmIV (NRPS-2), BlmIV (NRPS-2), and TlmIV (NRPS-2) compared to the β-Ala code; 100% for ZbmX (NRPS-8), BlmX (NRPS-8), and TlmX (NRPS-8) compared to the L-His code).
In the case of l-OH-valine, the situation presents itself very differently. In analogy to the NRPS-6 module of the BLM and TLM cluster, which has been proposed to incorporate l-threonine, the NRPS-6 module of the ZBM cluster would be expected to incorporate l- valine or l-OH-valine. However, ZbmVIIa exhibits an intact C and PCP domain, but completely lacks the respective A domain for amino acid activation, while BlmVII and TlmVII both contain an A domain with the predicted l-threonine specificity (Table 1, Table 2, and Figure 2A). A freestanding incomplete NRPS module, ZbmVIIb, composed of an A domain with predicted l-valine specificity and a PCP domain is proposed to complement the incomplete C-PCP module of NRPS-6 (ZbmVIIa).13 In-frame deletion of the zbmVIIb gene completely abolished ZBM production indicating that it is indeed involved in ZBM biosynthesis. ZBM production was restored in the ΔzbmVIIb mutant strain SB9017 to ~50% of wild-type level upon introduction of the complementation construct pBS9068. Cross-complementation with the corresponding complete NRPS module from the BLM biosynthetic gene cluster, NRPS-6 (BlmVII), did not recover the ZBM producing phenotype (Supporting Information). Potential reasons for this insufficient match for complementation may be represented by either the lack of protein-protein interaction or by unsuccessful processing of the respective non-natural intermediate. Biosynthesis and incorporation of l-OH-valine have been suggested to be completed via hydroxylation carried out by ZbmVIIc with homology to a cytochrome P450 enzyme from S. tubercidicus (CypLB, acc. no. AAT45286) and ZbmVIId with similarity to an acyltransferase from S. tubercidicus (TeLB, acc. no. AAT45287).25,26 Inactivation of both zbmVIIc and zbmVIId13 were found to abolish ZBM biosynthesis, thereby verifying their involvement in ZBM formation.
Freestanding A domain-containing partial NRPS modules can be found in various other microorganisms (Nostoc punctiforme, Stigmatella aurantiaca, Salinispora tropica, Pseudomonas syringae, Lyngbya majuscula) and some of them were reported to act in trans for the production of NRPS formed natural products.27–29 The existence of partial C-PCP modules with the PCP domain directly linked to the C domain is by far less common and only one module of the syringomycin NRPS30 resulted in full length alignment to ZbmVIIa. In syringomycin biosynthesis, a similar complementation mechanism for the missing A domain involving a transfer reaction catalyzed by an acyltransferase has been proposed as in ZBM biosynthesis.31 The replacement of an intact NRPS module such as BlmVII/TlmVII by a set of incomplete modules (ZbmVIIa and ZbmVIIb) accompanied by modifying (ZbmVIIc) and transferring (ZbmVIId) enzymes seems to be another strategy nature adopted to create structural diversity in a fashion very similar to combinatorial biosynthesis. One may speculate that a full NRPS module was evolved to lose an A domain while the incomplete A-PCP module, cytochrome P450, and acyltransferase encoding genes were simultaneously acquired from other microorganisms to imbue the NRPS biosynthetic machinery with the desired amino acid substitution.
Conclusion
This report compares and contrasts various aspects of three biosynthetic gene clusters for three structurally related natural products, BLM, TLM, and ZBM. In some respects, such as for the bithiazole (BLM, TLM) vs. thiazolinyl-thiazole (ZBM) formation, all three clusters look very similar yet are responsible for the formation of chemically different structures. This is also true for the genes zbmGFE and blmGFE encoding enzymes supposedly involved in NDP-6-deoxy-l-gulose (ZBM biosynthesis) and NDP-l-gulose (BLM biosynthesis) formation, respectively. Upon inactivation of the GDP-mannose-4,6-dehydratase gene zbmL, however, the remaining sugar biosynthetic enzymes, ZbmGFE, were not able to catalyze either NDP-l-gulose formation or attachment of the non-native substrate, NDP-d-mannose, to the aglycone. It remains to be seen whether BlmGFE will be able to catalyze such biosynthetic steps and prove useful for the generation of new ZBM analogues.
In contrast to these similarities, all three clusters also exhibit significant differences while forming very similar structural features. The BLM, TLM, and ZBM biosynthetic gene clusters all encode a different number of freestanding NRPS C domains which cannot be explained by any structural differences demanding more than one freestanding C domain, yet BlmXI and ZbmXI which do not have a counterpart in the TLM cluster were both proven to be required for BLM and ZBM formation, respectively. One further small difference on the structural level, that would have been expected to result in a simple change in amino acid specificity of an A domain plus the addition of a cytochrome P450 for l-valine hydroxylation, was apparently achieved by a much larger modification of the enzymatic machinery – one A domain was completely removed and replaced in trans by an A-PCP didomain plus acyltransferase plus cytochrome P450.
These examples once again demonstrate the flexibility of nature to achieve structural differences and similarities via various mechanisms and will surely inspire laboratory efforts to generate natural product structural diversity by combinatorial biosynthesis strategies.
Supplementary Material
Acknowledgement
We thank the Analytical Instrumentation Center of the School of Pharmacy, UW-Madison, for support in obtaining LC-MS data and the John Innes Center, Norwich, UK, for providing the REDIRECT Technology kit. This work was supported in part by the NIH grant CA94426. U.G. is a Postdoctoral Fellow of the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Dedicated to Dr. Koji Nakanishi of Columbia University for his pioneering work on bioactive natural products.
Supporting Information Available. Supporting Information includes experimental observation in support of this review, Tables S1, S2, S3 for plasmids, bacterial strains, and oligonucleotides, respectively, used in this study and Figure S1 and S2 for schematic presentations of gene inactivation carried out in the BLM and ZBM biosynthetic gene clusters, isolation and confirmation of the resultant mutant strains, and HPLC analysis of their phenotypes. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Newman DJ, Cragg GM, Snader KM. J. Nat. Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 2.Strohl WR. Metab. Eng. 2001;3:4–14. doi: 10.1006/mben.2000.0172. [DOI] [PubMed] [Google Scholar]
- 3.Cane DE, Walsh CT, Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 4.Walsh CT. ChemBioChem. 2002;3:125–134. doi: 10.1002/1439-7633(20020301)3:2/3<124::AID-CBIC124>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 5.Thomas MG, Bixby KA, Shen B. In: Antitumor Agents from Natural Products. Kingston DGI, Cragg GM, Newman DJ, editors. Boca Raton, Florida: CRC Press; 2005. pp. 519–551. [Google Scholar]
- 6.Shen B. Sci. STKE. 2004;2004:e14. doi: 10.1126/stke.2252004pe14. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez C, Zhu L, Brana AF, Salas AP, Rohr J, Mendez C, Salas JA. Proc. Natl. Acad. Sci. U.S.A. 2005;102:461–466. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Lanen SG, Oh TJ, Liu W, Wendt-Pienkowski E, Shen B. J. Am. Chem Soc. 2007;129:13082–13094. doi: 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, Nonaka K, Nie L, Zhang J, Christenson SD, Bae J, Van Lanen SG, Zazopoulos E, Farnet CM, Yang CF, Shen B. Chem. Biol. 2005;12:293–302. doi: 10.1016/j.chembiol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 10.Liu W, Christenson SD, Standage S, Shen B. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 11.Tao M, Wang L, Wendt-Pienkowski E, George NP, Galm U, Zhang G, Coughlin JM, Shen B. Mol. Biosyst. 2007;3:60–74. doi: 10.1039/b615284h. [DOI] [PubMed] [Google Scholar]
- 12.Du L, Sanchez C, Chen M, Edwards DJ, Shen B. Chem. Biol. 2000;7:623–642. doi: 10.1016/s1074-5521(00)00011-9. [DOI] [PubMed] [Google Scholar]
- 13.Galm U, Wendt-Pienkowski E, Wang L, George NP, Oh TJ, Yi F, Tao M, Coughlin JM, Shen B. Mol. Biosyst. 2009;5:77–90. doi: 10.1039/b814075h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steffensky M, Mühlenweg A, Wang ZX, Li SM, Heide L. Antimicrob. Agents Chemother. 2000;44:1214–1222. doi: 10.1128/aac.44.5.1214-1222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pojer F, Li SM, Heide L. Microbiology. 2002;148:3901–3911. doi: 10.1099/00221287-148-12-3901. [DOI] [PubMed] [Google Scholar]
- 16.Wang ZX, Li SM, Heide L. Antimicrob. Agents Chemother. 2000;44:3040–3048. doi: 10.1128/aac.44.11.3040-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh CT, Chen H, Keating TA, Hubbard BK, Losey HC, Luo L, Marshall CG, Miller DA, Patel HM. Curr. Opin. Chem. Biol. 2001;5:525–534. doi: 10.1016/s1367-5931(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 18.Du L, Chen M, Zhang Y, Shen B. Biochemistry. 2003;42:9731–9740. doi: 10.1021/bi034817r. [DOI] [PubMed] [Google Scholar]
- 19.Du L, Sanchez C, Chen M, Shen B. FEMS Microbiol. Lett. 2000;189:171–175. doi: 10.1111/j.1574-6968.2000.tb09225.x. [DOI] [PubMed] [Google Scholar]
- 20.Schneider TL, Shen B, Walsh CT. Biochemistry. 2003;42:9722–9730. doi: 10.1021/bi034792w. [DOI] [PubMed] [Google Scholar]
- 21.Bergendahl V, Linne U, Marahiel MA. Eur. J. Biochem. 2002;269:620–629. doi: 10.1046/j.0014-2956.2001.02691.x. [DOI] [PubMed] [Google Scholar]
- 22.Kelly WL, Hillson NJ, Walsh CT. Biochemistry. 2005;44:13385–13393. doi: 10.1021/bi051124x. [DOI] [PubMed] [Google Scholar]
- 23.Tao M, Wang L, Wendt-Pienkowski E, Zhang N, Yang D, Galm U, Coughlin JM, Xu Z, Shen B. Mol. BioSyst. 2010;6:349–356. doi: 10.1039/b918106g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang N, Zhu X, Yang D, Cai J, Tao M, Wang L, Duan Y, Shen B, Xu Z. Appl. Microbiol. Biotechnol. 2010;86:1345–1353. doi: 10.1007/s00253-009-2406-9. [DOI] [PubMed] [Google Scholar]
- 25.Jungmann V, Molnar I, Hammer PE, Hill DS, Zirkle R, Buckel TG, Buckel D, Ligon JM, Pachlatko JP. Appl. Environ. Microbiol. 2005;71:6968–6976. doi: 10.1128/AEM.71.11.6968-6976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molnar I, Hill DS, Zirkle R, Hammer PE, Gross F, Buckel TG, Jungmann V, Pachlatko JP, Ligon JM. Appl. Environ. Microbiol. 2005;71:6977–6985. doi: 10.1128/AEM.71.11.6977-6985.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaillancourt FH, Yin J, Walsh CT. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10111–10116. doi: 10.1073/pnas.0504412102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly WL, Li MT, Yeh E, Vosburg DA, Galoni Cacute DP, Kelleher NL, Walsh CT. Biochemistry. 2007;46:359–368. doi: 10.1021/bi061930j. [DOI] [PubMed] [Google Scholar]
- 29.Chang Z, Flatt P, Gerwick WH, Nguyen VA, Willis CL, Sherman DH. Gene. 2002;296:235–247. doi: 10.1016/s0378-1119(02)00860-0. [DOI] [PubMed] [Google Scholar]
- 30.Guenzi E, Galli G, Grgurina I, Gross DC, Grandi G. J. Biol. Chem. 1998;273:32857–32863. doi: 10.1074/jbc.273.49.32857. [DOI] [PubMed] [Google Scholar]
- 31.Singh GM, Vaillancourt FH, Yin J, Walsh CT. Chem. Biol. 2007;14:31–40. doi: 10.1016/j.chembiol.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Stachelhaus T, Dootz HD, Marahiel MA. Chem. Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 33.Challis GL, Ravel J, Townsend CA. Chem. Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 34.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson D. Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.