Abstract
Phylogenetic analyses of 16S rRNA support close relationships between the Gammaproteobacteria Sodalis glossinidius, a tsetse (Diptera: Glossinidae) symbiont, and bacteria infecting diverse insect orders. To further examine the evolutionary relationships of these Sodalis-like symbionts, phylogenetic trees were constructed for a subset of putative surface-encoding genes (i.e. ompA, spr, slyB, rcsF, ycfM, and ompC). The ompA and ompC loci were used towards examining the intra- and interspecific diversity of Sodalis within tsetse, respectively. Intraspecific analyses of ompA support elevated nonsynonymous (dN) polymorphism with an excess of singletons, indicating diversifying selection, specifically within the tsetse Glossina morsitans. Additionally, interspecific ompC comparisons between Sodalis and Escherichia coli demonstrate deviation from neutrality, with higher fixed dN observed at sites associated with extracellular loops. Surface-encoding genes varied in their phylogenetic resolution of Sodalis and related bacteria, suggesting conserved versus host specific roles. Moreover, Sodalis and its close relatives exhibit genetic divergence at the rcsF, ompA and ompC loci, indicative of initial molecular divergence. The application of outer membrane genes as markers for further delineating the systematics of recently diverged bacteria is discussed. These results increase our understanding of insect symbiont evolution, while also identifying early genome alterations occurring upon integration of microbes with eukaryotic hosts.
Keywords: symbiosis, insect, phylogeny, Sodalis
Introduction
Symbiosis enables the utilization of environments that would otherwise be rendered inhospitable and as such, is recognized as an important source of biological innovations particularly in regards to the radiation of the Class Insecta (Blochmann, 1887; Buchner 1965). The evolutionary trajectory of symbiosis towards obligate mutualism may develop through a parasitism to mutualism continuum through processes such as the attenuation of host fitness penalties (Jeon, 1972) and the conversion of horizontal transmission to a purely vertical mode (Ewald, 1987). Such a route is exemplified by ancient endocellular symbionts of various insect hosts, such as Buchnera aphidicola in aphids (Homoptera: Aphididae), which are thought to have evolved from less specialized but more prevalent microbial relations such as those involving general insect pathogens (Dale et al., 2001; Hosokawa et al., 2010).
The Gammaproteobacterium, Sodalis glossinidius, is the secondary symbiont of the tsetse fly (Diptera: Glossinidae). Tsetse flies have medical significance as obligate vectors of the parasitic Trypanosoma brucei subspp., the etiological agents of African trypanosomiasis. In contrast to the primary symbiont Wigglesworthia glossinidia, which has a strict localization to the tsetse bacteriome and an extensive co-evolutionary history with its host (Chen et al., 1999), Sodalis exhibits a wider tissue tropism including the host midgut, hemolymph and muscle (Cheng & Aksoy, 1999) with the symbiosis being of relatively recent origin (Weiss et al., 2006). The functional role of Sodalis within tsetse remains relatively unknown, although influences on enhancing host life longevity (Dale & Welburn, 2001) and vector competency (Farikou et al., 2010; Welburn et al., 1993) have been demonstrated.
Recent studies have shown that symbionts harbored within several host insect orders including Diptera, Coleoptera, Phthiraptera, and Hemiptera are highly related to Sodalis based on 16S rRNA sequences (Toju et al., 2010; Grunwald et al., 2010; Kaiwa et al., 2010; Fukatsu et al., 2007; Novakova & Hyspa, 2007; Weiss et al., 2006). These analyses indicate that this group of bacteria shares a recent common ancestor, despite now infecting a broad taxonomic range of hosts.
Selection pressures unique to ecological niches drive evolutionary diversification, with genomic alterations facilitating the adaptation to new habitats by bacteria. Outer membrane proteins, with known immunogenic properties, represent initial points of interspecific contact. Moreover, symbiont cell surfaces have been shown to be pivotal towards the homeostasis of host-bacterial relations (Weiss et al., 2008; Nyholm et al., 2009). Among related microorganisms, genes encoding surface-associated proteins are likely to represent preliminary examples of divergence due to host background differences and consequential symbiont adaptation. We believe that surface encoding genes, often representing hyper variable genes (Wimley, 2003; Zheng et al., 2003), may prove to be significant markers not only in deciphering the evolutionary distance between recently diverged microorganisms such as the Sodalis-allied bacteria, but also towards identifying preliminary molecular alterations associated with inhabiting diverse hosts.
For this study, we extend molecular phylogenetic analyses for this specific clade of Sodalis-like insect symbionts, particularly focusing on the symbionts of the tsetse fly species Glossina morsitans, G. brevipalpis, G. fuscipes and G. pallidipes, the slender pigeon louse Columbicola columbae (Phthiraptera: Philopteridae), and the bloodsucking hippoboscid fly Craterina melbae (Diptera: Hipposboscidae). We aim to further our understanding of their relatedness and identify initial effects associated with the colonization of different host species. The goals of the current study are; to assess intra/interspecies diversity of Sodalis, to provide 16S rRNA phylogenetic analysis of all ‘Sodalis-allied’ microbes described to date, and to compare the ability of surface encoding genes to systematically resolve relationships within this symbiont lineage.
Materials and Methods
Insects
Tsetse flies, G. morsitans and G. brevipalpis, were maintained at West Virginia University within the Department of Biology insectary as previously described (Snyder et al. 2010).
Interspecific diversity analyses
DNA isolation (Cr. melbae, G. morsitans, G. fuscipes, G. pallidipes, and G. brevipalpis) was performed using the Holmes-Bonner protocol (Holmes & Bonner, 1973). Nucleic acid extraction for Co. columbae was performed using the QIAamp tissue mini kit (Qiagen, Valencia CA). All samples were resuspended in 1X Tris-EDTA following DNA isolation. DNA samples were subjected to PCR amplification of genes encoding putative outer membrane components; specifically ompA, the outer membrane protein A, ompC, the osmoporin protein C, and rcsF, ycfM, slyB and spr, producing various outer membrane lipoproteins. PCR annealing temperatures, primers, and respective amplicon sizes are included in Supplemental File 1. Notably, amplification reactions of ycfM from Co. columbae and Cr. melbae and rcsF and slyB from Co. columbae were not successful. Negative controls were included in each set of amplification reactions. The amplification products were analyzed by agarose gel electrophoresis and visualized with Kodak 1D image analysis software. The amplicons were purified using QIAquick PCR purification kit (Qiagen) and subject to DNA sequencing at the West Virginia University Department of Biology Genomics Center on an ABI 3130xl analyzer (Applied Biosystems, Foster City CA) using a 3.1 BigDye protocol (Applied Biosystems). For each sample, three to five amplicons were sequenced in both directions and contigs were assembled using Ridom Trace Edit (RidomGmbH, Wurzburg Germany).
Assessing Sodalis intraspecies diversity within tsetse
The Sodalis ompA gene was amplified from two G. morsitans, G. fuscipes, G. brevipalpis and G. pallidipes individuals. Amplicons were ligated into pGEM-T vector (Promega) and Escherichia coli JM109 cells were transformed. Four colonies per individual tsetse were verified for an ompA insertion and sequenced as described above.
Molecular phylogenetic analyses
All analyses included sequence data collected in this study or publicly available at NCBI GenBank. DNA sequences were aligned using the Clustal X algorithm with default settings, and refined manually when necessary. Maximum parsimony [MP] and neighbor joining [NJ] analyses were performed with 1000 replicates in PAUP 4.0 (Swofford 2002). MP heuristic searches utilized the tree-bisection-reconnection (TBR) branch-swapping algorithm with 200 Max trees and starting trees were created using stepwise additions. All MP analyses were performed twice, where gaps were treated either as “missing data” or as a “5th character state”, with no differences noted between the results. NJ analyses implemented Kimura’s two parameter model (Kimura, 1980). Lineage support was measured by calculating nonparametric bootstrap (BS) values (n =1000) (Felsenstein, 1985).
The evolutionary models used for Bayesian analyses were determined using the Akaike Information Criterion (AIC) in MrModeltest 2.3 (Nylander, 2004). Bayesian analyses were performed in MrBayes 3.1.2 (Ronquist & Huelsenbeck, 2003), and the number of categories used to approximate the gamma distribution was set at four. Additionally, six Markov chains (Larget & Simon, 1999) were run for 3,000,000 generations for 16S rRNA and for 1,000,000 generations for surface-encoding genes. Posterior probability (PP) values were subsequently calculated. Stabilization of model parameters (burn-in) occurred around 2,400,000 and 800,000 generations for 16S rRNA and surface-encoding genes, respectively. Every 100th tree after stabilization (burn-in) was sampled to calculate a 50% majority-rule consensus tree. All trees were constructed using the program FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
Genetic divergence analyses
DnaSP (Librado and Rozas, 2009) was used to calculate synonymous (dS) and nonsynonymous (dN) rates and two common measures of nucleotide variation, π and θW, for determining ompA intraspecies variation within Glossina. Neutrality tests were also performed in DnaSP. The McDonald-Krietman test and Neutrality index (NI) were calculated by comparing the ratio of dS to dN mutations within either individual Glossina species for ompA, or among Glossina isolates for ompC, and an E.coli outgroup. The outgroup was composed of ecologically diverse E. coli representatives NC_000913, NC_008253 and NC_002655. These adaptive evolution tests have been shown to be most powerful when taxa are closely related (Clark et al., 2003). We chose E. coli as our representative outgroup because it is a close relative of Sodalis, and has a wide representation of publicly available genome strains.
Nucleotide sequence accession numbers
The nucleotide sequences determined in this study have been deposited in the NCBI GenBank database under accession numbers HM626140–HM826149.
Results and discussion
Phylogenetic placement of tsetse fly secondary symbionts (Sodalis) based on 16S rRNA analyses
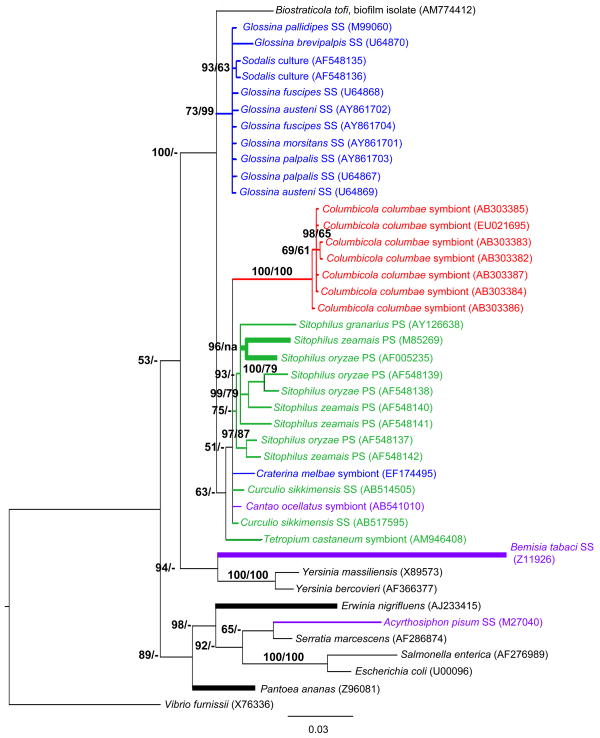
To examine the evolutionary relationships of the newly identified Sodalis-like symbionts, we constructed phylogenetic trees based on 16S rRNA sequences. Bayesian analysis supports the monophyly of Gammaproteobacteria symbionts isolated from diverse insect orders (i.e. Diptera, Coleoptera, Hemiptera and Phthiraptera) (Fig. 1). In general, there is a tight clustering of symbionts with respective insect host Order. Our Bayesian analysis also suggests the closer relationship of hippoboscid symbionts to weevil and pigeon louse symbionts, rather than to Sodalis, despite a common ancestry of their respective hosts within the Hippoboscoidea (Petersen et al., 2007), thus further substantiating a previous hypothesis of independent symbiont acquisition events by these hosts (Novakova & Hyspa, 2007). However, there is only moderate Bayesian support for this relationship (PP = 77, data not shown) that is further decreased (PP= 51) when symbionts of the recently reported chestnut weevil Cu. sikkimensis (Toju et al., 2010) and the stinkbug Ca. ocellatus (Kaiwa et al., 2010) are included in the analyses. Analyses were unable to resolve the relationships of the symbionts harbored within the hippoboscid, chestnut weevil, and stinkbug indicative of relatively recent establishments and inadequate time for 16S rRNA diversification, or alternatively the transfer of these symbionts within these insect orders. With Bayesian analysis, symbiont relationships within the Sitophilus clade are highly resolved in comparison to that of Sodalis, where the scattering of host species (i.e. not reflective of Sitophilus speciation (Conord et al. 2008)) suggests independent acquisition within species. It is possible that horizontal transmission, in addition to the previously described vertical route (Heddi et al. 1999), may also contribute to this phylogenetic patterning of symbionts; this warrants further study. Interestingly, although bacterial endosymbiosis is believed to be old within weevils (dating back approximately 125 Myr), symbiont replacement is believed to have occurred multiple times in Sitophilus weevils with causative factors remaining speculative (Conord et al. 2008).
Fig 1.
Molecular phylogenetic tree of 16S rRNA gene sequences from Sodalis and allied bacteria. A Bayesian analysis tree created from 1509 aligned nucleotides is shown; NJ analyses gave essentially identical results (data not shown). Branches in bold were constrained with MP analysis. PP (shown as %, i.e. 95% represents a PP value of 0.95) and BS values > 50% are indicated at the nodes (- = < 50% BS), respectively. The branch lengths are measured in expected substitutions per site. Sequence accession numbers are provided. Host species are indicated for symbiotic bacteria, with colors representing insect orders. PS; primary symbiont, SS; secondary symbiont.
Sodalis isolated from in vitro culture maintained through serial passage formed its own monophyletic clade, supporting diversification from current Glossina isolates. While culture isolates were grouped together based on 16S rRNA, Sodalis obtained from the same host species did not follow this pattern (i.e. symbionts within G. fuscipes, G. austeni and G. palpalis) suggesting either no diversity between tsetse fly isolates or the lack of resolution due to the conserved nature of this locus. Distance analyses of the 16S rRNA gene also support the higher similarity of bacteria within the Sodalis clade, relative to that housing the Sitophilus symbionts (data not shown), which may explain why analyses were unable to further resolve these relations (Fig. 1). Importantly, many branches could not be robustly resolved warranting the need for additional inquiries utilizing genes that are typically associated with higher evolutionary rates such as those encoding surface exposed molecules.
Phylogenetic placement of Sodalis-like symbionts based on surface encoding proteins
To further our understanding of the divergence of ‘Sodalis-allied’ bacteria, particularly those found within various Glossina spp., Co. cumbicola and Cr. melbae, and to also assess the application of these surface encoding genes in future analyses extending into other related symbionts, we reconstructed their phylogeny using six putative outer membrane encoding genes: rcsF, slyB, ompA, spr, ompC, and ycfM. With only a few exceptions (all spr and Glossina versus Cr. melbae slyB comparisons), the genetic distances of surface encoding loci between symbionts localized within hosts of different orders were greater in comparison to 16S rRNA.
In regards to the spr, slyB, and ycfM loci, although sufficient sequence similarities resulted in the Sodalis-like isolates forming a monophyletic clade within the Gammaproteobacteria distinct from many free-living members of this group, deeper taxonomic resolution was lacking (data not shown). The low phylogenetic signal provided by these loci suggests that they may not be involved in adapting to particular host species and/or may be structurally constrained. For example, comparative analyses of the spr lipoprotein amino acid sequence demonstrated the conservation of residues that form a unique Cys-His-His catalytic triad which is believed to form a substrate-binding cleft within the active site of this protein (Aramini et al., 2008) between examined Sodalis isolates, Cr. melbae and Co. columbae symbionts.
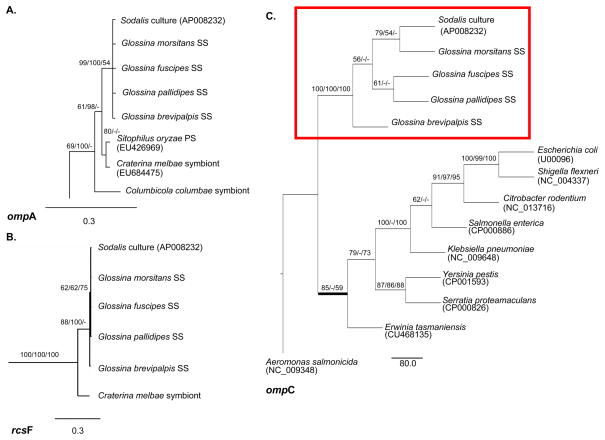
The ompA, ompC, and rcsF loci (Fig. 2) appear to be more informative towards the phylogenetic resolution of the Sodalis-like symbiont clade. With rcsF, sufficient phylogenetic signal was provided to enable clustering of the Glossina symbionts, with strong support, separate from the Cr. melbae symbiont (Fig. 2B). Interestingly, rcsF in E. coli has been shown to be involved in signaling transduction of perturbations and/or environmental cues from the cell surface (Majdalani et al., 2005). Diversification between Sodalis and Cr. melbae isolates may indicate functional adaptations, such as differences in the type of signaling encountered within the host species background. The Sodalis symbionts also formed a distinct clade with ompC phylogeny, with most mutations noted outside of the seven putative extracellular loops (Basle et al., 2006) of the different Glossina isolates. The one exception occurred in extracellular loop 4 where host interspecies diversity was observed with Sodalis isolates.
Fig. 2.
Molecular phylogenetic analyses of putative outer membrane encoding gene sequences from Sodalis-allied symbionts which support diversification. Bayesian trees inferred from (A) 1164 unambiguously aligned nucleotides of the ompA gene and (B) 426 nucleotides of the rcsF gene. Significance values are indicated in Bayesian PP/MP BS/NJ BS Branch lengths are measured in expected substitutions per site and depicted under each tree. (C) MP tree inferred from 1227 nucleotides of the ompC genes are shown with support values in the order of MP BS/Bayesian PP/NJ BS. Branch lengths depict the number of substitutions. Bold lines indicate discrepancies in tree renditions between analyses. Accession numbers are provided in parentheses. Host species are indicated for symbiotic bacteria; SS= secondary symbiont.
Relative to the other surface encoding genes analyzed in this study, the ompA gene exhibited the greatest diversity among symbionts due to a combination of point mutations and indels. The best-studied ompA gene variant, that of Escherichia coli K-12, encodes a 325 amino acid polypeptide (Chen et al., 1980). The N-terminal domain forms an eight-stranded β-barrel in the outer membrane, creating four surface-exposed loops (Pautsch & Schulz, 1998), while the C-terminus is periplasmic (Klose et al., 1988). Amino acid variations within outer membrane proteins mainly occur in the domains located in the extracellular regions while interspaced residues making up the β-strands tend to be conserved. In our analyses, relative to Glossina symbionts, a total of nine nonsynonymous mutations were observed among Cr. melbae, Co. columbae and Sitophilus (i.e. Sitophilus oryzae primary symbiont, SOPE) symbionts occurring in loops 1–4 of the OmpA protein. Differences noted in the ompA sequence between the Glossina symbionts were localized outside of the extracellular regions, similar to our observations with ompC. In relation to ompA, the Co. columbae symbiont exhibited the greatest nucleotide divergence resulting in its sister taxon placement relative to the other symbionts of interest with strong MP BS support. MP, Bayesian, and NJ analyses all grouped Glossina symbionts within their own clade indicative of diversification potentially arising from host adaptation processes.
Molecular evolution of Sodalis-like symbionts
The Sodalis ompA gene demonstrated a wide nucleotide variation (π) within tsetse species (Table 1), with the highest π exhibited within G. morsitans (π = 0.11) and the lowest within G. brevipalpis (π = 0.001). This observation is not unprecedented as evidence of endosymbiont genomes (e.g. Wolbachia) undergoing either purifying or diversifying selection when examined from different host species has also been described with cell envelope component genes (Brownlie et al. 2007).
Table 1.
Sodalis omp A nucleotide diversity within tsetse species and tests for neutral models of evolution. The neutrality index (NI), the ratio of synonymous to nonsynonymous mutations (dN/dS), was calculated using the McDonald-Kreitman test. Neutrality was examined within tsetse isolates (Tajima’s D, Fu and Li’s D*, and Fu and Li’s F*) and also compared to the outgroup Escherichia coli (accession number NC_000913) using Fu and Li’s D and Fu and Li’s F.
| Symbiont host species | πtotal | θw | ω(dN/dS) | NI | Tajima’s D | Fu and Li’s D* | Fu and Li’s F* | Fu and Li’s D | Fu and Li’s F |
|---|---|---|---|---|---|---|---|---|---|
| Glossina morsitans | 0.112 | 0.161 | 2.03 | 4.331*** | −1.75*** | −1.86** | −2.04** | −2.39** | −2.72** |
| Glossina pallidipes | 0.046 | 0.068 | 0.828 | 0.627 | −1.83*** | −1.97** | −2.16** | −2.19* | −2.56** |
| Glossina fuscipes | 0.004 | 0.006 | 0 | 1.000 | −1.13 | −1.3 | −1.37 | −1.49 | −1.65 |
| Glossina brevipalpis | 0.001 | 0.001 | 0 | 0.311 | −1.05 | −1.13 | −1.20 | −1.26 | −1.41 |
average pairwise nucleotide diversity; θw, segregating sites per haploid genome.
Statistical significance:
P<0.05;
P<0.02;
P<0.01.
Tests of neutrality (Tajima’s D, Fu and Li’s D* and F*, and Fu and Li’s D and F) indicate a significant excess of young, rare alleles for Sodalis ompA within G. morsitans and G. pallidipes. In summation, three indices (π, dN/dS and NI) support diversifying selection due to an abundance of low frequency Sodalis ompA haplotypes within G. morsitans. These observations may reflect the well-supported phenomenon of enhanced sequence evolution in endosymbiotic bacteria (Clark et al., 1999; Canback et al., 2004; Fry & Wernegreen, 2005). Similar to other endosymbionts, the small effective population size of Sodalis, a consequence of severe population bottlenecks during maternal transmission (Rio et al., 2006), predicts a larger proportion of nonsynonymous mutations due to drift that will generate higher dN to dS ratios (Ohta, 1972; Woolfit & Bromham, 2003).
Deviation from neutrality was also observed with Sodalis ompC isolates, as supported by a significant MK test (G=13.42, P=0.00025) when compared with E. coli. A high abundance of fixed dN substitutions within all Sodalis isolates provides strong evidence for positive selection at particular sites of the ompC gene. Notably, upon comparison of Sodalis with E. coli isolates, greater ompC amino acid sequence variation was observed at putative surface exposed loops suggesting their significance in adaptive evolution towards ecological niches.
Here we describe early genetic modifications likely involved in host adaptation within Sodalis-allied bacteria, specifically divergence in symbiont surface encoding genes. In general, this particular class of loci exhibited greater genetic distances among Sodalis-like bacteria than the 16S rRNA gene traditionally employed in phylogenetic analyses. Nevertheless, not all the surface encoding genes examined in this study proved equivalent in their ability to resolve phylogenetic relations. Differences in selective pressures arising from distinct host physiologies and feeding lifestyles (Rio et al., 2003; Toh et al., 2006) as well as the influence of other host microbiota members (Snyder et al., 2010) have been shown to affect symbiont genome evolution. Future studies should extend the phylogenetics of these surface-encoding loci, specifically rcsF, ompC and ompA, to other recently identified Sodalis-related symbionts to enhance phylogenetic resolution. Functional assays should also be pursued to examine the relevance of surface-encoding loci towards the process of endosymbiotic adaptation and to determine whether the described differences are sufficient to constrict host species colonization.
Supplementary Material
Acknowledgments
We thank Baneshwar Singh and Dr. Mariam Lekveishvili for technical assistance. We are grateful to Drs. Pierre Bize, Vaclav Hyspa and Takema Fukatsu for providing Cr. melbae and Co. columbae samples, respectively. We thank the Slovak Academy of Science and IAEA for tsetse pupae. We acknowledge the funding support of WVU Research Corporation, NASA NNX07AL53A and NIH R03AI081701.
References
- Aramini JM, Rossi P, Huang YJ, Zhao L, Jiang M, Maglaqui M, Xiao R, Locke J, Nair R, Rost B, Acton TB, Inouye M, Montelione GT. Solution NMR Structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli: Structural evidence for a novel cysteine peptidase catalytic triad. Biochemistry. 2008;47:9715–9717. doi: 10.1021/bi8010779. [DOI] [PubMed] [Google Scholar]
- Basle A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J. Mol. Biol. 2006;362:933–942. doi: 10.1016/j.jmb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Blochmann F. Uber das regel massige Vorkommen von backterienahnlichen Geblin in den Geweben und Eiem vershiedemer Inseckten. Z Biol. 1887;24:6. [Google Scholar]
- Brownlie JC, Adamski M, Slatko B, McGraw EA. Diversifying selection and host adaptation in two endosymbiont genomes. BMC Evol Biol. 2007;7:68. doi: 10.1186/1471-2148-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P. Endosymbiosis of animals with plant microorganisms. John Wiley and Sons; 1965. [Google Scholar]
- Canback B, Tamas I, Andersson SG. A phylogenomic study of endosymbiotic bacteria. Mol Biol Evol. 2004;21:1110–1122. doi: 10.1093/molbev/msh122. [DOI] [PubMed] [Google Scholar]
- Chen R, Schmidmayr W, Kramer C, Chen-Schmeisser U, Henning U. Primary structure of major outer membrane II (OmpA protein) of Escherichia coli K-12. Proc Natl Acad Sci USA. 1980;77:4592–4596. doi: 10.1073/pnas.77.8.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li S, Aksoy S. Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome associated endosymbiont, Wigglesworthia glossinidia. J Mol Evol. 1999;48:49–58. doi: 10.1007/pl00006444. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Aksoy S. Tissue tropism, transmission, and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol Biol. 1999;8:125–132. doi: 10.1046/j.1365-2583.1999.810125.x. [DOI] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, Ferreira S, Wang G, Zheng X, White TJ, Snisky JJ, Adams MD, Cargill M. Inferring non-neutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Clark MA, Moran NA, Baumann P. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol Biol Evol. 1999;16:1586–1598. doi: 10.1093/oxfordjournals.molbev.a026071. [DOI] [PubMed] [Google Scholar]
- Conord C, Despres L, Vallier A, Balmand S, Miquel C, Zundel S, Lemperiere G, Heddi A. Long-term evolutionary stability of bacterial endosymbiosis in Curculionidae: Additional evidence of symbiont replacement in the Dryopthoridae family. Mol Biol Evol. 2008;25:859–868. doi: 10.1093/molbev/msn027. [DOI] [PubMed] [Google Scholar]
- Dale C, Young SA, Haydon DT, Welburn SC. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc Natl Acad Sci USA. 2001;98:1883–1888. doi: 10.1073/pnas.021450998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale C, Welburn SC. The endosymbionts of tsetse flies- manipulating host-parasite interactions. Int J Parasitol. 2001;31:628–631. doi: 10.1016/s0020-7519(01)00151-5. [DOI] [PubMed] [Google Scholar]
- Ewald P. Transmission modes and the evolution of the parasitism-mutualism continuum. Ann NY Acad Sci. 1987;503:295–306. doi: 10.1111/j.1749-6632.1987.tb40616.x. [DOI] [PubMed] [Google Scholar]
- Farikou O, Njiokou F, Mdiba Mdiba JA, Njitchouang GR, Djeunga HN, Asonganyi T, Simarro PP, Cuny G, Geiger A. Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes- An epidemiological approach in two historical human African trypanosomiasis foci in Cameroon. Infect Genet Evol. 2010;10:115–121. doi: 10.1016/j.meegid.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fry AJ, Wernegreen JJ. the roles of positive and negative selection in the molecular evolution of insect endosymbionts. Gene. 2005;355:1–10. doi: 10.1016/j.gene.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Fukatsu T, Koga R, Smith WA, Tanaka K, Nikoh N, Sasaki-Fakatus K, Yoshizawa K, Dale C, Clayton DH. Bacterial endosymbiont of the slender pigeon louse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Appl Environ Microbiol. 2007;73:6660–6668. doi: 10.1128/AEM.01131-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald S, Pilhofer M, Holl W. Microbial associations in guts systems of wood- and bark- inhabiting longhorned beetles [Coleoptera: Cerambyicidae] Syst Appl Microbiol. 2010;33:25–34. doi: 10.1016/j.syapm.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Heddi A, Grenier A, Khatchadourian C, Charles H, Nardon P. Four intracellular genomes direct weevil biology: Nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc Natl Acad Sci USA. 1999;96:6814–6819. doi: 10.1073/pnas.96.12.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DS, Bonner J. Preparation, molecular weight, base composition, and secondary structure of giant ribonucleic acid. Biochemistry. 1973;12:2330–2338. doi: 10.1021/bi00736a023. [DOI] [PubMed] [Google Scholar]
- Hosokawa T, Koga R, Kikuchi Y, Meng XY, Fukatsu T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Appl Environ Microbiol. 2010;107:769–774. doi: 10.1073/pnas.0911476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon K. Development of cellular dependence on infective organism: microsurgical studies on amoebas. Science. 1972;179:1122–1123. doi: 10.1126/science.176.4039.1122. [DOI] [PubMed] [Google Scholar]
- Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, Ito M, Fukatsu T. Primary gut symbiont and secondary, Sodalis-allied symbiont of the scutellerid stinkbug Cantao ocellatus. Appl Environ Microbiol. 2010;76:3486–3494. doi: 10.1128/AEM.00421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Klose M, Schwarz H, MacIntyre S, Freudl R, Eschbach ML, Henning U. Internal deletions in the gene for an Escherichia coli outer membrane protein define an area possibly important for recognition of the outer membrane of this polypeptide. J Biol Chem. 1988;263:13291–13296. [PubMed] [Google Scholar]
- Larget B, Simon DL. Markov chain monte carlo algorithms for the bayesian analysis of phylogenetic trees. Mol Biol Evol. 1999;16:750–759. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Heck M, Stout V, Gottesman S. role of RcsF in signaling to the Rcs phosphorelay pathway in Escherichia coli. J Bac. 2005;187:6770–6778. doi: 10.1128/JB.187.19.6770-6778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova E, Hyspa V. A new Sodalis lineage from the bloodsucking fly Craterina melbae (Diptera: Hippoboscoidea) originated independently of the tsetse flies symbiont Sodalis glossinidius. FEMS Microbiol Lett. 2007;269:131–135. doi: 10.1111/j.1574-6968.2006.00620.x. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol. 2009;11:483–493. doi: 10.1111/j.1462-2920.2008.01788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. MrModeltest v2. Evolutionary biology centre. Uppsala University; Uppsala, Sweden: 2004. [Google Scholar]
- Ohta T. Population size and the rate of evolution. J Mol Evol. 1972;1:305–314. [PubMed] [Google Scholar]
- Pautsch A, Schulz GE. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- Petersen FP, Meier R, Narayananan Kutty S, Wiegmann BM. The phylogeny and evolution of host choice in the Hippoboscoidea (Diptera) as reconstructed using four molecular markers. Mol Phylogenet Evol. 2007;45:111–122. doi: 10.1016/j.ympev.2007.04.023. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rio RVM, Wu Y, Filardo G, Aksoy S. Dynamics of multiple symbiont density regulation during host development: tsetse fly and its microbial flora. Proc R Soc B. 2006;273:805–814. doi: 10.1098/rspb.2005.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio RV, Lefevre C, Heddi A, Aksoy S. Comparative genomics of insect-symbiotic bacteria: influence of host environment on microbial genome composition. Appl Environ Microbiol. 2003;69:6825–32. doi: 10.1128/AEM.69.11.6825-6832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Snyder AK, Deberry JW, Runyen-Janecky L, Rio RVM. Nutrient provisioning facilitates homeostasis between tsetse fly (Diptera: Glossinidae) symbionts. Proc R Soc B. 2010;277:2389–2397. doi: 10.1098/rspb.2010.0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP 4.0-Phylogenetic analysis using parsimony. Version 4. Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- Thompson JD, Higgins DG, Gibson JJ. Clustal W: improving the sensitivity of progressive multiple alignments through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Weiss BL, Perkin SA, Yamashita A, Oshima K, Hattori M, Aksoy S. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149–56. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toju H, Koga R, Nikoh N, Meng XY, Kimura N, Fukatsu T. “Candidatus Curculioniphilus buchneri” a novel clade of bacterial endocellular symbionts from weevels of the genus Curculio. Appl Environ Microbiol. 2010;76:275–282. doi: 10.1128/AEM.02154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss BL, Mouchotte RM, Rio RVM, Wu Y, Wu Z, Heddi A, Aksoy S. Interspecific transfer of bacterial endosymbionts beween tsetse fly species: infection establishment and effect on host fitness. Appl Environ Microbiol. 2006;72:7013–7021. doi: 10.1128/AEM.01507-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss BL, Wu Y, Schwank JJ, Tolwinski NS, Aksoy S. An insect symbiosis is influenced by bacterium-specific polymorphisms in outer-membrane protein A. Proc Natl Acad Sci USA. 2008;105:15088–15093. doi: 10.1073/pnas.0805666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn SC, Arnold K, Maudlin I, Goday GW. Rickettsia-like organisms and chitinase production in relation to transmission of trypanosomes by tsetse flies. Parasitology. 1993;107:141–145. doi: 10.1017/s003118200006724x. [DOI] [PubMed] [Google Scholar]
- Wimley WC. The versatile beta-barrel membrane protein. Curr Opin Struct Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- Woolfit M, Bronham L. Increased rates of sequence evolution in endosymbiotic bacteria and fungi with small effective population sizes. Mol Biol Evol. 2003;20:1545–1555. doi: 10.1093/molbev/msg167. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Roberts RJ, Kasif S. Identification of genes with fast-evolving regions in microbial genomes. Nucleic Acids Res. 2003;278:24825–24830. doi: 10.1093/nar/gkh935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.