Abstract
Retinal analogues have been used to probe the chromophore binding pocket and function of the rod visual pigment, rhodopsin. Despite the high homology between rod and cone visual pigment proteins, conclusions drawn from rhodopsin studies should not necessarily be extrapolated to cone visual pigment proteins. In this study, the effects of full-length and truncated retinal analogues on the human red cone opsin’s ability to activate transducin, the G protein in visual transduction, were assessed. The result with beta-ionone (6) confirms that a covalent bond is not necessary to deactivate the red cone opsin. In addition, several small compounds were found able to deactivate this opsin. However, as the polyene chain is extended in a trans configuration beyond the 9-carbon position, the analogues became agonists up to all-trans-retinal (3). The 22-carbon analogue (2) appeared to be neither agonist nor inverse agonist. Although the all-trans-C17 (5) analogue was an agonist, the 9-cis-C17 (11) compound was an inverse agonist, a result that differs from that with rhodopsin. These results suggest that the red cone opsin has a more open structure in the chromophore binding region than rhodopsin and its activation or deactivation as a G-protein receptor may be less selective than rhodopsin.
The highly conjugated nature of retinoids make these compounds amenable to spectroscopic studies.1, 2 Since the days of Hubbard and Wald3 and their demonstration that non-native cis isomers of retinal were able to generate pigments with rod opsin, the apoprotein of rhodopsin, Nakanishi and others have created a vast library of retinal analogues to probe the structure and function of rhodopsin.4–14 Rhodopsin is the well-studied visual pigment in rod photoreceptor cells and is used for dim light detection. However, everyday vision relies on cone photoreceptors that contain cone visual pigments homologous to rhodopsin and contain the same 11-cis-retinal chromophore. Although there are similarities between rod and cone pigments, cone pigments regenerate, activate, and inactivate faster than rhodopsin.15 Furthermore, some cone pigments readily release their chromophore in darkness unlike with rhodopsin.16–18
The protein component of visual pigments (opsin) is a G protein-coupled receptor. 11-cis-Retinal (1) both serves as the chromophore of the opsins and as an inverse agonist that holds the visual pigment protein in an inactive conformation; light photoisomerizes the chromophore to the all-trans form converting the ligand into an agonist. While retinal analogues have been shown to modulate the spectral properties of cone pigments,12, 19 less is known about their effects on cone opsin activity. Our group has shown that some retinal analogues can also modulate the effectiveness of the cone opsin’s ability to activate its G protein transducin and this activity can differ from that of the rhodopsin apoprotein.14, 20 In this report, these studies have been extended, and retinal analogues of varying lengths have been screened for their ability to modulate the activity of the human red cone opsin in a ligand-dependent manner.
Results and Discussion
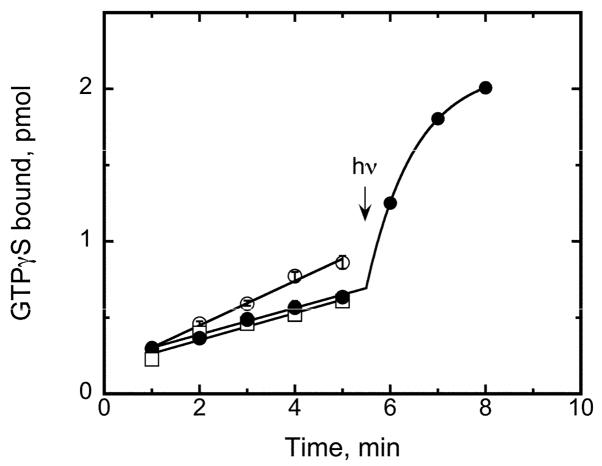
Previous studies from different groups have utilized point mutations or chimeric proteins to assess protein-ligand differences between rhodopsin and cone pigments. 21–29 Several full-length retinal analogues have been incorporated into cone opsins to assess their absorption properties, photochemistry, and Schiff base pK values to further characterize protein-ligand interactions.12, 19, 30–34 This study was focused on the effects of retinal analogues on the human red cone opsin’s ability to activate the G protein transducin in a ligand-dependent manner. Figure 1 illustrates the action of 11-cis-retinal (1) on the red cone opsin’s ability to activate transducin in the dark and with light, as has been shown previously where it was shown that at pH 6.4, 11-cis-retinal (1) acts as an inverse agonist.20 Also shown in Figure 1 is the basal transducin activity in the absence of opsin, which is indistinguishable from that by the 11-cis-retinal-treated sample. Thus 11-cis-retinal (1) does indeed deactivate the human red cone opsin. Unless stated otherwise, the subsequent reported activities have been corrected for this basal level and normalized to opsin activity to assess the relative effects of retinal and retinal analogues on their abilities to activate or deactivate the human red cone opsin.
Figure 1.
Transducin activation by the human red cone opsin and pigment. A radioactive filter binding assay is used to follow uptake of GTPγS as a function of time. The pH of the reaction is 6.4 where the opsin activity (open circles) is readily distinguishable from the 11-cis-retinal-bound form (filled circles). A 12 s pulse of >530 nm light at 5.5 min converts the 11-cis inverse agonist to the all-trans agonist and seen as an exponential rise due to the rapid rise and decay of the photoactivated species. Basal transducin activation was also measured (open squares) as a function of time by replacing red cone opsin-containing membranes with sham-transfected COS-1 cell membranes in the reaction.
Retinal analogues have long been used to study ligand-protein relationships to better understand rhodopsin. Despite the high sequence homology among all of the rod and cone opsins,15, 35 the effects of retinal analogues on rhodopsin structure and function does not necessarily translate directly to how they interact with cone opsins. For example, 9-demethyl retinal has been shown to inhibit formation of the active Meta II conformation of rhodopsin;36 in contrast, in cone pigments generated with this analogue, we showed that there is no such inhibition.34 Furthermore, the decay of the active species is prolonged in only the middle/long wavelength-sensitive cone pigment, and not in either of the two shorter wavelength-sensitive cone pigments. Another example of a retinal analogue having a different effect on rod and cone opsins is beta-ionone (7), which has been shown to be an agonist to the rhodopsin apoprotein,14, 37–39 but an inverse agonist to the long wavelength-sensitive cone opsin.14 Thus, specific interactions between ligand and opsin cannot always be generalized based on results with a single ligand and one opsin type. We have ascribed some of the differences to functional priorities demanded by rods and cones. The rod pigment rhodopsin requires a very inactive conformation in the dark to be exquisitely sensitive as a photon detector in dim light, and as a result the tolerance for steric changes from either the ligand or protein could be functionally detrimental. Cones, on the other hand, function under bright light conditions and must cycle rapidly and as a consequence a highly constrained ligand would impede efficiency.
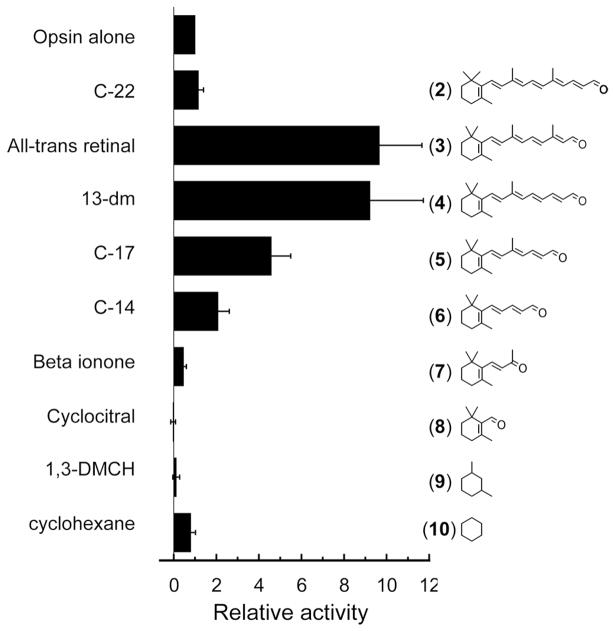
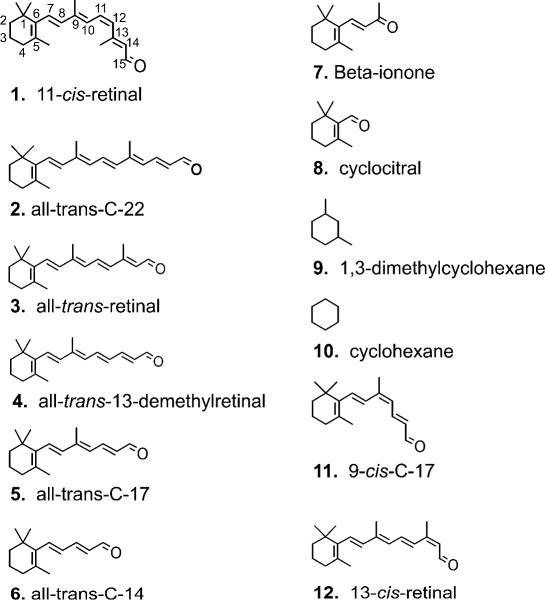
A series of “trans” retinal analogues has been tested. The assumption was made that the shorter compounds would not form pigments with measurable spectra but would affect the ability of the protein component to activate transducin if bound in the retinal binding pocket. The term “trans” as used here implies that the polyene chain does not have cis bonds, but these compounds also include shorter analogues that have either little or no polyene chain extending from the 6-carbon position of the ring. Figure 2 summarizes the relative effect of these analogues on the opsin’s ability to activate transducin. The results appear to set an upper and lower boundary for ligands that act as agonists and inverse agonists for these “trans” analogues. As shown previously with the rod and red cone opsins, the full-length all-trans retinal was an agonist. The longer C-22 compound (2) does not appear to alter the ability of opsin to activate transducin appreciably, suggesting a limit to the size of compounds that can complex with the red cone opsin to activate transducin. As the polyene chain becomes shorter than the all-trans-retinal C-20 compound (3), the agonistic behavior decreases. For “trans” compounds that do not extend beyond the 9-carbon position, they act as inverse agonists. Beta-ionone (7) has been shown previously to be able to resensitize bleached salamander red cone cells40 and act as an inverse agonist to salamander red cone opsin.14 Smaller compounds like cyclocitral (8) and 1,3-dimethylcyclohexane (9) were also inverse agonists, but required higher concentrations to obtain similar effects, likely due to their small size and lower affinity (see Experimental Section) although solubility of the different compounds may also contribute to this effect. While 1,3-dimethylcyclohexane (9) deactivates the red cone opsin, cyclohexane (10) itself does not. Similarly, an earlier study on the inhibition of rhodopsin pigment regeneration by small cyclohexyl derivatives concluded that at least 2 methyl groups were necessary to compete with 11-cis-retinal for the chromophore binding site of rhodopsin.41
Figure 2.
Normalized transducin activation by the human red cone opsin in the presence of the retinals and retinal analogues illustrated in the figure. Activity is shown after correcting for the basal activity and normalized to transducin activation by the red cone opsin alone. Each data point represents the mean of at least three measurements; error bars reflect the standard error of the means of opsin activity with the retinoid, opsin activity alone, and basal activity, which were propagated through the normalization calculations. All-trans-retinal is the native photoisomerized form of the chromophore and is a C-20 compound. C-22, C-17, and C-14 are the 22, 17, and 14 carbon aldehyde analogues of retinal; 13-dm is the all-trans 13-demethylretinal; 1,3-DMCH is 1,3-dimethylcyclohexane. The concentration of test compound in the reaction was 20 μM except for beta-ionone (7) and cyclocitral (8) (200 μM); and for 1,3-dimethylcyclohexane (9) and cyclohexane (10) (2 mM).
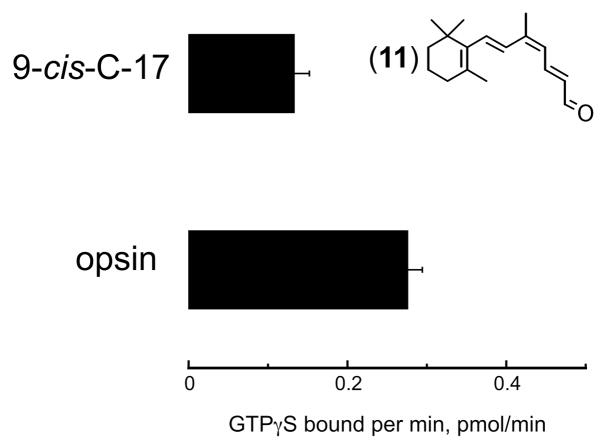
The all-trans-C-17 aldehyde (5) extends the polyene chain beyond the 9-carbon position. Since 9-cis-retinal can substitute for 11-cis-retinal as a chromophore, the 9-cis-C-17 compound (11) was assessed for its effect on red cone opsin activity. Figure 3 shows that this compound is an inverse agonist to the red cone opsin. It should be noted that no correction was made for the basal transducin activity and the data were not normalized. This result differs from the 9-cis-C-17 compound (11) action on rod opsin, where both the 9-cis- (11) and all-trans-C17 (5) compounds are agonists.37
Figure 3.
Transducin activation by the human red cone opsin in the absence and presence of the 9-cis-C-17 (11) aldehyde compound. Activity is not normalized nor corrected for the basal activity of transducin. The lowered activity in the presence of the 9-cis form of the C-17 aldehyde indicates that it is an inverse agonist and contrasts the findings of the same compound on the rod opsin.37 Error bars reflect standard error of the mean from at least three measurements.
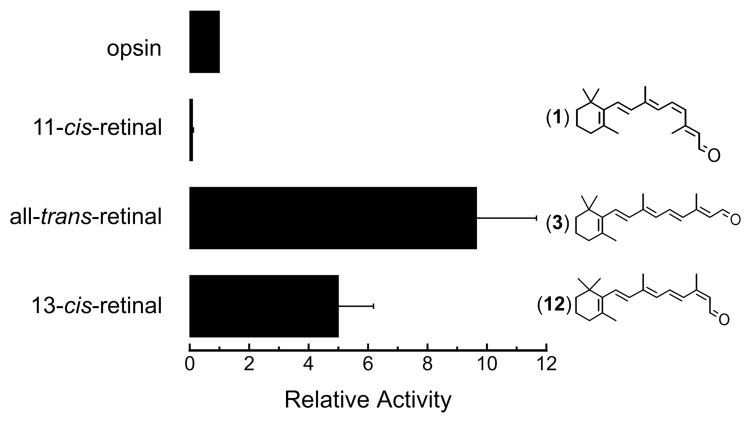
13-cis-Retinal (12), a full-length isomer of retinal, was also tested for its on human red cone opsin activity and found to be an agonist (Figure 4). The non-covalently binding 11-cis-retinol has also been shown by our group to deactivate red cone opsin, a result that again is opposite to that found for rhodopsin where it is an agonist.42 All-trans-retinol was an agonist for both rod and cone opsins tested.20 Thus, these results indicate that the red cone opsin can accommodate the beta-ionone ring and a 3-carbon polyene extension to be deactivated by the ligand, but beyond the 9-carbon position, there needs to be a cis bond. A covalent bond between the ligand and the opsin is not required to either deactivate or activate the opsin.
Figure 4.
Transducin activation by the human red cone opsin in the absence and presence of full-length retinal isomers. 11-cis-Retinal (1) is an inverse agonist, but all-trans- (3) and 13-cis- (12) retinals are agonists. Activity is shown after correcting for the basal activity and normalized to transducin activation by the red cone opsin alone. Each data point represents the mean of at least three measurements; error bars reflect the standard error of the means of opsin activity with the retinoid, opsin activity alone, and basal activity, which were propagated through the normalization calculations.
In 1991, Nakanishi43 asked and discussed why Nature selected 11-cis retinal as the isomeric form of the chromophore for visual pigments. This was addressed mostly in terms of the resultant spectral properties of the pigments formed, which are optimal for our natural light environment.43 While a covalent bond is not necessary to deactivate or activate the red cone opsin, the increased polyene chain length in compounds longer that C-14 (6) decreases the effective concentration required for the ligand to be an inverse agonist, perhaps due to better affinity to the chromophore binding site. In the native pigment, the inverse agonist is bound covalently in the dark and agonist activity occurs within 200 fs44 as photoisomerization of the bound retinal results in the rapid conversion to a bound agonist. Therefore 11-cis-retinal is the ideal molecule for vision from its effects on absorption, activity, and rapid conversion to an agonist within the rod and cone pigments.
Experimental Section
Materials
11-cis-Retinal (1) was synthesized as described previously.34, 45 All-trans-Retinal (3), beta-ionone (7), cyclocitral (8), 1,3-dimethylcyclohexane (9), cyclohexane (10), 13-cis-retinal (12) were purchased from Sigma/Aldrich (St. Louis, MO). The synthesis of and extinction coefficients for C-22- (2), all-trans-13-demethylretinal (4), C-17- (trans (5) and 9-cis (11)), and C-14- (6) retinal analogue compounds were previously reported.37 Retinoids and analogues were dissolved in ethanol at concentrations ranging from 1 to 100 mM.
Expression and Membrane Preparations of Human Red Cone Opsin
Human red cone opsin was transiently expressed in COS-1 cells as described previously.20 On the third day after transfection, COS-1 cells were harvested, pelletted, and stored at −80°C until needed. Membrane preparations from these frozen cell pellets were isolated from a discontinuous sucrose gradient.20, 46
Transducin Activation Assays
Rod transducin was purified from bovine retinas as described previously.47, 48 The ability of opsins to activate transducin was measured using a radioactive filter-binding assay as described previously.20, 46 The retinal/analogue in ethanol at a 1:50 dilution or ethanol alone (as a control) was added to the mixture of the opsin and transducin 1 min prior to the addition of the radioactive GTPγS, which corresponded to time zero. Ligand-dependent activation of transducin by the red cone opsin was linear over a 5-min time period; thus, for some measurements single 5 min data points were obtained. Unless stated otherwise, the activities were corrected for basal transducin activation at pH 6.4 and then normalized to transducin activation by opsin alone. The concentration of retinal analogue in the reaction was generally 20 μM except for beta-ionone (7) and cyclocitral (8), it was 200 μM; and for 1,3-dimethylcyclohexane (9) and cyclohexane (10), it was 2 mM.
Acknowledgments
We thank Patrice Goletz for technical assistance. This work was supported in part by NIH grants EY04939 (RKC) and EY019515 (MK); an unrestricted grant to the Department of Ophthalmology at the Medical University of South Carolina from Research to Prevent Blindness (RPB, New York); RKC is an RPB Senior Scientific Investigator.
Footnotes
Dedicated to Dr. Koji Nakanishi of Columbia University for his pioneering work on bioactive natural products.
References and Notes
- 1.Honig B, Ebrey TG. Annu Rev Biophys Bioeng. 1974;3:151–177. doi: 10.1146/annurev.bb.03.060174.001055. [DOI] [PubMed] [Google Scholar]
- 2.Honig B, Greenberg AD, Dinur U, Ebrey TG. Biochemistry. 1976;15:4593–4599. doi: 10.1021/bi00666a008. [DOI] [PubMed] [Google Scholar]
- 3.Hubbard R, Wald G. J Gen Physiol. 1952;36:269–315. doi: 10.1085/jgp.36.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakanishi K, Yudd AP, Crouch RK, Olson GL, Cheung H-C, Govindjee R, Ebrey TG, Patel DJ. J Am Chem Soc. 1976;98:236–238. doi: 10.1021/ja00417a040. [DOI] [PubMed] [Google Scholar]
- 5.Kropf A, Whittenberger BP, Goff SP, Waggoner AS. Exp Eye Res. 1973;17:591–606. doi: 10.1016/0014-4835(73)90088-2. [DOI] [PubMed] [Google Scholar]
- 6.Chan WK, Nakanishi K, Ebrey TG, Honig B. J Am Chem Soc. 1974;96:3642–3644. doi: 10.1021/ja00818a045. [DOI] [PubMed] [Google Scholar]
- 7.Crouch R, Purvin V, Nakanishi K, Ebrey T. Proc Natl Acad Sci USA. 1975;72:1538–1542. doi: 10.1073/pnas.72.4.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeGrip WJ, Liu RSH, Ramamurthy V, Asato A. Nature. 1976;262:416–418. doi: 10.1038/262416a0. [DOI] [PubMed] [Google Scholar]
- 9.Yoshizawa T, Fukada Y. Biophys Struct Mech. 1983;9:245–258. doi: 10.1007/BF00535660. [DOI] [PubMed] [Google Scholar]
- 10.Shichida Y, Nakamura K, Yoshizawa T, Trehan A, Denny M, Liu RSH. Biochemistry. 1988;27:6495–6499. doi: 10.1021/bi00417a044. [DOI] [PubMed] [Google Scholar]
- 11.Koutalos Y, Ebrey TG, Tsuda M, Odashima K, Lien T, Park MH, Shimizu N, Derguini F, Nakanishi K, Gilson HR, Honig B. Biochemistry. 1989;28:2732–2739. doi: 10.1021/bi00432a055. [DOI] [PubMed] [Google Scholar]
- 12.Fukada Y, Okano T, Shichida Y, Yoshizawa T, Trehan A, Mead D, Denny M, Asato AE, Liu RSH. Biochemistry. 1990;29:3133–3140. doi: 10.1021/bi00464a033. [DOI] [PubMed] [Google Scholar]
- 13.Lou J, Tan Q, Karnaukhova E, Berova N, Nakanishi K, Crouch RK. Methods Enzymol. 2000;315:219–237. doi: 10.1016/s0076-6879(00)15846-x. [DOI] [PubMed] [Google Scholar]
- 14.Isayama T, Chen Y, Kono M, DeGrip WJ, Ma J-X, Crouch RK, Makino CL. Vis Neurosci. 2006;23:899–908. doi: 10.1017/S0952523806230256. [DOI] [PubMed] [Google Scholar]
- 15.Ebrey T, Koutalos Y. Prog Retin Eye Res. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- 16.Crescitelli F. Vision Res. 1984;24:1551–1553. doi: 10.1016/s0042-6989(84)80004-8. [DOI] [PubMed] [Google Scholar]
- 17.Kefalov VJ, Estevez ME, Kono M, Goletz PW, Crouch RK, Cornwall MC, Yau K-W. Neuron. 2005;46:879–890. doi: 10.1016/j.neuron.2005.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto H, Tokunaga F, Yoshizawa T. Biochim Biophys Acta. 1975;404:300–308. doi: 10.1016/0304-4165(75)90337-2. [DOI] [PubMed] [Google Scholar]
- 19.Chen JG, Nakamura T, Ebrey TG, Ok H, Konno K, Derguini F, Nakanishi K, Honig B. Biophys J. 1989;55:725–729. doi: 10.1016/S0006-3495(89)82871-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ala-Laurila P, Cornwall MC, Crouch RK, Kono M. J Biol Chem. 2009;284:16492–16500. doi: 10.1074/jbc.M109.004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merbs SL, Nathans J. Science. 1992;258:464–466. doi: 10.1126/science.1411542. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Asenjo AB, Oprian DD. Biochemistry. 1993;32:2125–2130. doi: 10.1021/bi00060a001. [DOI] [PubMed] [Google Scholar]
- 23.Asenjo AB, Rim J, Oprian DD. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 24.Imai H, Kojima D, Oura T, Tachibanaki S, Terakita A, Shichida Y. Proc Natl Acad Sci USA. 1997;94:2322–2326. doi: 10.1073/pnas.94.6.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babu KR, Dukkipati A, Birge RR, Knox BE. Biochemistry. 2001;40:13760–13766. doi: 10.1021/bi015584b. [DOI] [PubMed] [Google Scholar]
- 26.Fasick JI, Applebury ML, Oprian DD. Biochemistry. 2002;41:6860–6865. doi: 10.1021/bi0200413. [DOI] [PubMed] [Google Scholar]
- 27.Kuwayama S, Imai H, Hirano T, Terakita A, Shichida Y. Biochemistry. 2002;41:15245–15252. doi: 10.1021/bi026444k. [DOI] [PubMed] [Google Scholar]
- 28.Kono M, Crouch RK, Oprian DD. Biochemistry. 2005;44:799–804. doi: 10.1021/bi047898f. [DOI] [PubMed] [Google Scholar]
- 29.Kuwayama S, Imai H, Morizumi T, Shichida Y. Biochemistry. 2005;44:2208–2215. doi: 10.1021/bi047994g. [DOI] [PubMed] [Google Scholar]
- 30.Crescitelli F, Liu RSH. Proc R Soc Lond B. 1988;233:55–76. doi: 10.1098/rspb.1988.0012. [DOI] [PubMed] [Google Scholar]
- 31.Liang J, Govindjee R, Ebrey TG. Biochemistry. 1993;32:14187–14193. doi: 10.1021/bi00214a018. [DOI] [PubMed] [Google Scholar]
- 32.Liang J, Steinberg G, Livnah N, Sheves M, Ebrey TG, Tsuda M. Biophys J. 1994;67:848–854. doi: 10.1016/S0006-3495(94)80544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis JW, Liang J, Ebrey TG, Sheves M, Livnah N, Kuwata O, Jäger S, Kliger DS. Biochemistry. 1997;36:14593–14600. doi: 10.1021/bi9712908. [DOI] [PubMed] [Google Scholar]
- 34.Das J, Crouch RK, Ma J-x, Oprian DD, Kono M. Biochemistry. 2004;43:5532–5538. doi: 10.1021/bi036097u. [DOI] [PubMed] [Google Scholar]
- 35.Yokoyama S. Prog Retin Eye Res. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- 36.Ganter UM, Schmid ED, Perez-Sala D, Rando RR, Siebert F. Biochemistry. 1989;28:5954–5962. doi: 10.1021/bi00440a036. [DOI] [PubMed] [Google Scholar]
- 37.Buczylko J, Saari JC, Crouch RK, Palczewski K. J Biol Chem. 1996;271:20621–20630. doi: 10.1074/jbc.271.34.20621. [DOI] [PubMed] [Google Scholar]
- 38.Han M, Groesbeek M, Sakmar TP, Smith SO. Proc Natl Acad Sci USA. 1997;94:13442–13447. doi: 10.1073/pnas.94.25.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kefalov VJ, Cornwall MC, Crouch RK. J Gen Physiol. 1999;113:491–503. doi: 10.1085/jgp.113.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin J, Crouch RK, Corson DW, Katz BM, MacNichol EF, Cornwall MC. Neuron. 1993;11:513–522. doi: 10.1016/0896-6273(93)90155-k. [DOI] [PubMed] [Google Scholar]
- 41.Crouch RK, Veronee CD, Lacy ME. Vision Res. 1982;22:1451–1456. doi: 10.1016/0042-6989(82)90209-7. [DOI] [PubMed] [Google Scholar]
- 42.Kono M, Goletz PW, Crouch RK. Biochemistry. 2008;47:7567–7571. doi: 10.1021/bi800357b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakanishi K. Amer Zool. 1991;31:479–489. [Google Scholar]
- 44.Schoenlein RW, Peteanu LA, Mathies RA, Shank CV. Science. 1991;254:412–415. doi: 10.1126/science.1925597. [DOI] [PubMed] [Google Scholar]
- 45.Ahmad R, Wheedon BCL. J Am Chem Soc. 1953;1953:3299–3315. [Google Scholar]
- 46.Robinson PR. Methods Enzymol. 2000;315:207–218. doi: 10.1016/s0076-6879(00)15845-8. [DOI] [PubMed] [Google Scholar]
- 47.Wessling-Resnick M, Johnson GL. J Biol Chem. 1987;262:3697–3705. [PubMed] [Google Scholar]
- 48.Yu H, Kono M, McKee TD, Oprian DD. Biochemistry. 1995;34:14963–14969. doi: 10.1021/bi00046a002. [DOI] [PubMed] [Google Scholar]