Abstract
Natural Killer T (NKT) cells respond to a variety of CD1d-restricted antigens (Ags), although the basis for Ag discrimination by the NKT cell receptor (TCR) is unclear. Here we describe NKT TCR fine specificity against several closely related Ags, termed altered glycolipid ligands (AGLs), which differentially stimulate NKT cells. The structures of five ternary complexes all revealed similar docking. Acyl chain modifications did not affect the interaction, but reduced NKT cell proliferation, indicating an affect on Ag processing or presentation. Conversely, truncation of the phytosphingosine chain caused an induced fit mode of TCR binding that affected TCR affinity. Modifications in the glycosyl head group had a direct impact on the TCR interaction and associated cellular response, with ligand potency reflecting the t1/2 life of the interaction. Accordingly, we have provided a molecular basis for understanding how modifications in AGLs can result in striking alterations in the cellular response of NKT cells.
Introduction
Natural Killer T (NKT) cells express specific αβ T cell receptors (TCRs) that recognize glycolipid antigens (Ag) presented by the non-classical MHC class I molecule, CD1d (Godfrey et al., 2004). Most human NKT cells express an invariant Vα24-Jα18 TCR α-chain, paired with a Vβ11 TCR β-chain, while murine NKT cells similarly express an invariant Vα14-Jα18 TCR α-chain, paired with 1 of 3 different TCR β-chain V genes (Vβ8, Vβ7, Vβ2) of which Vβ8.2, the homologue of Vβ11, is the most common (Bendelac et al., 2007; Godfrey et al., 2008). Upon activation, NKT cells rapidly produce an array of cytokines and can influence immune outcomes in a broad range of settings, including microbial immunity, tumor immunity, autoimmunity and allergy (Godfrey et al., 2004). α-galactosylceramide (α-GalCer) (Kawano et al., 1997), a synthetic α-glycolipid widely used for activating NKT cells, is considered as a surrogate for a largely unknown repertoire of natural NKT ligands and is now used experimentally in preclinical and clinical translational studies as a potent NKT cell agonist (reviewed in (Brutkiewicz, 2006)).
The structures of human and mouse NKT TCRs, unliganded, and in complex with CD1d-α-GalCer have been determined, as well as the mouse NKT TCR in complex with α-galactosyl-diacylglycerol (α-GalDAG) (Borg et al., 2007; Kjer-Nielsen et al., 2006; Li et al., 2010; Pellicci et al., 2009). These ternary complexes showed a conserved docking strategy that differs from all known TCR-peptide-MHC interactions (Godfrey et al., 2008). The invariant NKT TCR α-chain dominates this interaction, with the CDR1α loop interacting solely with the lipid Ag, whereas the CDR3α loop plays a central role, contacting CD1d and the lipid Ag (Borg et al., 2007; Pellicci et al., 2009). The role of the human Vβ11, and the homologous mouse Vβ8.2-chain is essentially restricted to the CDR2β loop which interacts with CD1d. Comparison of the mouse Vβ8.2 and Vβ7 NKT TCRCD1d-α-GalCer complexes shows similar docking, although conformational changes alters some of the contacts between the TCR α-chain with CD1d-α-GalCer, indicating that differential Vβ-chain usage could impact on CD1d-restricted Ag specificity (Pellicci et al., 2009).
NKT TCRs can bind an array of different lipid-based Ags in complex with CD1d, including bacteria-derived lipid Ags such as α-glycuronosylceramides, α-galactosyl-diacylglycerols and phosphatidyl-inositol mannosides, and mammalian lipid molecules, including the ceramides: iGb3, GD3 and β-glucosylceramide (reviewed in (Brutkiewicz, 2006; Godfrey et al., 2010)). Structural studies have provided insight into how CD1d can present natural and synthetic Ags (reviewed in (Godfrey et al., 2010)), although it is mostly unknown at the structural level how alterations in the CD1d-Ag landscape affect NKT TCR recognition. Some insight into the adaptability of the NKT TCR to recognise distinct CD1d-restricted Ags arose from NKT TCR mutagenesis experiments that underscored the importance of the CDR1α, CDR3α and CDR2β loops, yet also showed that some NKT TCR residues that contact Ag were not essential (Borg et al., 2007; Florence et al., 2009; Mallevaey et al., 2007; Pellicci et al., 2009; Scott-Browne et al., 2007; Wun et al., 2008). Further, glycolipids with different lipid tail lengths can alter the affinity for the NKT TCR, with the mechanism speculated to occur through alterations of the F′-pocket of CD1d, a major point of contact with the NKT TCR (McCarthy et al., 2007; Sullivan et al., 2010). While the NKT TCR α-chain is invariant, the β-chain plays a role in determining thresholds of Ag reactivity, and that this effectively enables some NKT TCRs to differentiate between Ags (Mallevaey et al., 2009). Furthermore, the composition of the CDR3β loop might determine CD1d autoreactivity to self Ags (Matulis et al., 2010)(Mallevaey et al. 2011, Immunity in press). Nevertheless, how the NKT TCR can recognize distinct self and non-self CD1d-Ags and possesses the capacity to discriminate between closely-related Ags, remains unclear (Gapin, 2010).
Altered Peptide Ligands (APLs) can dramatically affect MHC-restricted T-cell mediated biological outcomes (Sloan-Lancaster and Allen, 2003), yet cause only slight conformational readjustments at the TCR-pMHC-I interface (Baker et al., 2000; Degano et al., 2000). Paralleling this, numerous synthetic agonist analogues of α-GalCer have been described that differ in the structure of the glycosyl head group and/or lipid tails (reviewed in (Venkataswamy and Porcelli, 2009)). These represent the glycolipid equivalent of APLs, and have remarkably distinct effects on NKT cell function. Some analogues (such as OCH and C20:2) have the ability to promote T helper 2 (Th2) cell biased responses (Yu et al., 2005), whereas others (such as α-C-glycoside (Schmieg et al., 2003)) promote T helper1 (Th1) cell-biased responses. Thus, studies using glycolipid analogues demonstrate the feasibility of using these to manipulate the NKT cell response, which can translate to more tailored NKT cell based therapies (reviewed in (Cerundolo et al., 2009)). However, to achieve more targeted NKT-based therapeutics requires a greater understanding of the molecular basis of antigenic modulation of the NKT cell response. Here we report the structural and functional correlates of NKT TCR recognition of a series of closely related CD1d-restricted Altered Glycolipid Ligands (AGLs). Our findings provide a systematic analysis of the mechanisms by which NKT TCR interactions with closely related glycolipid ligands presented by CD1d influences fine specificity and functional responses.
Results
Affinity hierarchy for the glycolipid antigens
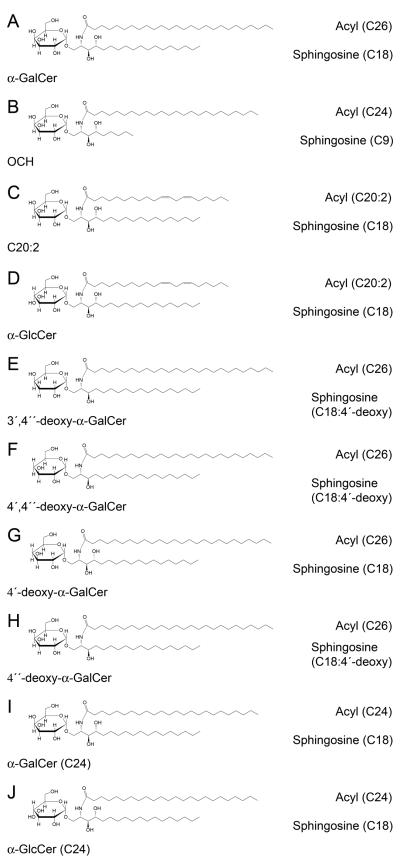
α-GalCer, also known as KRN7000, is the prototypical agonist for NKT cells that possesses an 18C phytosphingosine and a 26C acyl chain (Kawano et al., 1997). Because many analogues of this Ag have been generated that promote distinct NKT cell responses (reviewed in (Venkataswamy and Porcelli, 2009)), a critical question is whether the altered functionality is determined by their differential impact on TCR binding. We have investigated a panel of different AGLs (Figure 1A-J): (a) α-GalCer (b) OCH - with C9 phytosphingosine and C24:0 acyl chains (Miyamoto et al., 2001); (c) C20:2 - a C20 acyl chain and cis-diunsaturation at C11 and 14 (Yu et al., 2005); (d) α-GlcCer - identical ceramide component to C20:2, but α-linked glucosyl head group (Jervis et al., 2010); (e) 3′,4″-deoxy-α-GalCer – both the 3′-OH of the sugar, and the 4′-OH of the sphingoid base, are removed (Raju et al., 2009); (f) 4′,4″-deoxy-α-GalCer – both the 4′-OH of the sugar, and the 4′-OH of the sphingoid base, are removed (Raju et al., 2009). To control for the dual modifications separately, we synthesised a new AGL (g) 4′-deoxy-α-GalCer, with a 4′-deoxy-sugar but retaining the 4′-OH group on the sphingoid base. For functional studies (below) we also analysed (h) the 4″-deoxy-α-GalCer AGL (with an α-Gal sugar, but lacking the 4′-OH on the sphingoid base on the sphingoid base). We also included (i) α-GalCer (C24) and (j) α-GlcCer (C24), both with 24C acyl and 18C sphingosine chains, further enabling a direct comparison of the effect of modifying the orientation of the 4′-OH position of the sugar.
Fig. 1. Schematic of α-GalCer and the AGLs.
(A) α-GalCer (B) OCH (C) α-GalCer (C20:2) (D) α-GlcCer (C20:2) (E) 3′4″-deoxy-α-GalCer (F) 4′4″-deoxy-α-GalCer (G) 4′-deoxy-α-GalCer (H) 4″-deoxy-α-GalCer (I) α-GalCer (C24) (J) α-GlcCer (C24)
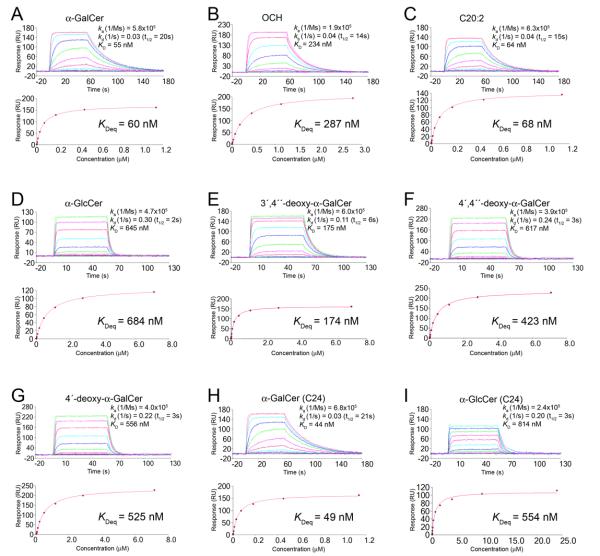
First, we determined the affinity of the interaction between the mouse Vβ8.2 NKT TCR and the CD1d-AGL complexes using surface plasmon resonance (SPR). Purified mouse CD1d was loaded with the eight individual AGLs, or α-GalCer as a positive control. The affinity as determined by responses at equilibrium (equilibrium dissociation constant, KDeq) of the TCR for mCD1d-α-GalCer was ≈ 0.059 μM, similar to previous reports. In contrast, the NKT TCR affinity for some, but not all, of the CD1d-AGL complexes deviated from that of CD1d-α-GalCer (Figure 2A-I, Supplementary Table 1). For example, the affinity of the NKT TCR-CD1d-α-GalCer (C20:2) interaction, at 0.068 μM (Figure 2C), and the NKT TCR-CD1d-α-GalCer (C24) interaction (KDeq = 0.049 μM) (Figure 2H) was essentially identical to that of the positive control, indicating that the modifications to the extent of acyl chain saturation and truncation did not impact on NKT TCR affinity. This result is consistent with earlier studies based on α-GalCer-CD1d tetramer versus C20:2-CD1d tetramer dissociation from cells (Im et al., 2009). The CD1d-OCH complex exhibited ≈ 0.3 μM affinity for the NKT TCR (Figure 2B), approximately 20% of the affinity of the NKT TCR-CD1d-α-GalCer interaction. While this is broadly consistent with previous avidity studies using CD1d tetramer dissociation, this affinity was still higher than expected from earlier studies (Im et al., 2009; Sullivan et al., 2010). This discrepancy may reflect differences between molecular affinity and tetramer avidity. Nonetheless, given that composition of the solvent exposed sugar is identical between OCH and α-GalCer, these data suggested that the modified sphingosine chain, which was buried within the F′-pocket, can indirectly affect head group presentation or CD1d conformation and therefore NKT TCR recognition. The affinity of the NKT TCR-CD1d-3′,4″-deoxy-α-GalCer interaction, at ≈ 0.2 μM (Figure 2E), was only approximately 30% of the affinity compared to the positive control, suggesting that the 3′-OH moiety of the galactosyl headgroup influenced, but was not critical for, the interaction. In contrast, the α-GlcCer (C24) and the α-GlcCer (C20:2) exhibited a much reduced affinity (KDeq of 0.55 μM and 0.68 μM respectively), suggesting that the 4′-OH moiety was a key determinant in the NKT TCR interaction (Figure 2D & I). Consistent with this, the two 4′-deoxy-α-GalCer AGLs (Figure 2F & G), which also differ from α-GalCer via modifications at the 4′-OH position on the sugar, also exhibited a much reduced affinity, at ≈ 0.5 μM. Thus, the 4′-OH moiety on the galactose headgroup was a key determinant in the NKT TCR interaction, and furthermore, modification of the 4″-OH group on the sphingosine chain did not impact on the lower affinity NKT TCR interaction with these AGLs.
Fig. 2. Analysis of the interaction between Vβ8.2 NKT TCR and the CD1d-analogues as assessed by surface plasmon resonance (SPR).
SPR sensograms showing the interaction between Vβ8.2 NKT TCR and CD1d loaded with (A) α-GalCer (B) OCH (C) C20:2 (D) α-GlcCer (E) 3′4″-deoxy α-GalCer (F) 4′4″-deoxy-α-GalCer (G) 4′-deoxy-α-GalCer (H) α-GalCer (C24) (I) α-GlcCer (C24). NKT TCR were injected over streptavidin immobilised CD1d-Ag and simultaneously over a control cell containing unloaded CD1d. (Top) Sensograms show the binding (response units, RU) of increasing concentrations of TCR to CD1d-Ag following baseline subtraction, displaying data points overlaid with curve fits. (Bottom) Saturation plots demonstrating equilbrium binding of NKT TCR to immobilised CD1d-Ag. The affinities derived by equilibrium analysis (KDeq) were equivalent to those derived by kinetic analysis.
Given that the affinity of the NKT TCR-CD1d-Ag interaction may not necessarily correlate with ligand potency, we also determined the kinetic rate constants of these interactions (Figure 2A-I). There was a very good correlation between the KD calculated by kinetic and equilibrium analyses, and the residual plots and low Chi2 values indicated that the kinetic rate constants were measured accurately. In comparison to the association and dissociation rate constants typical of TCR-pMHC interactions (van der Merwe and Davis, 2003), the NKT TCR engages CD1d-AGL complexes with fast on rates (ka) and slow off rates (kd), with a long t1/2 life of approximately 20s for the NKT TCRCD1d-α-GalCer positive control interaction. AGLs with modifications in the acyl tail did not appreciably affect the ka or kd of the interaction, whereas OCH impacted negatively on the association constant, which is suggestive of an induced fit mechanism for TCR-CD1d-OCH ligation. Interestingly, the AGLs with modifications in the glycosyl headgroup had a greater effect on the off-rate than the on-rate, with the off-rate increasing by up to a factor of 10, and as such the t1/2 life of these interactions were markedly shorter. Thus, these AGLs did not impact on the formation of the NKT TCR-CD1d-AGL complex, but resulted in less stable complexes being formed.
In summary, the structural modifications represented by these CD1d-AGL complexes showed different effects on the affinity of the NKT TCR interaction, with the hierarchical order: α-GalCer = α-GalCer (C20:2) = α-GalCer (C24) > 3′,4″-deoxy > OCH > 4′-deoxy =4′,4″-deoxy = α-GlcCer (C20:2) = α-GlcCer (C24). When the t1/2 life was used as a guide, this order was preserved, with the exception of OCH, which had a markedly longer half life when compared to that of the 3′,4″-deoxy AGL.
NKT cell proliferative response shows a contrasting hierarchy to affinity measurements
To investigate the relationship between the affinity of the AGLs and their ability to induce NKT cell proliferation, the AGLs were tested in an in vitro assay using carboxyfluorescein diacetate succiniimidyl ester (CFSE) labelled, NKT cell-enriched thymocytes co-cultured with splenocytes derived from Tcra-Jtm1Tg mice (Jα18−/−) as a source of Ag presenting cells (APC) (Figure 3A and B). Structural variations within the ceramide component had the greatest impact on NKT cell proliferation, especially evident at the low dose of 1 ng/ml. At this concentration, the AGLs with non-truncated sphingosine and acyl chains all produced robust proliferative responses. OCH also induced proliferation at 1 ng/ml; however, this was consistently weaker in comparison to the non-truncated AGLs. The major difference between OCH and α-GalCer is the truncated C9 instead of C18 phytosphingosine chain, although OCH also has a slightly shorter C24 acyl chain. The reduced NKT cell proliferation with OCH appeared to be due to the phytosphingosine chain truncation because the C24 acyl chain variant of α-GalCer drove proliferation to a similar extent to C26 α-GalCer. In contrast, the AGLs containing two double bonds within the acyl chain (C20:2 and α-GlcCer C20:2) barely induced NKT cell proliferation at 1 ng/ml. They were, however, able to induce proliferation at a higher dose (100 ng/ml) to a similar degree as their fully saturated counterparts. This suggested that modifications in the degree of saturation within the acyl chain can have an impact on the efficacy of glycolipid ligands as NKT cell agonists. In further support of this, α-GlcCer with a C24 saturated acyl chain induced similar NKT cell proliferation to α-GalCer with the C24 acyl chain. Given that the hierarchy in terms of proliferative response is clearly different from the affinity data, this suggested that the potency of the AGLs for inducing proliferation of NKT cells did not correlate precisely with the affinity or kinetics of the NKT TCR interactions. This implied that other factors besides the equilibrium binding affinity or kinetics of the NKT TCR can contribute to the biological responses of NKT cells. This is consistent with reports showing that APC processing and presentation can vary for different glycolipid ligands, with some, such as α-GalCer, being more dependent on CD1d internalisation, lysosomal processing, and re-expression in the context of plasma membrane lipid rafts, than others, such as OCH and C20:2 (Bai et al., 2009; Im et al., 2009; Sullivan et al., 2010). Therefore, we also established a plate bound CD1d assay to test the ability of the different analogues in the absence of APCs. This approach was used to assess the activation of an NKT cell hybridoma, as well as proliferation of freshly isolated NKT cells (Figure 3C). In both cases, the hierarchy of the response was clearly distinct to that observed when APCs were used, and correlated more closely with the affinity and t1/2 life of the ligands as determined by our SPR experiments. Collectively, these studies support the concept that while TCR affinity for different AGLs can influence NKT cell proliferation, differential Ag-presentation of these ligands appears to be a major factor in determining the threshold of the proliferative response.
Fig. 3. NKT cell proliferative response to AGLs.
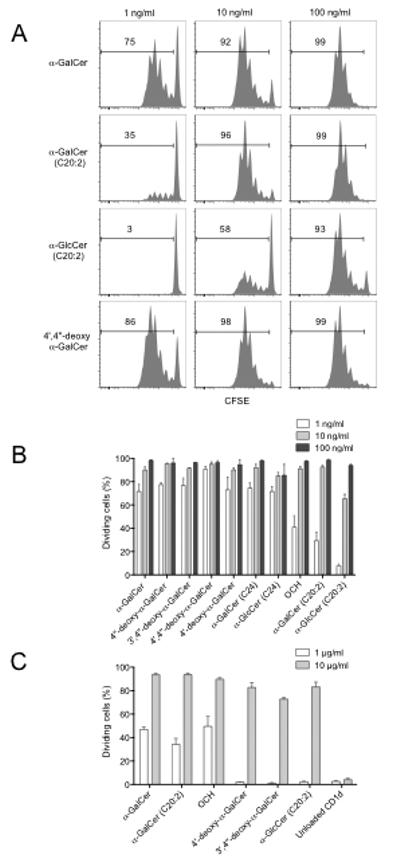
For (A) and (B), CFSE labelled thymocytes enriched for NKT cells were cultured for 3d with glycolipid-pulsed Tcra-Jtm1Tg (Jα18−/−) splenocytes. (A) A selection of representative flow cytometry profiles showing percent proliferated cells based on decreased CFSE intensity of α-GalCer-CD1d tetramer+ NKT cells at each AGL dose. (B) Graph depicts percentage divided cells (mean ± SEM) from 2-6 independent experiments carried out as above. (C) MACS enriched CD1d-α-GalCer tetramer+ thymic NKT cells (2×104) were CFSE-labelled and cultured in plates pre-coated with either 10 or 1 μg/ml CD1d-glycolipid complex. After 72h CFSE dilution was measured by flow cytometry. Graphs depict the percentage of divided NKT cells ± SEM of n=3 replicates per group from a single experiment.
Cytokine Response correlates with affinity hierarchy
While NKT cells are capable of producing a broad range of cytokines in response to α-GalCer (reviewed in (Godfrey et al., 2004), the amounts and ratio of the different cytokines produced may be modulated in response to different glycolipid AGLs (Im et al., 2009). Therefore, we tested cells and cell supernatants from similar cultures to those shown in Figures 3A and B for cytokine production. IFN-γ production by NKT cells was assessed by intracellular cytokine staining (ICS) while the production of IFNg, IL-4, IL-13 and IL-17 within the culture supernatant was determined using a cytometric bead array (CBA) assay after 8h stimulation with each of the AGLs (Figures 4A and 4B). Cytokine assays were tested following stimulation with the higher dose (100 ng/ml) at which proliferative responses to the AGLs were all comparable (Figures 3A and B). Interestingly, cytokine production by NKT cells did not correlate with proliferative potential for the AGLs under similar culture conditions. For example, the AGLs with unmodified galactose residues (α-GalCer, 4″-deoxy-α-GalCer, C20:2, OCH and α-GalCer (C24)) induced the highest percentages of IFN-γ producing NKT cells, whereas α-GlcCer (C20:2) repeatedly induced the least IFN-γ production of all and α-GlcCer (C24) was only marginally better at inducing IFN-γ production. The AGLs with the modified galactose moieties (3′,4″-deoxy, 4′,4″-deoxy-α-GalCer and 4′-deoxy-α-GalCer) were also less potent inducers of IFN-γ production from NKT cells in comparison to α-GalCer, despite a comparable capacity to induce NKT cell proliferation (Figure 4A). A similar pattern of cytokine production was observed for IFN-γ, IL-4, IL-13 and IL-17 in the culture supernatants in response to the different AGLs as measured by CBA (Figure 4B). While some AGLs, such as OCH and C20:2, are considered to be Th2-biasing Ags, this was not apparent in our assays. However, the Th2 biasing capacity of these agents is considered to be more a reflection of the differential ability of these ligands to promote downstream IFN-γ production by bystander cells, such as NK cells (Sullivan et al 2010) rather than a direct effect on NKT cell cytokine production. Indeed, direct analysis of cytokine production by NKT cells by ICS at 2 and 6 hours after in vivo challenge with OCH fails to reveal a Th2 bias by the NKT cells directly, when compared to α-GalCer (Sullivan et al 2010). When we compared serum cytokines 2 and 20 hours after in vivo treatment with α-GalCer, OCH and C20:2, the ratio of IL-4 to IFN-γ was similar to previous reports, showing that α-GalCer provided a more sustained IFN-γ response at 20h compared to C20:2 and OCH (Supplementary Figure 1).
Fig. 4. NKT cell cytokine response to AGLs.
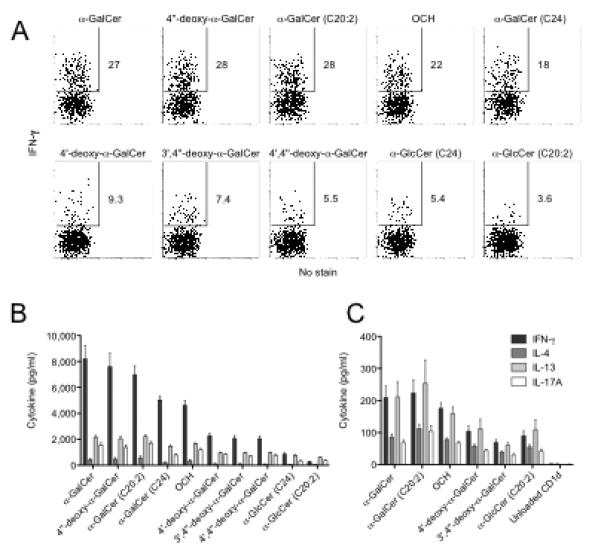
For (A) and (B), thymocytes enriched for NKT cells were cultured for 8h with Tcra-Jtm1Tg (Jα18−/−) splenocytes previously pulsed with glycolipid (100ng/ml). GolgiStop (BD Biosciences) was added for the last 4h (A) The percentage IFN-γ+ α-GalCer-CD1d tetramer+ NKT cells was detected by ICS. Data is representative of 4 similar experiments. (B) Culture supernatants were collected at 8h and cytokine amounts were quantified using CBA (BD Biosciences). Data is taken from one of at least 3 representative experiments, (n=5 replicates), graphs depict mean and SEM. (C) Supernatants from cells stimulated by plate bound CD1d-glycolipid complex, as described in Fig. 3C, were harvested at 72hr and cytokines analysed by CBA. Graphs depict the concentration of cytokines ± SEM of n=3 replicates per group from a single expt.
These data demonstrated that, in contrast to the proliferation of fresh NKT cells in response to the different AGLs presented by APCs, cytokine production by NKT cells in this assay broadly reflected the TCR affinity of these ligands. A similar pattern of cytokine production was detected in culture supernatants that were harvested from a plate bound CD1d-AGL stimulation assay (Figure 4C).
Structures of the NKT TCR-CD1d-AGL complexes
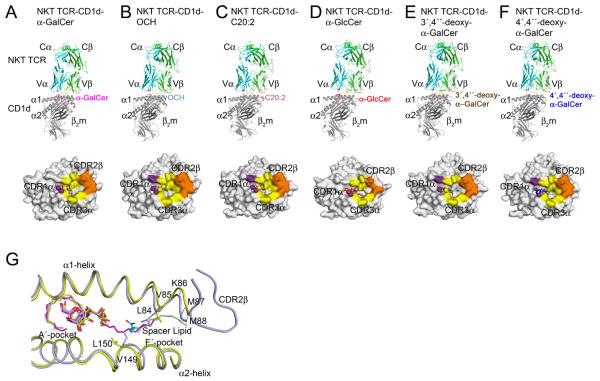
To address the structural basis of the fine specificity of the NKT TCR, we determined the structure of five NKT TCR-CD1d complexes containing different AGLs, namely OCH, C20:2, α-GlcCer, 3′,4″-deoxy-α-GalCer and 4′,4″-deoxy-α-GalCer (Supplementary Table 2, Supplementary Figure 2, Figure 5). These complexes all crystallized in the same space group with similar unit cell dimensions, and thus any structural differences observed can be attributed to the impact of the specific AGL.
Fig. 5. Structure of the NKT TCR in complex with different α-GalCer AGLs.
Ribbon representation of the NKT TCR-CD1d-Ag structures with the corresponding footprints shown underneath. (A) NKT TCR in complex with CD1d-α-GalCer. (B) NKT TCR-CD1d-OCH. (C) NKT TCR-CD1d-C20:2. (D) NKT TCR-CD1d-α-GlcCer. (E) NKT TCR-CD1d-3′4″-deoxy-α-GalCer. (F) NKT TCR-CD1d-4′4″-deoxy-α-GalCer. Footprints are color-coded based on the CDR loop contributions. TCRα, cyan; TCRβ, green; CDR1α loop, purple; CDR3α loop, yellow; CDR2β loop, orange; CD1d heavy chain, grey; β2m, dark grey; α-GalCer, magenta; C20:2, pink; α-GlcCer, red; 3′4″-deoxy-α-GalCer, brown; 4′4″-deoxy-α-GalCer, blue; OCH, light blue; Spacer lipid, light green. (G) NKT TCR induced fit of CD1d-OCH within the F′-pocket. Comparison of the binary CD1d-OCH (yellow; spacer lipid, cyan) with the ternary NKT TCR-CD1d-OCH complex(light blue; spacer lipid, light green). α-GalCer is shown in magenta.
In all 5 complexes, the NKT TCR adopted the characteristic docking mode initially observed for the NKT TCR-CD1d-α-GalCer complex (Borg et al., 2007; Pellicci et al., 2009), indicating a conserved footprint without any re-positioning of the NKT TCR caused by the AGLs (Figure 5A - F). For 4 of the complexes (OCH, C20:2, 3′,4″-deoxy- & 4′,4″-deoxy-α-GalCer), the buried surface area (BSA) upon ligation was 740-790 Å2, which compares closely to the BSA value of 760 Å2 at the NKT TCR-CD1d-α-GalCer interface (Figure 5A, B, C, E & F). Within the footprints of these 4 complexes the α-chain contributed approximately three times more BSA than the β-chain, in which the Vα14 and Jα18-encoded interactions were mediated by the CDR1α and CDR3α respectively (approximately 18% versus 57% BSA respectively). Further, for all 5 complexes, the Vβ8.2 interactions were mediated solely via the CDR2β loop interacting with the α1-helix of CD1d (approximately 24% BSA). This conserved set of CDR2β-mediated interactions included Tyr 48β and Tyr 50β interacting with Glu 83 and Lys 86 of CD1d, the latter of which formed vdw interactions with Glu 56β (Supplementary Table 3). This is comparable to what was previously observed for the NKT TCR-CD1d-a-Galcer interaction.
For the NKT TCR-CD1d-α-GlcCer complex, the BSA was only ≈ 600 Å, with the CDR1α, CDR3α and CDR2β-mediated interactions contributing 19%, 47%, and 33% BSA respectively (Figure 5D). The lower BSA was attributable to the increased mobility of the CDR3α loop within this complex, in which Leu 99α was not resolved in the electron density, and thus was not included in the final refined model. In the NKT TCR-CD1d-α-GalCer complex, Leu 99α made specificity-governing contacts with Arg 79 and Val 149 of CD1d (Supplementary Table 3). Thus, the increased mobility of Leu 99α in the α-GlcCer complex would contribute towards the lower affinity of this interaction.
To evaluate if the 5 AGLs caused any structural perturbations within CD1d, their respective CD1d Ag-binding clefts were compared to the NKT TCR-CD1d-α-GalCer complex and non-liganded counterparts if available (Figure 5G and not shown). Minimal root-mean-square-deviations (r.m.s.d.) (range 0.26 - 0.33 Å) were observed, indicating that the AGLs do not noticeably alter the structure of the CD1d-Ag binding cleft when ligated to the NKT TCR. Despite truncation and un-saturation of the acyl tails of C20:2 and α-GlcCer, the first 7 carbons of their acyl tails adopted a similar conformation to that observed in CD1d-α-GalCer. For CD1d-α-GalCer, the tip of the 26C acyl tail curled back on itself to form stabilising intra-vdw interactions. As only 7 of the 20 acyl tail carbons were observed for the C20:2 ternary complex, it suggests that the majority of the acyl chain lacks these stabilising intra-van der Waal (vdw) interactions and hence is mobile within the A′-pocket. Nevertheless, the A′-pocket of CD1d, when bound to the C20:2 and α-GlcCer, maintained a near identical conformation when compared to CD1d-α-GalCer. Previous studies involving α-GalCer analogues with modifications on the acyl tails reported the presence of a spacer lipid in the A′-pocket (Schiefner et al., 2009) however this was not observed in the NKT TCR-CD1d-C20:2 and α-GlcCer complexes. We suggest that this is attributable to the lengthier acyl chain analogues used in our studies, which could be expected to sterically hinder the binding of a spacer lipid.
While the 4″-deoxy modification on the sphingosine tail of the 3′,4″-deoxy- and 4′,4″-deoxy α-GalCer AGLs did not affect the conformation of CD1d-Ag binding cleft, the 3′-OH on the sphingosine tail shifted slightly (≈ 0.7 Å) to accommodate for this modification (Figure 6). The acyl and sphingosine chains of the 3′,4″-deoxy- and 4′,4″-deoxy AGLs, when bound to CD1d, adopted a similar conformation to that observed in the CD1d-α-GalCer complex.
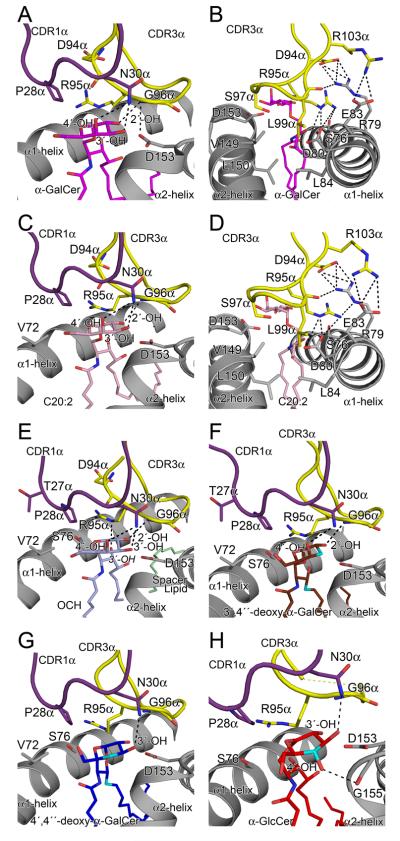
Fig. 6. Comparison of the glycsoyl head group NKT TCR interactions.
(A) α-GalCer (B) α-GalCer. Additional view of CDR3α loop interactions with CD1d. (C) C20:2 (D) C20:2. Additional view of CDR3α loop interactions with CD1d. (E) OCH. (F) 3′4″-deoxy-α-GalCer (G) 4′4″-deoxy-α-GalCer. (H) α-GlcCer. CDR1α loop, purple; CDR3α loop, yellow; CD1d α-helices, grey; α-GalCer, magenta; C20:2, pink; OCH, light blue; spacer lipid, light green; 3′4″-deoxy-α-GalCer, brown; 4′4″-deoxy-α-GalCer, blue; α-GlcCer, red; site of modification to analogues, cyan. H-bonds shown as dashed black lines.
Within the NKT TCR-CD1d-OCH complex, a spacer lipid was observed to occupy “the void” in the F′-pocket, essentially mimicking the end of the sphingosine tail as observed in CD1d-α-GalCer. However, the spacer lipid extended beyond this limit such that it almost protruded out of the F′-pocket and adopted a different conformation to the spacer lipid observed in the binary mouse CD1d-OCH structure (Sullivan et al., 2010) (Figure 5G). Interestingly, when the OCH complex was compared to α-GalCer complex, the truncated sphingosine tail of OCH did not cause a repositioning of the F′-roof of CD1d (not shown) (McCarthy et al., 2007; Sullivan et al., 2010). However, a comparison of the CD1d-Ag binding clefts of the binary CD1d-OCH complex (Sullivan et al., 2010) and the NKT TCR-CD1d-OCH complex, showed notable displacements (> 1 Å) for Leu 84, Val 85, Lys 86, Met 87, Met 88 and Val 149. Further, Leu 84 and Leu 150 in the binary CD1d-OCH structure adopted very different conformations when compared to that observed in the NKT TCR-CD1d-OCH complex (Figure 5G). While we cannot discount that these conformational differences between CD1d-OCH and NKT TCR-CD1d-OCH complexes were attributable to the spacer lipid, our results nevertheless indicated that the NKT TCR can operate via an induced-fit mechanism, whereby the NKT TCR moulds the F′-pocket of the CD1d-Ag binding cleft to enable optimal engagement.
The bases of NKT TCR fine specificity
Three of the α-GalCer AGLs, for which structures were determined (α-GlcCer, 3′,4″-deoxy- and 4′,4″-deoxy-α-GalCer), contained glycosyl modifications – the only moiety of the CD1d-restricted Ag that is surface exposed and thus directly contacted by the NKT TCR. The C20:2 and OCH AGLs possessed modifications in the acyl and sphingosine tails respectively, and the interaction between the α-galactosyl head group of these latter two AGLs and the NKT TCR was essentially identical to that observed in the NKT TCR CD1d-α-GalCer interaction (Figure 6A-E). Namely, for the C20:2 and OCH AGLs, the galactose ring sat underneath the CDR1α loop and abuts the CDR3α loop, forming vdw contacts on one face of the sugar ring with Arg 95α, Gly 96α and Pro 28α. Arg 95α also made vdw contacts with the 3′-OH of the sphingosine chain. Gly 96α H-bonds to the 2′-hydroxyl, while Asn 30α H-bonds to both the 3′- and 4′- OH groups of the galactose ring (Figure 6C & E). Further, within the NKT TCR-CD1d-OCH and C20:2 complexes, the CDR3α loop interacted with mCD1d. Arg 95α and Arg 103α flanked Arg 79 from CD1d, and these positive charges were dissipated by neighbouring acidic groups of Asp 94α, Glu 83 and Asp 80 (Figure 6D). Collectively, these CDR3α loop and CD1d residues provided a focussed network of polar and salt-bridging interactions.
As a direct result of modifications in the glycosyl head groups of α-GlcCer, 3′,4″-deoxy- and 4′,4″-deoxy-α-GalCer AGLs, the interactions with the NKT TCR were modified to varying degrees, although there was no notable juxta-positioning in the side chain conformations of the NKT TCR to compensate for these modifications (Supplementary Table 3). In the NKT TCR-CD1d-3′,4″-deoxy-α-GalCer AGL complex, the loss of the 3′-OH resulted in a loss of a H-bond to Asn 30α from the CDR1α loop and the vdw contacts with Gly 96α of the CDR3α loop; in addition, a H-bond to Asp153 of CD1d was lost that could potentially have an impact on the stability of the CD1d-3′,4″-deoxy-α-GalCer complex (Figure 6F). Collectively, the loss of these interactions also caused a slight tilting of the galactosyl head group, such that a maximal displacement of 0.7 Å with respect to the NKT TCR-CD1d-C20:2 complex was observed at the 5′-C position.
In the NKT TCR-CD1d-4′4″-deoxy-α-GalCer AGL complex, there was a greater shift of the galactosyl head group, with the entire ring being displaced down towards CD1d by approximately 0.9 Å. As a consequence, not only did the lack of the 4′-OH moiety result in a loss of a H-bond to Asn 30α, but also resulted in a loss of vdw contacts between the 3′-OH and CDR3α loop, and a lengthening of the H-bond between this 3′-OH moiety and Asn 30α (Figure 6G). Similarly, in the NKT TCR-CD1d-α-GlcCer AGL complex, the glucosyl head group was also displaced by approximately 0.9Å, but in this instance it repositioned along the roof of the A′-pocket, which also resulted in the 3′-OH moiety being too far away to interact with Gly 96α of CDR3α, as well as Asp 153 of CD1d (Figure 6H). Within this complex, the 4′-OH of the glucosyl ring pointed in a different direction to the 4′-OH of the galactosyl ring, resulting in a lost H-bond with Asn 30α of the TCR, and formation of an H-bond to the main chain of Gly 155 of CD1d.
Accordingly the effects of modification of the 4′-OH were more marked than for the 3′-OH, thereby providing a basis for understanding the greater dependency of the 4′-OH in NKT TCR recognition.
The contribution of TCR β-chain to AGL recognition
We have previously identified differences in the NKT TCR-CD1d-α-GalCer contacts depending on the use of Vβ8.2 or Vβ7 β-chain (Pellicci et al., 2009). In particular, vdw contacts with the 4′-OH on the galactosyl ring are observed for Vβ8.2 but not for Vβ7 NKT TCR. We therefore wanted to explore NKT TCR Vβ8.2 versus Vβ7 usage in response to the AGLs used in this study, particularly those that varied at the 4′-OH moiety. The percentage of NKT cells expressing Vβ7 or Vβ8.1 or 8.2 were analysed after 3 days of in vitro proliferation in the presence of the different AGLs (Figure 7A and 7B). While all AGLs tested were capable of activating both Vβ7 and Vβ8.1 and 8.2 NKT cells, those with an altered 4′-OH on the sugar (α-GlcCer, α-GlcCer (C24), 4′4″-deoxy-α-GalCer and 4′-deoxy-α-GalCer) induced higher percentages of Vβ7+ NKT cells than cultures stimulated with α-GalCer, α-GalCer (C24), C20:2 and OCH. We also determined the degree of Vβ usage with cell division by gating in individual cell division peaks (Figure 7C and data not shown). The bias toward Vβ7 proliferation was dependent on the altered glycosyl head group, because both 4′4″-deoxy-α-GalCer and 4′-deoxy-α-GalCer showed a Vβ7 bias, whereas 4″-deoxy α-GalCer did not. Similarly, both α-GlcCer (C20:2) and α-GlcCer (C24) showed a Vβ7 bias, while α-GalCer (C24) and α-GalCer (C20:2) did not. Thus, for AGLs with an altered 4′-OH sugar moiety, especially α-GlcCer, that Vβ7+ NKT cells were progressively enriched with each division cycle. Conversely, other AGLs, especially OCH, resulted in a loss of Vβ7+ NKT cells with each progressive division cycle, a result that is consistent with an earlier study suggesting that OCH is preferentially recognised by Vβ8+ NKT TCRs (Stanic et al., 2003). These data support the concept that AGLs not only have a direct impact on NKT cell function at the level of individual cells, but also at the population level where different NKT cell clones may be engaged depending on their TCR β-chain usage. With this in mind, we engineered an NKT cell hybridoma to express the same TCR β-chain that was used in our SPR and crystallography studies above. This hybridoma was challenged with the AGLs presented by in vitro generated bone marrow dendritic cells, and the response measured by IL-2 production after 24 hours (Figure 7D). These experiments showed a hierarchy of IL-2 production that broadly reflected the affinity of the different AGLs for this particular NKT TCR. Namely, C20:2 and α-GalCer provided a clear IL-2 response at lower doses, whereas α-GlcCer did not stimulate at the same lower doses and, even at higher doses, it induced less IL-2 than α-GalCer and C20:2, whereas 3′4″-deoxy-α-GalCer, 4′4″-deoxy-α-GalCer, 4′-deoxy-α-GalCer and OCH showed responses that were in between α-GalCer and α-GlcCer. This response was also distinct from the proliferation hierarchy observed for freshly isolated NKT cells (Figure 3A and B) but was largely consistent with the proliferation of fresh NKT cells in response to glycolipids presented by plate bound CD1d (Figure 3C), and with the hierarchy observed for the cytokine profiles derived from fresh NKT cells (Figure 4).
Fig. 7. Vβ7 usage and the CD1d-AGLs.
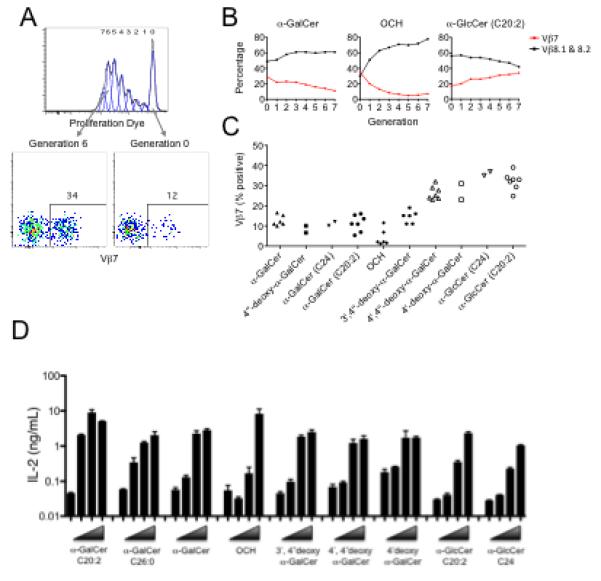
For (A-C) CFSE or eFluor 670 labelled thymocytes enriched for NKT cells were cultured for 3d with glycolipid-pulsed Tcra-Jtm1Tg (Jα18−/−) splenocytes and Vβ expression of α-GalCer-CD1d tetramer+ NKT cells was analysed by flow cytometry at distinct division cycles. (A) an example of the approach to determine the percentage of undivided, or division 6 cells that were Vβ7+ after culture in the presence of glycolipid. (B) percentage of Vβ7 or Vβ8.1-8.2+ cells within each division peak for 3 representative glycolipids. (C) Summary of results showing the percentage of Vβ7+ NKT cells within the most proliferated cells (from the last major generation) from 2-6 independent experiments. (D) Dose response analysis of NKT cell hybridoma, engineered to express the NKT TCR used in SPR and crystallography studies (see methods), stimulated overnight with BMDCs plus the various AGLs at concentrations from 125 nM to 125 pM. Each data point represents the mean of triplicate IL-2 readings ± SD from a single experiment. BMDCs derived from Cd1d−/− mice used in the same conditions did not stimulate the hybridoma (data not shown).
Discussion
NKT cells can recognise a variety of CD1d-restricted glycolipid Ags, yet can discriminate between closely related CD1d-restricted ligands (reviewed in (Godfrey et al., 2010; Venkataswamy and Porcelli, 2009)). α-GalCer is a clinicial trial phase I therapeutic of which many derivatives have been synthesized with a view to modulate the NKT cell response (reviewed in (Venkataswamy and Porcelli, 2009)). There is great interest in the ability of AGLs to modulate the NKT cell response, although the molecular basis for such effects remains unclear. Various explanations for the differential effects of AGLs on NKT cell function include: differing affinities, pharmacokinetics, involvement of different APCs; distinct CD1d loading kinetics; and distinct intracellular processing and presentation on the cell surface (Im et al., 2009; Sullivan et al., 2010) (Venkataswamy and Porcelli, 2009). Our study has formally demonstrated the structural and biophysical basis for NKT TCR fine specificity against a set of closely related glycosylceramide analogues, which we have termed altered glycolipid ligands (AGLs). We have also related these molecular and biochemical observations to the NKT cell proliferative response, cytokine production and NKT TCR Vβ usage. We selected eight AGLs, thereby gaining a broad perspective on the range of effects such modifications could impact on the structural and functional aspects of the NKT response.
In MHC-restricted immunity, subtle alterations of the antigenic peptide can markedly modulate the immune response. However, despite the dramatically different biological outcomes such APLs can elicit, only slight conformational readjustments are observed at the TCR-pMHC interface (Baker et al., 2000; Degano et al., 2000). Our findings mirror the APL studies of the MHC-restricted immune response, in that none of the AGLs studied had a profound impact on the NKT TCRCD1d-Ag interaction. In MHC-restricted immunity, neither the affinity nor the half-life of the interaction has been an absolute predictor of ligand potency (Sloan-Lancaster and Allen, 2003), leading to other factors such as dwell-time, TCR-pMHC confinement models and aggregate t1/2 (ta) being proposed as more reliable indicators of ligand potency (Aleksic et al., 2010; Govern et al., 2010). To compound matters, recent studies have shown that solution-based affinity measurements may not be fully reflective of what occurs in situ (Huppa et al., 2010). Moreover, the myriad of TCR-pMHC docking modes, suggests that a unifying parameter defining ligand potency in MHC-restricted immunity may prove challenging. In contrast however, CD1d is monomorphic, the NKT TCR is semi-invariant, and at present the mode of NKT TCR-CD1d-Ag docking appears to be highly conserved. Thus, a priori, affinity or t1/2 values may represent a more reliable indicator with respect to NKT TCR ligand potency. A confounding issue relating to CD1d restricted Ags however is that modifications in the lipid tails can impact on ligand potency indirectly, and this certainly seems to be the case for AGLs with acyl chain modifications. Our data indicated that the altered potency arising from acyl chain modifications can only arise from alterations in Ag-processing and presentation, as they neither impacted on the NKT TCR-CD1d-AGL interaction nor on the affinity or kinetics of the interaction. Interestingly however, modifications within the sphingoid base, as exemplified by the OCH AGL, impacted on the affinity of the interaction, ostensibly by causing a slower on-rate, indicative of an induced-fit interaction, consistent with our structural data and recent observations in NKT TCR recognition of a microbial ligand (Li et al., 2010). This parallels similar observations in TCR-pMHC recognition, whereby buried alterations (polymorphisms) within the Ag-binding cleft of MHC can change the dynamics of the pMHC complex, which in turn alter the affinity of the TCR interaction (Archbold et al., 2009; Tynan et al., 2007). Thus while the NKT TCR is considered to be relatively rigid (Borg et al., 2007), it can nevertheless operate in an induced-fit manner by reshaping the CD1d-Ag landscape, which is speculated to occur for the recognition of bulkier CD1d-restricted ligands such as iGb3 (Florence et al., 2009).
Modifications of the glycosyl headgroup directly reduced the t1/2 of the interaction, with the NKT TCR exhibiting greater sensitivity for the 4′-OH position of α-GalCer than the 3′-OH position. The 3′-OH modification solely resulted in a loss of interactions at this position, while modifications at the 4′-OH, as observed for the α-GlcCer and 4′-deoxy AGLs, altered the bonding network at both the 4′- and 3′- positions, thereby providing a molecular basis for understanding the greater sensitivity of the NKT TCR for the 4′-OH position of α-GalCer. These headgroup modifications were directly related to potency, in which the α-GlcCer and 4′-deoxy AGLs showed the lowest potency when compared to the 3′-deoxy AGL. Different AGLs may also drive expansion of different NKT cell clones, depending on their TCR β-chain usage, which represents another means by which these ligands may differentially influence NKT cell activation. For example, the preferential Vβ8 usage by NKT cells proliferating in response to OCH further supports the notion that the NKT TCR-CD1d interaction is finely tuned to subtle alterations within the F′-pocket. Conversely, modifications to the 4′-OH position of the sugar (α-GlcCer and 4′4″-deoxy α-GalCer) appeared to favour the Vβ7+ NKT cells, which were more highly represented within more proliferated cells. These data indicate that the hierarchical NKT response with respect to Vβ usage maybe dependent on the fine-specificity requirements of the NKT TCR-CD1d interaction.
Our findings provide a molecular basis for our understanding of how different AGLs can promote distinct responses by CD1d-dependent NKT cells. Our findings simultaneously provide a rational basis for the design of tailor-made NKT-based therapeutics and a better understanding of the fine specificity of the NKT TCR.
Materials and Methods
Glycolipid Ags
The glycolipid analogues used in this study were synthesised as previously described (Jervis et al., 2010; Miyamoto et al., 2001; Raju et al., 2009; Yu et al., 2005). See supplementary information for synthesis of 4′-deoxy-α-GalCer.
Mice
C57BL/6 (B6) and Tcra-Jtm1Tg mice (Jα18−/−) (obtained from Dr. M. Taniguchi (RIKEN, Yokohama, Japan and backcrossed 10 times to B6 background) mice were maintained under SPF conditions in the animal facilities of the Dept of Microbiology and Immunology, The University of Melbourne. All experiments involving mice were approved by the University of Melbourne animal ethics committee and performed in accordance with ethics guidelines regarding the proper handling of animals.
CFSE Labelling and Proliferation Assay
Splenocytes from Tcra-Jtm1Tg mice (Jα18−/−) were pulsed overnight with glycolipid in the presence of IL-2 at 100 U/ml. Cells were harvested and overlayed on FCS, then centrifuged and resuspended in fresh media to remove excess glycolipid. Thymocytes were enriched for NKT cells by complement depletion of anti-HSA and anti-CD8 labelled cells. Dead cells were removed using Histopaque (Sigma) gradient centrifugation. Thymocytes were labelled with either 2 mM 5-, 6-carboxyfluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes) or 5 mM eFluor 670 (eBiosciences) and incubated for 10 min at 37C. Splenocytes and thymocytes were then combined at a ratio of 2:1 respectively and cultured in 96 well flat-bottom plates. For proliferative analysis cells were harvested after 72h and stained with anti-TCR-β (BD Biosciences) and α-GalCer (PBS-44)-loaded CD1d tetramer. For Vβ analysis, cells were harvested after 72h and stained with either anti-Vβ7 or Vβ8.1-8.2 (BD Biosciences) and α-GalCer-loaded CD1d tetramer. Generational divisions of proliferating NKT cells were determined using FlowJo software (Treestar).
Cytokine measurements
Using similar stimulation conditions to the proliferation assay, IFN-γ production was analysed by ICS on cells incubated with Golgistop (BD Biosciences) for the last 4h of an 8h culture. Cells were surface stained, fixed and permeabilized (Cytofix-Cytoperm Kit, BD Biosciences) prior to staining with either anti-IFN-γ–phycoerythrin or anti-IFN-γ–allophycocyanin. Culture supernatants were collected after 8h and analysed using CBA flex set for mice (BD Biosciences).
Protein expression, purification of the NKT TCR and CD1d
Cloning, expression and purification of the mouse NKT TCR (Vα14-Jα18; Vβ8.2) used in this study have been previously described (Pellicci et al., 2009). Cloning, expression, purification and loading of the mouse CD1d protein were performed as described previously (Matsuda et al., 2000; Pellicci et al., 2009).
Ternary complex purification, crystallization, structure determination, and refinement
Purified mouse NKT TCR and CD1d-glycolipids were mixed and the ternary complexes were isolated by gel filtration on a Superdex 200 10-300 column, concentrated to 8 mg/ml in 10 mM Tris, pH 8.0, and 150 mM NaCl. Large crystals grew in 22% polyethylene glycol 400, 0.1 M ammonium acetate, and 0.1 M BisTris, pH 5.6. Data were collected at the Australian Synchrotron Facility in Melbourne and processed and refined using standard software packages (see supplementary information).
Surface plasmon resonance measurements and analysis
The interaction between soluble NKT TCR and the CD1d-glycolipid complexes was analysed by SPR with a Bio-Rad ProteOn XPR36 instrument (Hercules, CA) essentially as described previously (Pellicci et al., 2009).
Cell lines and retroviral packaging
TCRα and β constructs were cloned into retroviral plasmids and expressed by retroviral transduction of the 5KC-78.3.20 TCRα- and TCRβ-negative hybridoma, as previously described (Scott-Browne et al., 2007).
Hybridoma stimulation
A total of 5×104 hybridomas were cultured for 20h with 5×104 BMDCs, in complete RPMI medium containing 10% FCS, with the indicated concentration of AGLs. BMDCs were generated from single-cell suspensions from femurs and tibia of C57BL/6 mice or CD1d−/− mice and cultured for 6 d in RPMI medium containing 10% FCS and 0.25% B78hi-conditioned medium (B78hi is a cell line that produces GMCSF). Hybridoma responses were measured by IL-2 ELISA in accordance with standard protocols.
Plate bound CD1d stimulation assay
Soluble mouse CD1d was loaded overnight with glycolipids at a 1:3 molar ratio, and flat-bottom 96 well plates were then coated with 60 μl of 10, 1 or 0.1 μg/ml CD1d-glycolipid solution for 3h at 37C and then washed twice. MACS-anti-PE-beads were used to enrich CD1d tetramer-PE labelled thymic NKT cells (95% pure) that were CFSE-labelled and 2×104 added to each well. After 48hr cells were placed into fresh (uncoated) plates to remove stimulus, and at 72hr cells were analysed by flow cytometry and cytokines in culture supernatant were analysed by CBA.
Supplementary Material
Acknowledgements
We thank the staff at the Australian synchrotron for assistance with data collection. We also thank Mr David Taylor, Dr Sumone Chakravarti and Mr Ken Field for assistance. We are grateful to Prof Paul Savage for generously providing α-GalCer (PBS44) for CD1d tetramer loading. This work was supported by the Cancer Council of Victoria, the National Health and Medical Research Council of Australia (NHMRC) and the Australian Research Council. GC is supported by CRI pre-doctoral scholarship; LCS by an NHMRC RD Wright Fellowship. SAP was supported by NIH grant AI45889. AH was supported by NIH grant R01GM087136. LG is supported by NIH grants (AI076463 and AI078246) and NIH training grant T32 AI07405 to M.H.Y. DIG is supported by an NHMRC Principal Research Fellowship; JR is supported by an ARC Federation Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The coordinates of the mouse NKT TCR-CD1d-OCH; 3′4″-α-GalCer; 4′4″-α-GalCer; C20:2; and α-GlcCer have been deposited in the Protein Data Bank under accession IDs: 3ARB, 3ARD, 3ARE, 3ARF, 3ARG respectively.
References
- Aleksic M, Dushek O, Zhang H, Shenderov E, Chen J-L, Cerundolo V, Coombs D, van der Merwe PA. Dependence of T Cell Antigen Recognition on T Cell Receptor-Peptide MHC Confinement Time. Immunity. 2010 ;32:163–174. doi: 10.1016/j.immuni.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archbold JK, Macdonald WA, Gras S, Ely LK, Miles JJ, Bell MJ, Brennan RM, Beddoe T, Wilce MC, Clements CS, et al. Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition. J Exp Med. 2009;206:209–219. doi: 10.1084/jem.20082136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Sagiv Y, Liu Y, Freigang S, Yu KO, Teyton L, Porcelli SA, Savage PB, Bendelac A. Lysosomal recycling terminates CD1d-mediated presentation of short and polyunsaturated variants of the NKT cell lipid antigen {alpha}GalCer. Proc Natl Acad Sci U S A. 2009;106:10254–10259. doi: 10.1073/pnas.0901228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BM, Gagnon SJ, Biddison WE, Wiley DC. Conversion of a T cell antagonist into an agonist by repairing a defect in the TCR/peptide/MHC interface: implications for TCR signaling. Immunity. 2000;13:475–484. doi: 10.1016/s1074-7613(00)00047-9. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The Biology of NKT Cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- Brutkiewicz RR. CD1d ligands: the good, the bad, and the ugly. J Immunol. 2006;177:769–775. doi: 10.4049/jimmunol.177.2.769. [DOI] [PubMed] [Google Scholar]
- Cerundolo V, Silk JD, Masri SH, Salio M. Harnessing invariant NKT cells in vaccination strategies. Nat Rev Immunol. 2009;9:28–38. doi: 10.1038/nri2451. [DOI] [PubMed] [Google Scholar]
- Degano M, Garcia KC, Apostolopoulos V, Rudolph MG, Teyton L, Wilson IA. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity. 2000;12:251–261. doi: 10.1016/s1074-7613(00)80178-8. [DOI] [PubMed] [Google Scholar]
- Florence WC, Xia C, Gordy LE, Chen W, Zhang Y, Scott-Browne J, Kinjo Y, Yu KO, Keshipeddy S, Pellicci DG, et al. Adaptability of the semi-invariant natural killer T-cell receptor towards structurally diverse CD1d-restricted ligands. EMBO J. 2009;28:3579–3590. doi: 10.1038/emboj.2009.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapin L. iNKT cell autoreactivity: what is 'self' and how is it recognized? Nat Rev Immunol. 2010;10:272–277. doi: 10.1038/nri2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what's in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Pellicci DG, Patel O, Kjer-Nielsen L, McCluskey J, Rossjohn J. Antigen recognition by CD1d-restricted NKT T cell receptors. Semin Immunol. 2010;22:61–67. doi: 10.1016/j.smim.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Rossjohn J, McCluskey J. The Fidelity, Occasional Promiscuity, and Versatility of T Cell Receptor Recognition. Immunity. 2008;28:304–314. doi: 10.1016/j.immuni.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Govern CC, Paczosa MK, Chakraborty AK, Huseby ES. Fast on-rates allow short dwell time ligands to activate T cells. Proceedings of the National Academy of Sciences. 2010;107:8724–8729. doi: 10.1073/pnas.1000966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa JB, Axmann M, Mortelmaier MA, Lillemeier BF, Newell EW, Brameshuber M, Klein LO, Schutz GJ, Davis MM. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im JS, Arora P, Bricard G, Molano A, Venkataswamy MM, Baine I, Jerud ES, Goldberg MF, Baena A, Yu KO, et al. Kinetics and cellular site of glycolipid loading control the outcome of natural killer T cell activation. Immunity. 2009;30:888–898. doi: 10.1016/j.immuni.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jervis PJ, Veerapen N, Bricard G, Cox LR, Porcelli SA, Besra GS. Synthesis and biological activity of [alpha]-glucosyl C24:0 and C20:2 ceramides. Bioorganic & Medicinal Chemistry Letters. 2010;20:3475–3478. doi: 10.1016/j.bmcl.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Borg NA, Pellicci DG, Beddoe T, Kostenko L, Clements CS, Williamson NA, Smyth MJ, Besra GS, Reid HH, et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–673. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Girardi E, Wang J, Yu ED, Painter GF, Kronenberg M, Zajonc DM. The Va14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. The Journal of Experimental Medicine. 2010;207:2383–2393. doi: 10.1084/jem.20101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallevaey T, Fontaine J, Breuilh L, Paget C, Castro-Keller A, Vendeville C, Capron M, Leite-de-Moraes M, Trottein F, Faveeuw C. Invariant and non-invariant Natural Killer T cells exert opposite regulatory functions on the immune response during murine schistosomiasis. Infect Immun. 2007;75:2171–2180. doi: 10.1128/IAI.01178-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallevaey T, Scott-Browne JP, Matsuda JL, Young MH, Pellicci DG, Patel O, Thakur M, Kjer-Nielsen L, Richardson SK, Cerundolo V, et al. T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31:60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang C-R, Koezuka Y, Kronenberg M. Tracking the Response of Natural Killer T Cells to a Glycolipid Antigen Using CD1d Tetramers. The Journal of Experimental Medicine. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulis G, Sanderson JP, Lissin NM, Asparuhova MB, Bommineni GR, Schumperli D, Schmidt RR, Villiger PM, Jakobsen BK, Gadola SD. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol. 2010;8:e1000402. doi: 10.1371/journal.pbio.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, Bossi G, Salio M, Denkberg G, Reddington F, et al. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007;204:1131–1144. doi: 10.1084/jem.20062342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing T(H)2 bias of natural killer T cells. Nature. 2001;413:531–534. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Sullivan LC, Kyparissoudis K, Brooks AG, Reid HH, Gras S, Lucet IS, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R, Castillo BF, Richardson SK, Thakur M, Severins R, Kronenberg M, Howell AR. Synthesis and evaluation of 3″- and 4″-deoxy and -fluoro analogs of the immunostimulatory glycolipid, KRN7000. Bioorganic & Medicinal Chemistry Letters. 2009;19:4122–4125. doi: 10.1016/j.bmcl.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefner A, Fujio M, Wu D, Wong C-H, Wilson IA. Structural Evaluation of Potent NKT Cell Agonists: Implications for Design of Novel Stimulatory Ligands. Journal of Molecular Biology. 2009;394:71–82. doi: 10.1016/j.jmb.2009.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieg J, Yang G, Franck RW, Tsuji M. Superior protection against malaria and melanoma metastases by a C-glycoside analogue of the natural killer T cell ligand alpha-Galactosylceramide. J Exp Med. 2003;198:1631–1641. doi: 10.1084/jem.20031192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne JP, Matsuda JL, Mallevaey T, White J, Borg NA, McCluskey J, Rossjohn J, Kappler J, Marrack P, Gapin L. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- Sloan-Lancaster J, Allen PM. ALTERED PEPTIDE LIGAND,ÄìINDUCED PARTIAL T CELL ACTIVATION: Molecular Mechanisms and Role in T Cell Biology. Annual Review of Immunology. 2003;14:1–27. doi: 10.1146/annurev.immunol.14.1.1. [DOI] [PubMed] [Google Scholar]
- Stanic AK, Shashidharamurthy R, Bezbradica JS, Matsuki N, Yoshimura Y, Miyake S, Choi EY, Schell TD, Van Kaer L, Tevethia SS, et al. Another view of T cell antigen recognition: cooperative engagement of glycolipid antigens by Va14Ja18 natural TCR. J Immunol. 2003;171:4539–4551. doi: 10.4049/jimmunol.171.9.4539. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Nagarajan NA, Wingender G, Wang J, Scott I, Tsuji M, Franck RW, Porcelli SA, Zajonc DM, Kronenberg M. Mechanisms for glycolipid antigen-driven cytokine polarization by Valpha14i NKT cells. J Immunol. 2010;184:141–153. doi: 10.4049/jimmunol.0902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan FE, Reid HH, Kjer-Nielsen L, Miles JJ, Wilce MC, Kostenko L, Borg NA, Williamson NA, Beddoe T, Purcell AW, et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–684. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- Venkataswamy MM, Porcelli SA. Lipid and glycolipid antigens of CD1d-restricted natural killer T cells. Semin Immunol. 2009;22:68–78. doi: 10.1016/j.smim.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wun KS, Borg NA, Kjer-Nielsen L, Beddoe T, Koh R, Richardson SK, Thakur M, Howell AR, Scott-Browne JP, Gapin L, et al. A minimal binding footprint on CD1dglycolipid is a basis for selection of the unique human NKT TCR. The Journal of Experimental Medicine. 2008;205:939–949. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KO, Im JS, Molano A, Dutronc Y, Illarionov PA, Forestier C, Fujiwara N, Arias I, Miyake S, Yamamura T, et al. Modulation of CD1d-restricted NKT cell responses by using N-acyl variants of {alpha}-galactosylceramides. Proc Natl Acad Sci U S A. 2005;102:3383–3388. doi: 10.1073/pnas.0407488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.