Abstract
Tellurite (Tel) resistant enterohaemorrhagic Escherichia coli (EHEC) O157:H7 is a global pathogen. In strain EDL933 Tel resistance (TelR) is encoded by duplicate ter cluster in O islands (OI) 43 and 48, which also harbour iha, encoding the adhesin and siderophore receptor Iha. We identified five EHEC O157:H7 strains that differentiate into large (L) colonies and small (S) colonies with high and low Tel minimal inhibitory concentrations (MICs) respectively. S colonies (Tel-MICs ≤ 4 µg ml−1) sustained large internal deletions within the TelR OIs via homologous recombination between IS elements and lost ter and iha. Moreover, complete excision of the islands occurred by site-specific recombination between flanking direct repeats. Complete excision of OI 43 and OI 48 occurred in 1.81 × 10−3 and 1.97 × 10−4 cells in culture, respectively; internal deletion of OI 48 was more frequent (9.7 × 10−1 cells). Under iron limitation that promotes iha transcription, iha-negative derivatives adhered less well to human intestinal epithelial cells and grew slower than did their iha-positive counterparts. Experiments utilizing iha deletion and complementation mutants identified Iha as the major factor responsible for these phenotypic differences. Spontaneous deletions affecting TelR OIs contribute to EHEC O157 genome plasticity and might impair virulence and/or fitness.
Introduction
Enterohaemorrhagic Escherichia coli (EHEC) O157:H7 is the predominant Shiga toxin (Stx)-producing pathogen of humans (Karch et al., 2005; Tarr et al., 2005). EHEC O157:H7 resists the highly toxic tellurium oxyanion, tellurite (TeO32−; Tel), and therefore grows in concentrations of Tel that inhibit most other E. coli (Zadik et al., 1993; Taylor et al., 2002; Bielaszewska et al., 2005; Orth et al., 2007). This characteristic, together with its inability to ferment sorbitol, has been exploited in selective strategies to isolate EHEC O157:H7 from faeces, food and the environment using Tel-containing media, such as cefixime-Tel sorbitol MacConkey agar (CT-SMAC) (Onoue et al., 1999; Van Duynhoven et al., 2002).
EHEC O157:H7 Tel resistance (TelR) is encoded by the chromosomal terZABCDEF cluster (Taylor et al., 2002; Bielaszewska et al., 2005), which is highly homologous to the ter cluster on plasmid R478 in Serratia marcescens (Whelan et al., 1995; Taylor et al., 2002). EHEC O157:H7 strain EDL933 contains two identical copies of the ter gene cluster within identical O islands (OI) 43 and 48, which are integrated in tRNA genes serW and serX, respectively (Perna et al., 2001). In contrast, O157 Sakai outbreak strain RIMD 0509952 harbours only a single ter cluster-containing island (SpLE1) integrated in serX (Hayashi et al., 2001). This OI is also termed the TelR and adherence-conferring island, based on its first description in EHEC O157:H7 strain 86–24, because it also encodes the iron-regulated gene A (IrgA) homologue adhesin (Iha) (Tarr et al., 2000).
TelR is a common, but not obligatory, feature of EHEC O157:H7. Tel-susceptible E. coli O157:H7 strains have been isolated in North America (Taylor et al., 2002) and Europe (Bielaszewska et al., 2005). Tel susceptibility (TelS) is related to lack of ter genes (Taylor et al., 2002; Bielaszewska et al., 2005), but mechanisms underlying the ter absence (i.e. loss of ter genes during infection or after shedding, or primary non-possession) are unknown. Here, we report the frequency of and mechanisms for loss of the ter gene cluster in EHEC O157:H7 clinical isolates during laboratory passage. We characterized phenotypes associated with loss of the ter cluster and adjacent genes, in particular iha. Our data suggest that full or partial deletions of the TelR island(s) diminish the virulence and/or fitness of this pathogen.
Results
Two different colony phenotypes in E. coli O157:H7 differ by susceptibility to tellurite
We identified five E. coli O157:H7 clinical isolates that produced two different colony morphologies during subculture on SMAC agar: typical large (L) and atypical small (S) colonies (Fig. 1). Both variants were confirmed to be E. coli O157:H7 by O:H serotyping and presence of rfbEO157 and fliCH7 (Table 1). Of the eight phenotypes we initially tested (Table 1), the L and S colonies (hereafter designated L and S strains, respectively, if they are derived from a single parent isolate) differed only in their susceptibilities to Tel. L strains grow well on CT-SMAC and have high (256–1024 µg ml−1) Tel minimal inhibitory concentrations (MICs). S strains do not grow on CT-SMAC and have low (≤ 4 µg ml−1) Tel-MICs (Table 1). All five L strains contain all seven ter genes (terZABCDEF), whereas the corresponding S strains lack these genes (Table 1). These results were corroborated by Southern hybridization with a terC probe (data not shown).
Fig. 1.
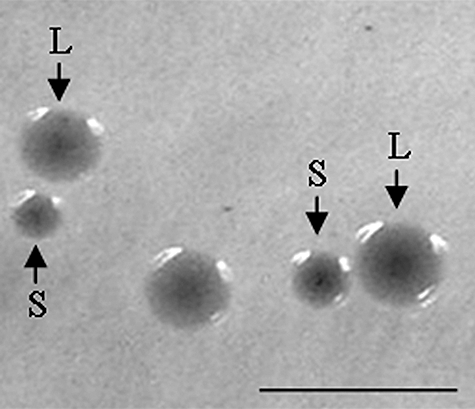
Different colony phenotypes of E. coli O157:H7 strains on sorbitol MacConkey agar. L, large colonies; S, small colonies. Bar represents 5 mm.
Table 1.
Characteristics of L and S colony variants of EHEC O157:H7 strains.
| Strain no.a | Serotype | SF/GUD | Phage type | EHEC-Hly | Stx titre | stx genotype | rfbEO157 | fliCH7 | terb | Tel-MIC (µg ml−1) | Growth on CT-SMACc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 81L | O157:H7 | −/− | 8 | + | 512 | stx2 | + | + | + | 256 | > 1000 |
| 81S | O157:H7 | −/− | 8 | + | 256 | stx2 | + | + | − | 2 | 0 |
| 95L | O157:H7 | −/− | 8 | + | 256 | stx1 | + | + | + | 512 | > 1000 |
| 95S | O157:H7 | −/− | 8 | + | 256 | stx1 | + | + | − | 4 | 1 |
| 134L | O157:H7 | −/− | 4 | + | 1024 | stx2 + stx2c | + | + | + | 256 | > 1000 |
| 134S | O157:H7 | −/− | 4 | + | 512 | stx2 + stx2c | + | + | − | 2 | 0 |
| 135L | O157:H7 | −/− | 31 | + | 512 | stx2 | + | + | + | 256 | > 1000 |
| 135S | O157:H7 | −/− | 31 | + | 512 | stx2 | + | + | − | 2 | 0 |
| 154L | O157:H7 | −/− | 8 | + | 128 | stx2c | + | + | + | 1024 | > 1000 |
| 154S | O157:H7 | −/− | 8 | + | 256 | stx2c | + | + | − | < 1 | 0 |
L, large colony variant, and S, small colony variant of each strain indicated.
All seven genes of the ter cluster (terZABCDEF) were tested; +, all were present; −, all were absent.
Number of colonies grown after overnight incubation on plates inoculated with 1 × 105 colony-forming units.
+, the characteristic was present; −, the characteristic was absent.
SF, sorbitol fermentation; GUD, production of β-D-glucuronidase; EHEC-Hly, EHEC haemolysin production; CT-SMAC, cefixime-tellurite sorbitol MacConkey agar.
The ter cluster in L strains is located in homologues of OI 43/OI 48
Analytical PCR (Fig. S1A, Table S1) demonstrated that all five L strains contain a complete ter cluster organized in the same order as in EDL933, and located within OI 43/OI 48 homologues (Fig. 2). However, in strains 81L, 134L and 154L we detected additional IS elements downstream and/or upstream of the ter cluster, which are not present in OI 43/OI 48 of EDL933 and which partially replaced island-specific genes (Fig. 2). Light cycler-based PCR demonstrated that strains 81L, 134L and 154L contain a single copy of TelR-encoding island, while strains 95L and 135L contain duplicate copies, as in EDL933 (Fig. S2).
Fig. 2.
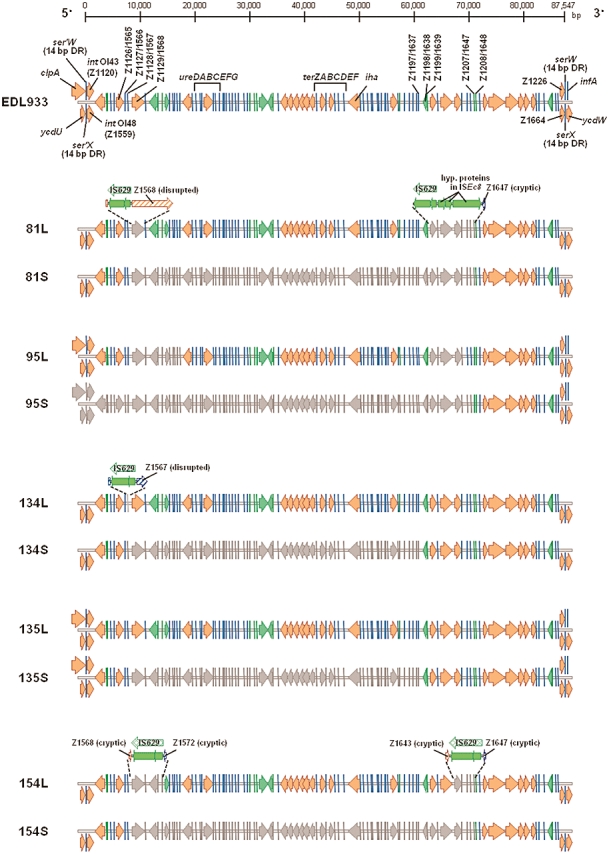
Tellurite resistance (TelR) islands and flanking structures in L and S strains determined by PCR mapping. Single arrows indicate ORFs and flanks of OI 48 in strains that harbour only this OI. Duplicated arrows at the 5′ and 3′ ends of the island indicate the presence of two OI copies and depict the respective ORFs at these positions in OI 43 (upper arrows) and OI 48 (lower arrows). Genes located directly upstream (clpA and ycdU, respectively) and downstream (serW/infA and serX/ycdW, respectively) of OI 43 and OI 48 are also indicated. Orange arrows and blue lines indicate ORFs that were present and grey arrows/lines indicate absent ORFs. An ORF was considered present if an amplicon of the same size as that elicited from O157 strains EDL933 and Sakai was produced in the corresponding PCR. If no amplicon was produced, the ORF was considered absent (in strains with OI 43 and OI 48, only regions absent in both OIs could be identified as missing). ORFs with similarity to mobile genetic elements (putative transposases and insertion sequences) are highlighted in green. Insertion of novel IS elements is depicted above the corresponding regions in L strains (analysis of similar deletions in strain 95L was hindered by the presence of two TelR–OI copies). Putative P4-family integrase genes (int), urease (ure) and tellurite resistance (ter) gene clusters, iha (encoding IrgA homologue adhesin), direct repeat regions (DR) flanking the OIs and intact/cryptic tRNA genes (serW/ser'W and serX/ser'X, respectively) are indicated. The scale (in bp) is above the graph. Results of PCR mapping of one of three independent S colonies derived from each L strain are shown (all three S derivates of the same L strain provided identical PCR results).
To identify the integration sites of the TelR-encoding islands in L strains, we determined the intactness of tRNA genes serW and serX (Fig. S1B, Table S1), into which OI 43 and OI 48, respectively, are integrated in EDL933 (Perna et al., 2001). In strains 95L and 135L, serW and serX are occupied (Fig. 3), suggesting the presence of OI 43 and OI 48 homologues, respectively, in these locations. In strains 81L, 134L and 154L, serX is occupied and serW is intact (Fig. 3); these strains possess a homologue of OI 48 only. The synteny of these OIs and the two O157 genome reference strains was confirmed by PCRs targeting the upstream (5′) and downstream (3′) junction of each OI with the core genome (Fig. 3, Table 2) (designations of the 5′ and 3′ ends of OI 43/OI 48 used in this paper are based on the orientation of these OIs in the sequenced genome of EDL933; GenBank Accession No. AE005174).
Fig. 3.
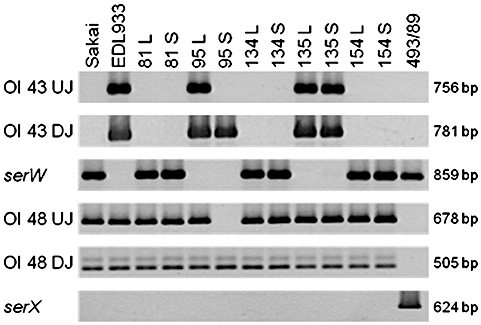
Amplification of serW, serX and the upstream (UJ) and downstream (DJ) junctions of OIs 43 and 48 in L and S variants of EHEC O157:H7 strains. Strains tested, PCR targets and lengths of PCR amplicons are listed across the top and to the left and right of the rows of amplicons respectively. Purified chromosomal DNA (20 ng) was used as a template in all PCRs. In PCRs targeting serW and serX, the presence of an amplicon of the same intensity as that from the positive control strain 493/89 (sorbitol-fermenting, ter-negative EHEC O157:NM that has intact serW and serX, as determined by sequence analysis in this study) indicates that the target locus is intact; the absence of an amplicon combined with amplification of UJ and DJ of the respective OI indicates that the locus is occupied by this OI. Amplification of UJs of OI 43 and OI 48 in strain 95S is hindered by the absence of the 5′ end of each OI in this strain (Fig. 2).
Table 2.
Copy numbers and genomic integration sites of TelR-OI in L and S strains.
| Status ofc | Genomic junctions of TelR-OIsd | |||||||
|---|---|---|---|---|---|---|---|---|
| Strain no.a | terb | TelR-OI copy no. | serW | serX | UJ/DJ OI 43 | UJ/DJ OI 48 | OI present | OI integrated in |
| 81L | + | 1 | I | O | −/− | +/+ | 48 | serX |
| 95L | + | 2 | O | O | +/+ | +/+ | 43 + 48 | serW, serX |
| 134L | + | 1 | I | O | −/− | +/+ | 48 | serX |
| 135L | + | 2 | O | O | +/+ | +/+ | 43 + 48 | serW, serX |
| 154L | + | 1 | I | O | −/− | +/+ | 48 | serX |
| 81S | − | 0 | I | O | −/− | +/+ | 48Te | serX |
| 95S | − | 0 | O | O | −/+ | −/+ | 43T + 48T | serW, serX |
| 134S | − | 0 | I | O | −/− | +/+ | 48T | serX |
| 135S | − | 0 | O | O | +/+ | +/+ | 43T + 48T | serW, serX |
| 154S | − | 0 | I | O | −/− | +/+ | 48T | serX |
| EDL933f | + | 2 | O | O | +/+ | +/+ | 43 + 48 | serW, serX |
| Sakaig | + | 1 | I | O | −/− | +/+ | 48 | serX |
| 493/89h | − | 0 | I | I | −/− | −/− | None | n.a. |
L, large, and S, small colony variant of each respective strain.
+, all seven genes of the ter cluster (terZABCDEF) were present; −, all seven genes were absent.
I, the gene is intact; O, the gene is occupied.
+, amplicon size was identical to that from strain EDL933; −, no amplicon was obtained.
T, truncated OI 43 and/or OI 48.
Control strains; data of PCR analyses are in agreement with complete genomic sequences of EHEC O157:H7 strains EDL933 (GenBank Accession No. AE005174), and Sakai RIMD 0509952 (GenBank Accession No. NC_002695); strain 493/89 (sorbitol-fermenting EHEC O157:NM) lacks the ter gene cluster (Bielaszewska et al., 2005) and has serW and serX intact as determined by sequence analysis in this study.
TelR-OI, tellurite resistance-encoding O islands; UJ, upstream (5′) junction; DJ, downstream (3′) junction; n.a., not applicable.
S strains contain truncated homologues of OI 43/OI 48
All five S strains lack the ter gene cluster (Table 1), but except for missing 5′ ends of both islands in strain 95S, the junctions between OI 43 and/or OI 48 and the core genome are intact in all S strains (Figs 2 and 3, Table 2). Therefore, we systematically PCR-mapped (Fig. S1A) these truncated island(s) in three independent S colonies derived from each L strain to determine the extent of deletions. In all cases, the PCR suggested that a single deletion in an L strain resulted in the observed S colonies (representative S colony shown in Fig. 2). Moreover, in pulsed-field gel electrophoresis of XbaI-digested genomic DNA, all three S colonies that were descended from the same L strain shared identical restriction patterns, which differed by two to nine bands from that of the respective L strain (Fig. S3). This further confirmed that the genomic changes resulting from the deletions in OI 43/OI 48 were highly similar or identical in the derivatives of the same L strain. Altogether, the data from the PCR mapping and pulsed-field gel electrophoresis suggested that each of the parental L strains was ‘pre-programmed’ to undergo a particular sort of OI 43/OI 48 degeneration.
A precise characterization of the deleted regions is difficult in strains 95S and 135S, because both parental L strains contain the nearly identical (at least in EDL933) OI 43 and OI 48. In strain 135S, an internal portion of each OI of at least 52 kb was lost, and at least 70 kb of both islands were lost from strain 95S (Fig. 2). Moreover, the absence of clpA and ycdU (upstream of OI 43 and OI 48, respectively) from strain 95S (Fig. 2) suggests that a region of the upstream backbone genome was co-deleted with a major part of each island. Primer walking along the regions upstream of OI 43 and OI 48 (Table S2) demonstrated deletions of ∼ 2.9 kb (ORFs Z1118 and clpA) and ∼ 145.9 kb (ORFs Z1399 up to ycdU) of the core chromosome respectively. Analysis of a 3711 bp amplicon connecting ORFs Z1398 and Z1650, which spans the internal deletion of OI 48 as well as deleted parts of the core chromosome demonstrated that strain 95S lost in total a 217 535 bp fragment extending from the coding region of ORF Z1398 to the intergenic region between ORFs Z1646 and Z1647 (Fig. S4). Scrutiny of the respective ORFs/intergenic regions demonstrated no elements that could be responsible for homologous or site-specific recombination. We assume that a novel IS629 integrated 596 bp downstream of the start codon of ORF Z1398, which rendered the gene cryptic, before the IS element recombined with a 259 bp cryptic IS629 overlapping with ORF Z1647 (Fig. S4; see also next paragraph). Efforts to produce an amplicon spanning the deletion upstream of OI 43 failed.
The other three S strains harboured a truncated homologue of OI 48 that retained the 5′ end of the island from integrase gene (Z1559) to ORF Z1566 (∼ 7 kb; strain 134S) or Z1567 (∼ 8 kb; strains 81S and 154S), and the 3′ end of the island from ORF Z1638 (strain 134S) or Z1648 (strains 81S and 154S) to the last gene (Z1664) (∼ 26 and 16 kb, respectively) (Fig. 2). Large (∼ 53–62 kb) internal OI 48 regions were absent (Fig. 2).
Analysis of deletions in truncated OI 48
To determine the extent and putative mechanism(s) of deletions in OI 48 of the three strains with single ter islands, we PCR-amplified regions spanning the deletions in the S strains and their supposed boundaries in the corresponding L strains and sequenced the amplicons. In strain 134S, a 53 773 bp deletion extends from ORF Z1567 to the region between ORFs Z1637 and Z1638 (Fig. 4A). In the parental strain 134L we detected a full-length IS629 element (1310 bp) that integrated 33 bp downstream of the start codon of ORF Z1567 (Figs 2 and 4A) and has 99% identity to ORFs Z1638/Z1639 (which have, in turn, 95% identity to IS629 of EHEC O157 Sakai); this IS element is not present at this position either in EDL933 or O157 Sakai. Deletions in strains 81S (62 678 bp) (Fig. 4B) and 154S (62 672 bp) (Fig. 4C) start within ORF Z1568 and end in ORF Z1647, a putative transposase that overlaps with a 259 bp cryptic IS629. In strain 81L, a full-length IS629 disrupted ORF Z1568 65 bp downstream of the start codon. In addition, ORFs Z1640 to Z1646 were substituted with a ∼ 3.7 kb mosaic structure composed of an additional IS629 and ORFs similar to hypothetical proteins in ISEc8 (Figs 2 and 4B). In strain 154L, a full-length IS629 occurs at the start point of the internal deletion between cryptic ORFs Z1568 and Z1572 replacing ORFs Z1569 to Z1571. At the end-point of the internal deletion we found a full-length IS629 between cryptic ORFs Z1643 and Z1647 replacing Z1644 to Z1646 (Figs. 2 and 4C). Taken together, integration of novel IS elements into the TelR-encoding islands of L strains and their subsequent homologous recombination with each other or with already existing IS elements such as ORFs Z1638/Z1639 and Z1647 (similar to IS629) caused the observed deletions (Fig. 4).
Fig. 4.
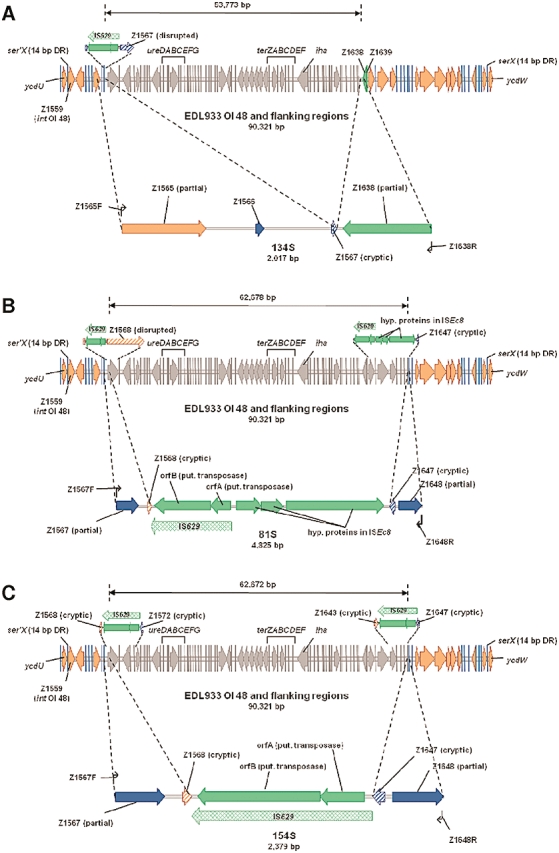
Deletions of OI 48 in strains 134S (A), 81S (B) and 154S (C). The upper part in each illustration shows the organization of OI 48 and its flanking regions in reference E. coli O157:H7 strain EDL933. Insertions of novel IS elements in the respective L strains are depicted above the island. The solid line above the genetic map depicts the extent of internal deletions in the three S strains (depicted in grey in the EDL933 sequence). In the lower part, the size (given in bp below the strain number) and genetic organization of the connecting fragment amplified after deletion of an internal part of OI 48 in each respective strain is shown (not drawn to scale). Small arrows symbolize primer binding sites for amplification of the connecting fragment (PCR primers and conditions are in Table S1). Outer dotted lines indicate homologies between OI 48 of EDL933 and the amplification product. The green arrows between the inner dotted lines depict sequences replacing the deleted region (IS, insertion sequence; hyp., hypothetical; put., putative). Indicated genes of OI 48 and flanking regions are int (putative P4-family integrase), ure (urease) and ter (tellurite resistance) clusters, iha, ORFs relevant to the deletion analyses, serX/ser'X (intact/cryptic tRNA gene) including direct repeats (DR) flanking the OI and ycdU/ycdW (chromosomal genes).
Recombination between flanking direct repeats deletes TelR-encoding OIs
In EDL933, OIs 43 and 48 are flanked by perfect 14 bp (TGGCGGTGAGGGGG) direct repeats (DRs), which encompass the last 14 bp of serW and serX, respectively, and are identical to the DRs of the Shigella Resistance Locus pathogenicity island (SRL-PAI) of Shigella flexneri 2a (Luck et al., 2001; Turner et al., 2004). Furthermore, each of these two OIs in EDL933 has an integrase gene that is 15 bp shorter than, but otherwise similar to, the integrase of the SRL-PAI (99% nucleotide identity). These observations suggest a mechanism whereby OI 43 and OI 48 could be lost by site-specific recombination between their DRs, similar to what is observed with SRL-PAI (Turner et al., 2001; 2004;) and other genomic islands (Rajanna et al., 2003; Middendorf et al., 2004; Sakellaris et al., 2004).
The five L and S E. coli O157:H7 strains were further analysed to test this hypothesis. Except for intOI43 of strain 95L and intOI48 of strain 135L, all other int genes in OIs 43 and 48 were identical in length and sequence to the corresponding genes in EDL933 (data not shown). intOI43 of 95L and intOI48 of 135L were of the same length as the int genes in EDL933, but contained six and seven mostly identical point mutations resulting in two and three amino acid changes, respectively (data not shown). Furthermore, in all but one of the L and S strains, the junctions between the core genome and the complete or truncated OI 43 and/or OI 48 are highly homologous (> 97%) to the boundaries of the corresponding OIs in EDL933 and contain the same 14 bp DRs. This suggests that both OI 43 and OI 48 could be deleted in their entirety by site-specific recombination. The only exception is strain 95S, which contains the 14 bp sequences corresponding to the DRs at the 3′ flank of OI 43 and OI 48, but the upstream junctions of these OIs and thus the 5′ DRs are missing (Figs 2 and 3); this makes complete excision of both OIs by site-specific recombination impossible.
Indeed, in strains 81L/81S, 95L, 134L/134S, 135L/135S and 154L/154S, which contain complete/truncated OI 43 and/or OI 48, intact serW and/or serX genes were amplified simultaneously with the upstream and downstream junctions of the islands using more template DNA (100 ng instead of 20 ng, data not shown). These dual amplicons indicate that a subpopulation of each outgrowth lost OI 43/OI 48 in their entirety. The same segment loss was observed in strains EDL933 and Sakai. Sequences of the intact serW and serX genes in each of these strains are identical to those in an OI 43- and OI 48-negative, in silico-generated derivative of EDL933 demonstrating that complete excision of OI 43 and/or OI 48 based on site-specific recombination between flanking DRs occurred in the strains. Taken together, these data demonstrate a complete excision of OI 43/OI 48, in addition to internal OI deletions, in E. coli O157:H7.
Instability of TelR-encoding OIs
The proportions of cells in overnight cultures that had sustained deletions were determined using quantitative real-time PCR. Complete and internal deletions of OI 48 occurred in 1.97 ± 0.75 × 10−4 (average data for all strains) and 9.70 ± 0.23 × 10−1 cells (average data for strains with OI 48 only) respectively. The complete excision of OI 43 occurred in a proportion of 1.81 ± 0.48 × 10−3 cells.
Adherence of L and S strains to human intestinal epithelial cells and role of Iha
We asked if deletions of OI 43 and OI 48 influence the capacity to adhere to human intestinal epithelial cells because these islands contain iha, encoding Iha (Fig. 2) (Tarr et al., 2000; Perna et al., 2001). We compared adherence of L (iha+) and S (iha-) strains to HCT-8 and Caco-2 cells using iron-limited conditions [Dulbecco's minimal essential medium (DMEM); iron < 0.05 µg ml−1] that significantly upregulate iha transcription compared with that in iron (10 µM FeCl2)-repleted DMEM (iron 0.50 µg ml−1) and Luria–Bertani (LB) broth (iron 0.59 µg ml−1) (Fig. S5). The DMEM-cultured L strains adhered significantly more efficiently than did their identically cultured iha- S derivatives to HCT-8 cells, as demonstrated by numbers of bacteria attached per cell (Fig. 5A). Moreover, in most cases, the morphological pattern of the adherence differed between the L and S strains. L strains usually displayed diffuse adherence with most bacteria attaching to cell peripheries (Fig. 5B), whereas S strains adhered mostly in small loose clusters (Fig. 5C) in a localized adherence-like pattern (Scaletsky et al., 1999) or as scarce single bacteria. All DMEM-cultured L strains also adhered to Caco-2 cells to greater extents than did their S derivatives (Fig. 5D), but there was no distinct morphological difference between adherence of L and S strains; all strains displayed localized adherence-like patterns, with bacterial clusters being larger and more frequent in L (Fig. 5E) than in S strains (Fig. 5F). In contrast to the strains cultured in DMEM, no apparent difference in adherence to any of the cell lines was observed in any of the L/S pairs when strains were cultured in iron-repleted DMEM and in LB broth where minimal or no iha transcription occurs (Fig. S5). Under these conditions, all strains adhered weakly (range, 1–3, mean, 1.7 ± 0.8; and range, 0–4, mean, 1.8 ± 1.1 bacteria per cell respectively). These data suggest an important role of Iha in the adherence of L strains grown under iron-limited conditions to cultured intestinal epithelial cells.
Fig. 5.
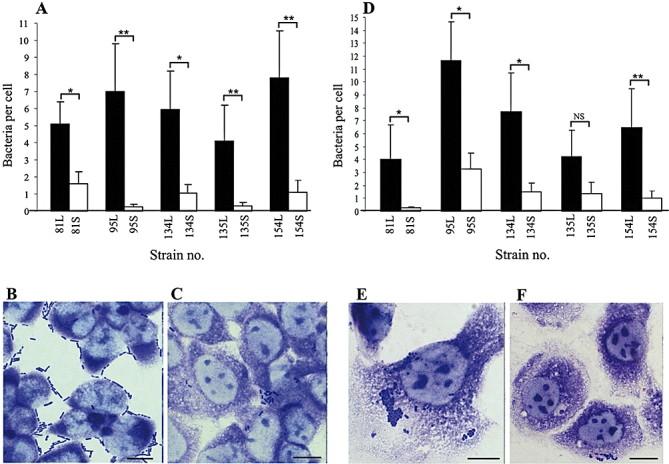
Adherence of iha+ L strains and their iha− S derivatives to human intestinal epithelial cells. Strains were grown overnight in DMEM (iron < 0.05 µg ml−1) without shaking and adherence assay was performed as described in Experimental procedures. All bacteria and cells were counted in 10 randomly selected fields and bacteria per cell were averaged. Quantitative differences between adherence of the respective L and S strains were determined using unpaired Student's t-test. Morphological pattern of adherence was evaluated according to Scaletsky et al. (1999).
A and D. Adherence of L and S strains to HCT-8 (A) and Caco-2 cells (D) quantified by numbers of bacteria attached per cell. *P < 0.05; **P < 0.001; NS, not significant. Data are expressed as mean ± standard deviations of number of bacteria attached per cell from three independent experiments.
B, C, E and F. Adherence patterns on HCT-8 (B and C) and Caco-2 cells (E and F) exemplified by strain pair 95L (B and E) and 95S (C and F). Bars represent 10 µm.
We confirmed that the L and S strains contained the same panel of other proven or putative EHEC O157:H7 adhesins and determined their expression in DMEM. Each of the L and S strains harboured eae encoding intimin (Donnenberg et al., 1993), lpfA1 and lpfA2 encoding major fimbrial subunits of long polar fimbriae 1 and 2, respectively (Torres et al., 2002; 2004;), and ehaA encoding the EHEC autotransporter A (EhaA) (Wells et al., 2008). However, in contrast to iha, whose transcription in L strains is significantly upregulated in DMEM, relative transcription of the non-iha adhesin genes was low (usually below 2.0), and comparable in L and S strains (Fig. S6). Hence, Iha appears to be the major adhesin involved in the adherence of L strains to HCT-8 and Caco-2 cells under iron-limited conditions.
To further establish the role of Iha in adherence, we constructed an isogenic iha deletion mutant of strain 154L (154LΔiha), and compared its adherence capacity with those of the parental strain 154L, iha-complemented 154LΔiha, strain 154S and iha-complemented strain 154S (all cultured in DMEM) (for constructs see Table S4). The adherence capacity of the mutant 154LΔiha to both cell lines was reduced nearly to the level of that of strain 154S (Fig. 6A, C and E). However, iha complementation of 154LΔiha and of strain 154S restored the adherence capacity of each respective complemented strain (154LΔiha/pWKS30iha and 154S glmS::iha) basically to that of strain 154L (Fig. 6A, C and E). To extend these findings, we compared the adherence of prototypic EHEC O157:H7 strain Sakai (RIMD 0509952), its isogenic iha deletion mutant (SakaiΔiha) and iha-complemented deletion mutant (SakaiΔiha glmS::iha). As in strain 154L, the iha deletion mutant of O157 Sakai adhered less well, whereas the iha complementation returned the adherence to the level of the wild-type strain (Fig. 6B, D and F). These experiments confirm a major contribution of Iha to the adherence of EHEC O157:H7 to cultured intestinal epithelial cells under iron-limited conditions. Notably, while completely restoring the adherence capacity quantitatively, the iha complementation of strains 154LΔiha and 154S restored only partially (Fig. 6C) the peripheral adherence pattern produced by L strains on HCT-8 cells. This suggests that an additional bacterial or HCT-8 cell factor is involved in this adherence phenotype, which is specific for HCT-8 cells.
Fig. 6.
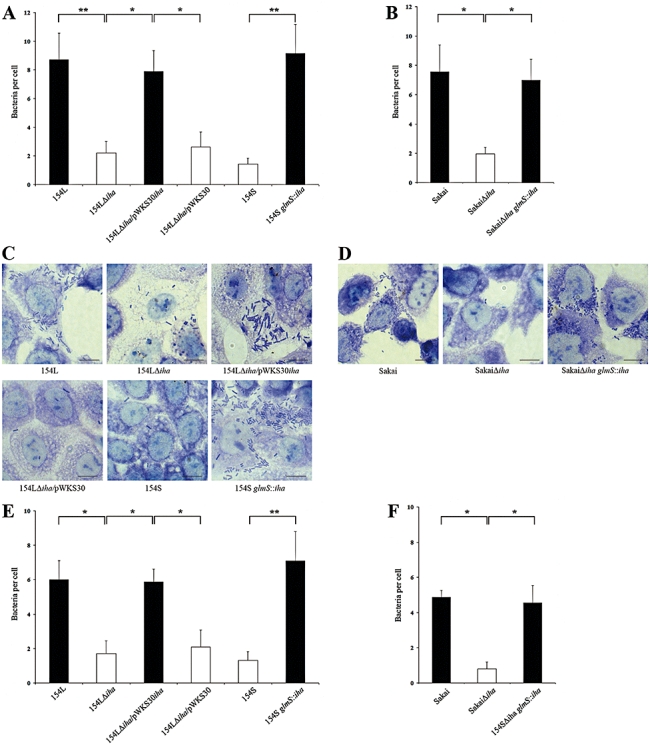
Role of Iha in the adherence of EHEC O157:H7 strains 154L and Sakai to human intestinal epithelial cells. Strains 154L, 154S and their iha deletion (154LΔiha) and iha complementation (154LΔiha/pWKS30iha and 154S glmS::iha) mutants (and 154LΔiha/pWKS30 vector control) were grown overnight in DMEM without shaking and adherence assay was performed as described in Experimental procedures. EHEC O157:H7 strain Sakai, and its iha deletion (SakaiΔiha) and iha complementation (SakaiΔiha glmS::iha) mutants were tested in parallel. To quantify the adherence, all bacteria and cells were counted in 10 randomly selected fields and bacteria per cell were averaged. Differences between adherence of iha+ and iha- strains were determined using unpaired Student's t-test.
A, B, E and F. Adherence of wild-type strains and their iha deletion and iha complementation mutants to HCT-8 (A and B) and Caco-2 cells (E and F) quantified by numbers of bacteria attached per cell. *P < 0.05; **P < 0.001. Data are expressed as mean ± standard deviations of number of bacteria attached per cell from three independent experiments.
C and D. Photomicrographs showing HCT-8 adherence patterns of strains analysed for quantitative adherence to these cells in A (C) and B (D). Bars represent 10 µm.
Impact of Iha on the growth of L and S strains under iron-limited conditions
Because Iha is a siderophore receptor in uropathogenic E. coli (Léveilléet al., 2006), we asked if the iha+ L strains and their iha- S derivatives grow differently under iron-limited conditions (DMEM) where iha transcription is upregulated (Fig. S5). In three of five strain pairs, L strains grew significantly more rapidly in DMEM than their corresponding S derivatives (Fig. 7). Repletion of DMEM with 10 µM FeCl2 remedied this growth impairment (Fig. 7). These data suggest that the lack of Iha contributes to reduced growth rates of S strains in low-iron-milieus.
Fig. 7.
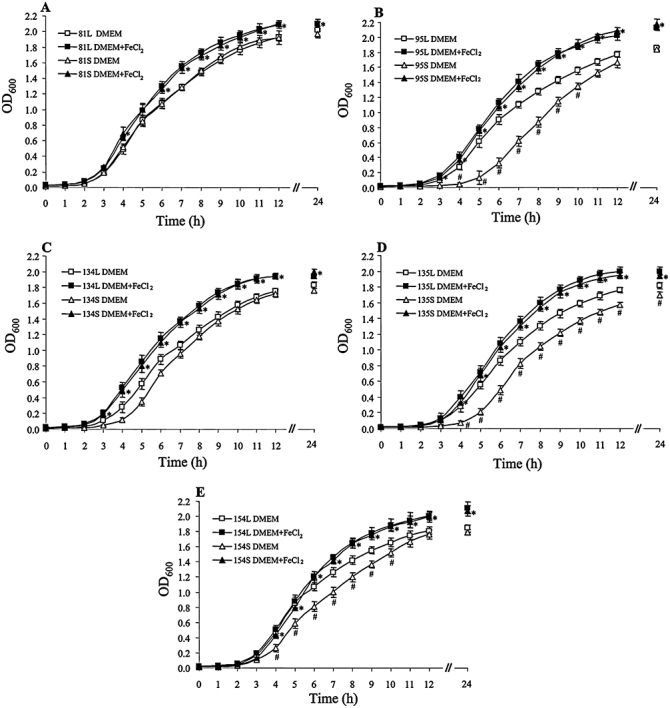
Growth of L (iha+) and S (iha−) variants of E. coli O157:H7 strains in iron-limited and iron-repleted conditions.
A–E. L and S strains were grown in DMEM without (iron < 0.05 µg ml−1) and with 10 µM FeCl2 (iron 0.5 µg ml−1) and bacterial growth was monitored by measuring optical density at 600 nm (OD600) at the time points indicated. #, the difference in OD600 between the corresponding L and S strain grown in DMEM was statistically significant (P < 0.05; unpaired Student's t-test); *, the difference in OD600 between the S strain cultured in DMEM and in DMEM with 10 µM FeCl2, respectively, was statistically significant (P < 0.05; unpaired Student's t-test). Data are presented as means ± standard deviations from three independent experiments.
To test this hypothesis, we compared growth rates of strains 154L, 154S and their respective iha deletion and iha complementation mutants in DMEM and in DMEM with 10 µM FeCl2 (Fig. 8). In DMEM, the iha deletion mutant 154LΔiha grew significantly slower than did the parental strain 154L, and comparably slow as did strain 154S. iha complementation of 154LΔiha and 154S restored the growth rate of each respective complemented strain (154LΔiha/pWKS30iha and 154S glmS::iha) to the level of strain 154L (Fig. 8A). The same impact of iha deletion and complementation on the growth in DMEM was observed in the O157 Sakai strain and its mutants SakaiΔiha and SakaiΔiha glmS::iha (Fig. 8B). Repletion of DMEM with 10 µM FeCl2 remedied the growth of iha deletion mutants 154LΔiha and SakaiΔiha (Fig. 8C and D), as also observed in strain 154S (Fig. 8C) and other S strains (Fig. 7). Taken together, these data suggest that Iha is essential for growth of EHEC O157:H7 under iron limitation and its absence in S strains impairs their growth under such conditions. The ability of S strains to grow, though slower, under iron deficiency can be explained by involvement of other (non-Iha) iron acquisition systems identified in EHEC (Torres and Payne, 1997; Kresse et al., 2007). The expression of such siderophore systems in all S strains is demonstrated by the ability of supernatants of overnight DMEM cultures to bind iron from a chrome azurol S/iron(III)/hexadecyltrimethylamonium bromide complex (Schwyn and Neilands, 1987) (data not shown).
Fig. 8.
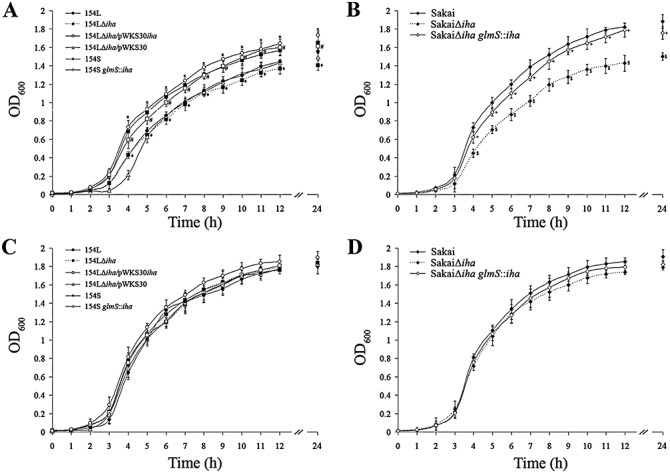
Impact of Iha on growth of EHEC O157:H7 strains 154L and Sakai under iron-limited and iron-repleted conditions.
A–D. Strains 154L, 154S, and their respective iha deletion and iha complementation mutants 154LΔiha, 154LΔiha/pWKS30iha, and 154S glmS::iha (and 154LΔiha/pWKS30 vector control) (A and C) and strains O157 Sakai, SakaiΔiha and SakaiΔiha glmS::iha (B and D) were grown in DMEM (A and B) and in DMEM with 10 µM FeCl2 (C and D) and bacterial growth was monitored by measuring OD600 at the time points indicated. The differences in OD600 values between 154L and 154LΔiha (*), 154LΔiha and 154LΔiha/pWKS30iha (#), 154S and 154S glmS::iha (x), O157 Sakai and SakaiΔiha (§), and SakaiΔiha and SakaiΔiha glmS::iha (+) grown in DMEM were statistically significant (P < 0.05; unpaired Student's t-test). Data are presented as means ± standard deviations from three independent experiments.
Influence of Iha on colony phenotype
We further determined whether iha loss contributes to the reduced size of S colonies by comparing the sizes of S and the respective L colonies on DMEM agar without (iron < 0.05 µg ml−1) and with 10 µM FeCl2 (iron 0.50 µg ml−1). On plain DMEM agar, S colonies of all five strains were significantly smaller than their parental L colonies (example in Fig. 9A, B and E). Supplementation of DMEM agar with 10 µM FeCl2 significantly increased the size of S (example in Fig. 9D and E), but not L (Fig. 9C and E) colonies. Next, we compared colony sizes of strains 154L, 154S, and their respective iha deletion and iha complementation mutants grown as above. On DMEM agar, colonies of the 154LΔiha mutant were significantly smaller than those of strain 154L, and of similar size to those of strain 154S (Fig. 10A and B). iha complementation of 154LΔiha and 154S increased colony size of each respective complemented strain (154LΔiha/pWKS30iha and 154S glmS::iha) to that of strain 154L (Fig. 10A and B). Similarly, iha deletion from the O157 Sakai strain significantly reduced colony size of the SakaiΔiha mutant, whereas iha complementation of this mutant (SakaiΔiha glmS::iha) returned the colony size to that of wild-type O157 Sakai (Fig. 10C and D). On DMEM agar with 10 µM FeCl2 colonies of all strains had similar sizes regardless of the presence or absence of iha (Fig. 10E–H). Thus, absence of Iha leads to atypical small colony phenotype in EHEC O157:H7, in particular on media with decreased iron content.
Fig. 9.
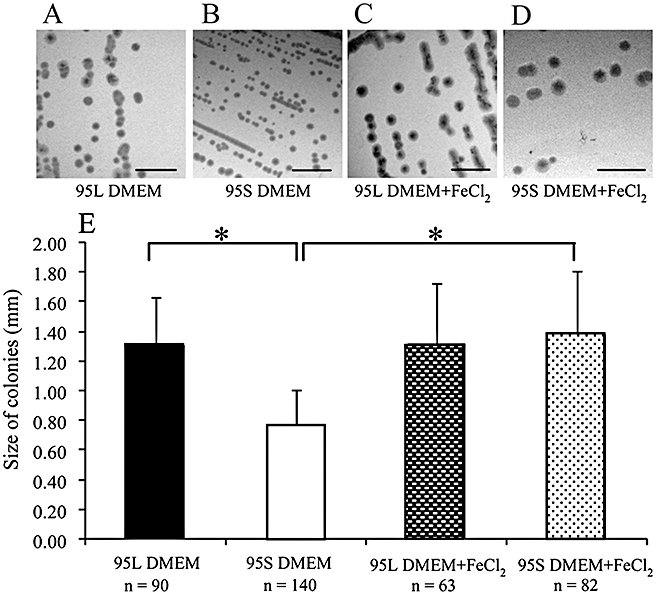
Influence of iron content on colony size of EHEC O157:H7 strains exemplified in strain pair 95L/95S. One L and one S colony of the strain was inoculated on a plate of DMEM agar and DMEM agar with 10 µM FeCl2 and incubated at 37°C for 26 h. After visual inspection, the plates were photographed and diameter of all or most well-separated colonies from each plate (n) was determined using a Power Point measuring tool. Differences between colony sizes under different conditions were calculated using unpaired Student's t-test.
A–D. Size of L and S colonies of strain 95 cultured on DMEM agar without (A and B, respectively) and with 10 µM FeCl2 (C and D, respectively) as determined by visual inspection. Bars correspond to 5 mm.
E. Size of L and S colonies on DMEM agar without and with 10 µM FeCl2 expressed as mean ± standard deviation of diameter of the indicated numbers (n) of colonies.*P < 0.05.
Fig. 10.
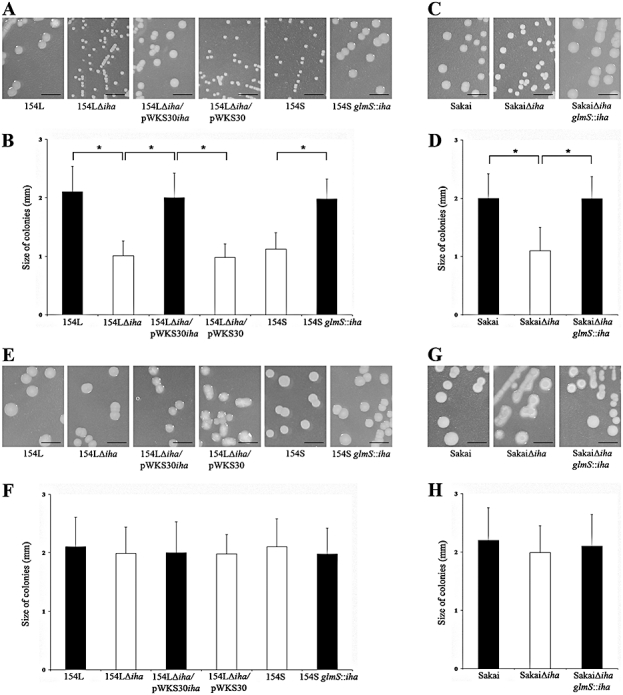
Influence of Iha on colony size. One colony of each strain 154L, 154S, O157 Sakai and their respective iha deletion and iha complementation mutants (as well as of 154LΔiha/pWKS30 vector control) was inoculated on a plate of DMEM agar and DMEM agar with 10 µM FeCl2 and incubated at 37°C for 26 h. After visual inspection, the plates were photographed and diameter of at least 50 well-separated colonies was determined using a Power Point measuring tool. Differences between colony sizes of corresponding iha+ and iha- strains were calculated using unpaired Student's t-test.
A and C. Colony sizes of strains 154L, 154S and their respective iha deletion and iha complementation mutants (A) and of O157 Sakai strain and its iha deletion and iha complementation mutants (C) cultured on DMEM agar without FeCl2 as determined by visual inspection. Bars correspond to 5 mm.
B and D. Colony sizes of the strains shown in A and C, respectively, expressed as mean ± standard deviation of diameter of at least 50 colonies.*P < 0.05.
E and G. Colony sizes of strains 154L, 154S and their respective iha deletion and iha complementation mutants (E) and of O157 Sakai strain and its iha deletion and iha complementation mutants (G) cultured on DMEM agar with FeCl2 as determined by visual inspection. Bars correspond to 5 mm.
F and H. Colony size of the strains shown in E and G, respectively, expressed as mean ± standard deviation of diameter of at least 50 colonies.
Discussion
We are increasingly recognizing the non-static nature of the genomes of bacterial pathogens (Maurelli et al., 1998; Dobrindt et al., 2004; Brzuszkiewicz et al., 2006; Ahmed et al., 2008; Kaper and Karmali, 2008; Mellmann et al., 2009; Waldor, 2010). A rapid change in the genomic architecture of EHEC O157, even within a single strain, has been attributed to loss of stx genes and their encoding bacteriophages (Murase et al., 1999; Feng et al., 2001; Mellmann et al., 2005; 2008; Bielaszewska et al., 2006a). Such an event might enable strains to survive in the guts of humans and animals by avoiding lysis via stx phage induction (Mellmann et al., 2005; 2008;). Here, we demonstrate spontaneous loss of large internal regions of genomic islands OI 43 and/or OI 48 via homologous recombination between novel and existing IS elements, which removes TelR and Iha from clinical E. coli O157:H7 isolates. Additionally, complete excision of both islands is observed via site-specific recombination between flanking DRs in a proportion of cells, resembling site-specific excision of other genomic islands (Bach et al., 1999; Turner et al., 2001; Tauschek et al., 2002; Bueno et al., 2004; Middendorf et al., 2004; Sakellaris et al., 2004). In contrast, internal deletions of OI 48 occurred in a substantially higher proportion of cells (average of 9.7 × 10−1) than those reported for other genomic islands (10−5–10−6) (Turner et al., 2001; Tauschek et al., 2002; Middendorf et al., 2004). Nevertheless, the frequency is similar in magnitude to that observed for the ‘magnetosome island’ of Magnetospirillum gryphiswaldense, where spontaneous mutants affected in magnetosome formation arise at a frequency of up to 10−2 after prolonged storage or exposure to oxidative stress, a process assumed to be also based on integration of new IS elements and subsequent homologous recombination (Schübbe et al., 2003; Ullrich et al., 2005). Although we observed the deletions of OI 43 and/or OI 48 in vitro (i.e. during laboratory passage), isolation of Tel-susceptible EHEC O157:H7 directly from patients' stools (Bielaszewska et al., 2005) suggests that TelR island excisions also occur during infection.
Besides the implications for microbial diagnosis (i.e. Tel-susceptible EHEC O157:H7 will not grow on CT-SMAC), the deletions in OI 43/OI 48 might have consequences for virulence and evolution of EHEC O157:H7. Several lines of evidence support the role of OI 43/OI 48 in the virulence and/or fitness of EHEC O157:H7 and involvement of iha and ter gene cluster in this process. Tarr et al. (Tarr et al., 2000) demonstrated that EHEC O157:H7 strain 86–24 with a deletion in iha adhered less well than the parental strain with functional iha to HeLa cells, corroborating the role of Iha as an adhesin. In another study, deletion of iha from strain 86–24 reduced adherence of the mutant to pig enterocytes in an iron-restricted milieu of a ligated ileal loop, but not in an in vitro adherence assay using strains cultured in iron-rich brain heart infusion broth, in which Iha expression might have been compromised (Yin et al., 2009). Johnson et al. demonstrated the role of Iha in a murine model of uropathogenesis (Johnson et al., 2005). Our findings add to these data, by demonstrating that an iha deletion in strain 154L reduced adherence to cultured human intestinal epithelial cells under iron limitation, i.e. when iha is most robustly transcribed. However, this effect is not seen in iron repletion when little iha transcription occurs. We also report for the first time that iha can be lost spontaneously by wild-type EHEC O157:H7 strains via partial or full excision of OI 43/OI 48, and that this process reduces adherence to intestinal epithelial cells under iron limitation. Moreover, our ability to restore adherence capacity of strain 154S by iha complementation confirms that loss of iha, and not of neighbouring genes, reduced adherence of this strain (and likely also of the other S strains) to human intestinal epithelial cells. Our data strengthen the case for Iha as an iron-regulated adhesin of EHEC O157:H7. Moreover, the colonizing ability (and perhaps intestinal survival in general) of the iha- derivatives might be further limited as a consequence of their decreased ability to compete for iron, as we demonstrated by reduced growth of iha- S strains as well as iha deletions mutants 154LΔiha and O157 SakaiΔiha under iron-limited conditions. Whether or not this effect is associated with the proposed role of Iha, which is absent in S strains, as a siderophore receptor (Léveilléet al., 2006) warrants further investigations.
The functional role of ter genes in bacteria is not known. In S. marcescens, the ter genes on plasmid R478 encode, in addition to TelR, resistance to pore-forming colicins (Whelan et al., 1995). Therefore, if the ter cluster in EHEC O157:H7 encodes a similar function, strains harbouring these loci might better compete in polymicrobial milieus. Consequently, the loss of the ter cluster might reduce virulence (or colonization capacity) because of diminished competitive potential. Our data do not allow us to evaluate the role of the ter cluster in adherence to human intestinal epithelial cells separate to that of the major contribution of Iha. However, currently available experimental data from our study and one other report (Yin et al., 2009) suggest that deletions within OI 43/OI 48 negatively influence virulence and/or fitness of EHEC O157:H7. This is of a particular importance considering the high frequency of such deletions.
Sorbitol-fermenting (SF) EHEC O157:NM (non-motile), a close relative of EHEC O157:H7 (Feng et al., 1998; Leopold et al., 2009), lacks complete or truncated OI 43/OI 48 integrated in serW/serX (Fig. 3). However, SF EHEC O157:NM strains possess a large mosaic island composed of fragments of SRL-PAI and ∼ 20 kb of the 3′ end of OI 43 of EDL933, which lacks ter and iha (Janka et al., 2005). Our finding of similar remnants of OI 43/OI 48 in TelS S variants of EHEC O157:H7 analysed in this study prompts speculation that SF EHEC O157:NM originally possessed a homologue of OI 43 that was subsequently truncated via genomic deletions and became a part of the mosaic island, probably during genomic rearrangements. Thus, deletions in OI 43/OI 48 might have played a role in the evolution of the EHEC O157 group. The finding of a hybrid island that contains segments of the 3′ end of OI 48 of EDL933 in EHEC O113:H21 (Shen et al., 2004) indicates the frequent occurrence of recombination events in this element. Moreover, a functional homologue of TelR-encoding island is found in various non-O157 EHEC (Tarr et al., 2000) and in enterotoxigenic E. coli (Parreira et al., 2008) suggesting that this segment can be assimilated by divergent genomes.
Different scenarios might explain how Tel-susceptible EHEC O157:H7 (Taylor et al., 2002; Bielaszewska et al., 2005) arise. Such strains might have never had the TelR-encoding islands. Alternatively, such strains might have originally possessed TelR island(s), which were subsequently completely excised from the chromosome by site-specific recombination, as demonstrated in our study. Indeed, intact serW and serX genes, potentially resulting from either of these scenarios, are found in some E. coli O157:H7 (Taylor et al., 2002). A third scenario based on our data, which appears to occur most frequently, is that TelS results from internal deletions in TelR-encoding islands that encompass the ter gene cluster.
In summary, OI 43/OI 48 deletions are another mechanism of genome plasticity in the EHEC 1 clade. The deletions are accompanied by phenotypic and functional changes. These changes reduce virulence and/or fitness, and are at least partially attributed to the loss of iha, which might play dual roles in the virulence of EHEC O157:H7, i.e. as an adhesin and a siderophore receptor. The frequency of excision in vivo, the biologic role of this process and the survival consequences of these mutations warrant further investigation.
Experimental procedures
Bacterial strains and their genotypic and phenotypic characterization
The five E. coli O157:H7 strains displaying morphological dissociation associated with loss of TelR-encoding islands were isolated during 6 years from five patients (four with haemolytic uraemic syndrome and one with bloody diarrhoea) living in five different cities in Germany, indicating epidemiological independence between the strains. The dissociation into L and S colonies was observed after two to four passages on SMAC agar (Becton Dickinson, Sparks, MD, USA). Between these passages the strains were stored between 1 and 5 days at 4°C. L and S colonies from each strain were biochemically confirmed as E. coli (API 20 E; bioMérieux, Marcy l'Etoile, Lyon, France), serotyped (Prager et al., 2003), phage typed (Liesegang et al., 2000) and tested by PCR for rfbEO157 (Nagano et al., 1998), fliCH7 (Eklund et al., 2006), stx genotype (Friedrich et al., 2002; Bielaszewska et al., 2006b) and the terZABCDEF cluster (Bielaszewska et al., 2005). Tel-MICs were determined using microdilution (Sahm and Washington, 1991). Each strain was tested in duplicate and in two independent experiments using 5 × 104 cfu per well and serial dilutions (from 1024 to 1 µg ml−1) of potassium tellurite (K2TeO3) (Sigma-Aldrich, Taufkirchen, Germany) in 100 µl of LB broth. The MIC was defined as the lowest concentration of K2TeO3 that completely inhibited growth after overnight incubation at 37°C. The ability to grow on CT-SMAC agar (K2TeO3 2.5 µg ml−1, cefixime 0.05 µg ml−1; Becton Dickinson) was determined on plates inoculated with 1 × 105 cfu after overnight incubation (Bielaszewska et al., 2005). Stx titres were determined in a Vero cell assay (Bielaszewska et al., 2006b) and defined as the reciprocal of the highest dilution of culture supernatant that was cytotoxic in 50% of cells after 3 days of incubation. Production of EHEC haemolysin was sought on enterohaemolysin agar (Sifin, Berlin, Germany) and β-D-glucuronidase activity was assessed using nutrient agar with 4-methylumbelliferyl-β-D-glucuronide (MUG) (Becton Dickinson).
PCR assays for mapping and analyses of deletions of TelR-encoding islands
Polymerase chain reaction primers and conditions are listed in Table S1. Positions of the PCR primers in OI 43/OI 48 of E. coli O157:H7 strain EDL933 and the flanking regions are depicted in Fig. S1 and primers used to analyse internal deletions in OI 48 are depicted in Fig. 4. PCRs for mapping of TelR-encoding islands were performed in the iCycler (version 1.259; Bio-Rad, München, Germany) using reagents from PEQLAB Biotechnologie (Erlangen, Germany) (Sonntag et al., 2004) and 2.5 µl of bacterial DNA purified with InstaGene Matrix (Bio-Rad) as a template. PCRs to detect integration sites of TelR-encoding islands, junctions between OI 43/OI 48 and the core genome, and to produce connecting fragments for sequence analysis of OI 43/OI 48 deletions were performed in a Biometra thermocycler using the RED Taq ReadyMix PCR Reaction Mix with MgCl2 (Sigma-Aldrich, München, Germany). The PCR master mix (20 µl) contained 20–100 ng of chromosomal DNA as a template and 10 pmol of each primer. Six microlitre aliquots of the reactions were analysed by electrophoresis in 1% (wt/vol) agarose gels. E. coli O157:H7 strain EDL933 (Perna et al., 2001), E. coli K-12 strain MG1655 (Blattner et al., 1997) and SF EHEC O157:NM strain 493/89 (Janka et al., 2005) were used as PCR controls.
Analysis of the core genome deletions in strain 95S
The extents of the core genome deletions upstream of OI 48 and OI 43, respectively, in strain 95S were investigated using primer walking along each respective region starting from ycdU and clpA, respectively (for PCR primers see Table S2). Connecting fragment spanning deletion upstream of OI 48 was produced using primers Z1398-1 and Z1650-2 (Tables S2 and S1, respectively) and sequenced as described below. To produce a connecting fragment spanning the deletion upstream of OI 43, ORFs Z1117, Z1116 and Z1115 found to be present upstream of OI 43 using the primer walking were PCR connected (primers Z1117-1, Z1116-1 and Z1115-1, respectively) (Table S2) with ORFs Z1210 and Z1211 downstream of the internal deletion in OI 43 (Fig. 2); primers Z1650-2 and Z1651-2 (Table S1) that target the identical ORFs Z1210 and Z1211, respectively, in OI 43 were used for this purpose.
Sequence analysis
Amplicons were sequenced using purified PCR products (PCR Purification Kit; Qiagen, Hilden, Germany), and an automated ABI Prism 3130xl Genetic Analyzer and the ABI Prism BigDye Terminator Ready Reaction Cycle Sequencing Kit (version 3.1, Applied Biosystems, Darmstadt, Germany). Sequences were analysed using the Vector NTI Advance 11 software (Invitrogen, Karlsruhe, Germany). Homology searches were performed using the EMBL-GenBank database (http://www.ncbi.nlm.nih.gov/BLAST).
Light cycler-based PCR quantification of terC
Genomic DNA was isolated using the DNeasy Kit (Qiagen). terC and gyrB (used as an internal standard) were amplified using the QuantiTect SYBR Green PCR Kit (Qiagen) and primer pairs TerC-F1/TerC-R1 and GyrB-F2/GyrB-R2, respectively (Taylor et al., 2002). The PCRs were performed in the LightCycler System (Roche Diagnostics, Mannheim, Germany) as described (Zhang et al., 2005). After the final cycle, a melting curve analysis was performed with continuous fluorescence reading from 65°C to 95°C. A standard curve for the determination of DNA concentration was prepared using 10-fold dilutions of the total genomic DNA from E. coli O157:H7 strain Sakai (RIMD 0509952) ranging from 10−1 (20 ng µl−1) to 10−5 (2 pg µl−1). The concentrations of terC and gyrB DNAs were determined using LightCycler Software 3 second derivative method analysis (Roche Diagnostics) and terC DNA was normalized to gyrB DNA. The terC/gyrB DNA ratio for each strain was expressed as a mean (standard deviation) of three independent experiments.
Quantitative real-time RT-PCR
Total RNA was isolated from L and S strains grown in LB broth and DMEM using the RNeasy Mini Kit (Qiagen). Co-purified DNA was removed using RNase-free DNase (Roche Diagnostics). A one-step quantitative real-time RT-PCR, performed with an iCycler iQ-5 (Bio-Rad) and the QuantiTect SYBR Green RT-PCR kit (Qiagen) measured the relative expression of mRNA of iha, eae, lpfA1, lpfA2 and ehaA. The PCR reactions were performed in 96-well plates using a 20 µl volume containing 1 µl of total RNA (100 ng), 10 µl of 2× QuantiTect SYBR Green RT-PCR master mix, 0.2 µl of QuantiTect RT mix and 200 nM of each primer (Blumer et al., 2005; Léveilléet al., 2006; Chen et al., 2007; Torres et al., 2009) (for primers see Table S3). The PCR included a reverse transcription step at 50°C for 30 min, and polymerase activation and preliminary denaturation at 95°C for 15 min, followed by 35 cycles of denaturation at 94°C for 10 s, annealing at 53°C to 60°C for 20 s and extension at 72°C for 20 s. A melting curve analysis to confirm the specificity of the amplification products was constructed with continuous fluorescence reading from 55°C to 95°C. Data were analysed using the Bio-Rad iQ5 standard edition optical system software V2.0. The iha, eae, lpfA1, lpfA2 and ehaA mRNAs were normalized to gapA mRNA. Each PCR was performed three times with three independent RNA preparations.
Determination of deletions affecting the TelR-encoding island
A real-time PCR approach with the StepOnePlus Real-Time PCR System (Applied Biosystems) was used to determine the proportion of intact serW and serX tRNA genes resulting from site-specific excision of OI 43 and OI 48, respectively, and of internal deletions in OI 48 in DNA extracted (DNAeasy kit; Qiagen) from overnight cultures of reference strains EDL933, Sakai, 493/89 as well as all L and S strains and adjusted to a concentration of 10 ng µl−1. All reactions were run in triplicate for 40 cycles and contained a mixture of 2–4 µl chromosomal DNA (10 ng µl−1), 2 µl of each primer (5 pmol µl−1) and 1xSYBR Green PCR Master Mix in a total volume of 20 µl according to the manufacturer's instructions. Post-experimentally, a melting curve analysis was performed (60°C to 95°C with 0.3°C increments) to verify product purity. All data were analysed with the StepOne Software v2.1.
To determine the amount of genome equivalents (GE) per µl of DNA solution (#GE), primer pair 131 (Table S1) was used to amplify an internal fragment of recA from all strains. As a standard 102–106 GEs of strain Sakai were used (calculation based on the published genome size) (Hayashi et al., 2001). Subsequently, 102–106 copies of strain 493/89, an OI 43/OI 48-negative derivative, were used as standard for PCRs 125 and 126, respectively (Table S1), to determine the amount of GEs with intact serW (Wi) and serX (Xi) genes. The proportion of cells with full excision of the respective island, i.e. OI 43-negative (43neg) and/or OI 48-negative (48neg), was calculated as the quotient of Wi and #GE and Xi and #GE respectively. Finally, an equimolar mixture of 102–106 copies of plasmid pTerE (3417 bp) (Table S4) and GEs of strain 493/89 was used as a standard in PCR 132 (Table S1) to amplify an internal fragment of terE from strains Sakai, 81L, 134L and 154L (each contains only a single TelR-encoding island) and determine the amount of terE-positive GEs (E+). The proportion of GEs with internal deletions of OI 48 (Eint−) was calculated according to the following formula: Eint− = [(#GE − E+) − 48neg]/#GE.
Southern blot hybridization
Genomic DNA was digested with BamHI and PstI (New England Biolabs, Frankfurt, Germany), separated in 0.6% agarose and transferred to a nylon membrane. The membrane was probed under stringent conditions with a digoxigenin-labelled terC probe (Taylor et al., 2002) using DIG DNA Labelling and Detection Kit (Roche Diagnostics) (Bielaszewska et al., 2005).
Pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis was performed using the PulseNet protocol (Hunter et al., 2005) except that the running time was prolonged to 40 h to achieve more distinct separation of smaller bands. The XbaI-digested DNA of Salmonella enterica serovar Braenderup strain H9812 was used as a standard (Hunter et al., 2005). Restriction patterns were analysed and the cluster analysis was performed with BioNumerics software, version 5.1 (Applied Maths BVBA, Sint-Martens-Latem, Belgium).
Construction of iha deletion and complementation mutants
The iha deletion mutants of EHEC O157:H7 strains 154L (154LΔiha) and Sakai (RIMD 0509952) (SakaiΔiha) were generated using lambda red-based recombineering (Datsenko and Wanner, 2000). Briefly, the chloramphenicol acetyltransferase gene (cat) cassette of plasmid pKD3 was amplified using primers del_iha_for and del_iha_rev (Table S5), with overhangs homologous to the 5′ and 3′ regions of the O157 Sakai iha gene. Purified PCR product was transformed into electrocompetent O157 Sakai or 154L cells carrying the plasmid pKD46. The cat cassette was cured upon transformation with plasmid pCP20 (Cherepanov and Wackernagel, 1995). iha mutants were screened using PCR and Southern blot.
For in trans complementation of iha mutants, a 2916 bp genomic fragment of strain O157 Sakai that contained the functional iha gene including its 400 bp upstream and 350 bp downstream region was amplified by PCR using the Phusion DNA polymerase (New England Biolabs) and the primers iha_for2 and iha_rev (Table S5). Purified PCR product was ligated into plasmid pWKS30 (Wang and Kushner, 1991) that had been linearized by restriction with SmaI and dephosphorylated with Antarctic phosphatase (both New England Biolabs). Screening for correct plasmid clones (pWKS30iha) was performed by PCR and the correct orientation of the insert was verified by sequencing. Strain 154LΔiha was transformed with pWKS30 and pWKS30iha respectively.
For chromosomal complementation of iha mutants, the functional iha gene including its 400 bp upstream and 350 bp downstream region was PCR-amplified using primers iha-pKD4_for and iha-pKD4_rev (Table S5). Following digestion with BstBI/HindIII (New England Biolabs), the resulting 2916 bp PCR product was purified and ligated into BstBI/HindIII-digested plasmid pKD4. Screening for correct plasmid clones (pKD4iha) was performed using PCR and verified by sequencing. The iha fragment together with the kanamycin resistance gene was then PCR-amplified using the Phusion DNA polymerase and primers iha-int_for and iha-int_rev (Table S5). The resulting 3971 bp PCR product was transformed into relevant iha mutants carrying plasmid pKD46. Selection of transformants in which the iha::kan fragment was chromosomally inserted downstream glmS was done on LB agar plates supplemented with kanamycin (30 µg ml−1). Screening for strains SakaiΔiha and 154S chromosomally complemented with iha was made by PCR and correct insertion of the iha::kan fragment was verified by sequencing. iha transcription in all iha+ constructs was confirmed by quantitative real-time RT-PCR as described above.
Iron content in culture media
Iron content in culture media was determined using atomic absorption spectroscopy. Briefly, LB broth, DMEM (Johnson et al., 2005) and DMEM supplemented with 10 µM FeCl2 were complemented with 1 ml of HNO3 per 100 ml. A triplicate of each solution was analysed in a Unicam Solaar 939 AA spectrometer with acetylene/air burner (split 10 cm) at 248.3 nm. Final iron content was calculated using a linear external calibration (0.1, 0.3, 0.5, 0.7, 0.9, 1.1, 1.3, 1.5 mg l−1) (DIN 38406–32, 2000). Based on this analysis, the media contained 0.59 µg ml−1, < 0.05 µg ml−1 and 0.50 µg ml−1 of iron respectively. DMEM agar and DMEM agar with 10 µM FeCl2 were prepared from liquid media by adding 1.5% (wt/vol) of agar–agar base (Carl Roth, Karlsruhe, Germany).
Cell cultures and adherence assay
Human ileocaecal adenocarcinoma epithelial cell line HCT-8 (ATCC CCL-244) and colonic carcinoma cell line Caco-2 (German collection of microorganisms and cell cultures, Braunschweig, Germany; ACC 169) were cultured as described (Sonntag et al., 2005; Aldick et al., 2007). For adherence assays, 105 cells per well were seeded in 24-well plates (Corning, Corning, NY, USA) containing coverslips and grown until they were ∼ 70% confluent. The cells were washed with phosphate-buffered saline (PBS), replenished with fresh medium with 0.5% D-mannose (Merck, Darmstadt, Germany), and infected with ∼ 1 × 108 cfu of overnight stationary cultures of L and S strains, O157 Sakai strain and their respective iha deletion and iha complementation mutants (Table S4) in DMEM, DMEM with 10 µM FeCl2 or LB broth (only L and S strains were grown in the latter two media). After 3 h of incubation with bacteria (37°C, 5% CO2), cells were washed three times with PBS, and incubated another 3 h in fresh culture medium. The cultures were 10 times washed with PBS, fixed (70% ethanol), stained (10% Giemsa) (Merck, Darmstadt, Germany) and mounted using Glycergel (DakoCytomation, Hamburg, Germany). Bacterial adherence was examined using light microscopy (Axio Imager A1; Zeiss, Jena, Germany) and the adherence patterns were photographed (AxioCam MRm camera) (Zeiss). Bacteria and cells were counted in 10 randomly selected fields on each coverslip and bacteria per cell were averaged. The enumerator was unaware of the identity of the cells being counted. Differences in quantitative adherence of iha+ and iha- strains were evaluated using unpaired Student's t-test (P < 0.05 considered significant).
Growth in DMEM
One colony of each L strain, S strain, O157 Sakai strain and the respective iha deletion and iha complementation mutants (Table S4) was grown overnight (37°C, 180 r.p.m.) in 2 ml of DMEM without or with 10 µM FeCl2. An aliquot of the overnight culture was inoculated in 20 ml of the same medium to produce an OD600 between 0.015 and 0.025 (the starting OD600 values of corresponding L and S strains and the respective iha mutants were identical). Bacterial growth (37°C, 180 r.p.m.) was monitored spectrophotometrically (OD600) hourly for 12 h and again at 24 h. Each strain was tested in each medium in triplicate and growth curves were constructed by plotting mean OD600 values (standard deviations) against time. Differences in growth kinetics of iha+ and iha- strains were evaluated using unpaired Student's t-test.
Influence of iha expression on colony size
One colony of each L strain, S strain, the O157 Sakai strain and their corresponding iha deletion and iha complementation mutants (Table S4) was inoculated on DMEM agar without or with 10 µM FeCl2. After incubation at 37°C for 26 h the plates were photographed and the sizes of all or most well-separated colonies on each plate were determined using a Power Point (Microsoft) measuring tool. Differences between sizes of iha+ and iha- colonies under different conditions were calculated using unpaired Student's t-test.
Siderophore expression
Siderophore expression was detected colorimetrically in supernatants of overnight cultures of S strains grown in DMEM using chrome azurol S/iron(III)/hexadecyltrimethylamonium bromide complex as an indicator of iron binding (Schwyn and Neilands, 1987).
Acknowledgments
This study was supported by grant from the Interdisciplinary Centre of Clinical Research (IZKF) Münster No. Me2/023/08, by grant EU Network ERA-NET PathoGenoMics II (No. 0315443), by a grant from the Medical Faculty of the University Muenster (No. BD9817044) and by NIH Grant R01 AI47499. The work was also supported by the German Research Foundation (SFB 479, TP A1). We thank Herbert Schmidt (University of Hohenheim, Stuttgart, Germany) for providing us with E. coli O157:H7 strain Sakai. We are also grateful to Anni Bommer for determination of iron content in culture media, and to Dagmar Mense, Margarete Junge, Nadine Brandt, Olena Mantel (Münster) and Barbara Plaschke (Würzburg) for skilful technical assistance.
Supporting Information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ahmed N, Dobrindt U, Hacker J, Hasnain SE. Genomic fluidity and pathogenic bacteria: applications in diagnostics, epidemiology and intervention. Nat Rev Microbiol. 2008;6:387–394. doi: 10.1038/nrmicro1889. [DOI] [PubMed] [Google Scholar]
- Aldick T, Bielaszewska M, Zhang W, Brockmeyer J, Schmidt H, Friedrich AW, et al. Hemolysin from Shiga toxin-negative Escherichia coli O26 strains injures microvascular endothelium. Microbes Infect. 2007;9:282–290. doi: 10.1016/j.micinf.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Bach S, Buchrieser C, Prentice M, Guiyoule A, Msadek T, Carniel E. The high-pathogenicity island of Yersinia enterocolitica Ye8081 undergoes low-frequency deletion but not precise excision, suggesting recent stabilization in the genome. Infect Immun. 1999;67:5091–5099. doi: 10.1128/iai.67.10.5091-5099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M, Tarr PI, Karch H, Zhang W, Mathys W. Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates. J Clin Microbiol. 2005;43:452–454. doi: 10.1128/JCM.43.1.452-454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M, Prager R, Zhang W, Friedrich AW, Mellmann A, Tschape H, Karch H. Chromosomal dynamism in progeny of outbreak-related sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl Environ Microbiol. 2006a;72:1900–1909. doi: 10.1128/AEM.72.3.1900-1909.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M, Friedrich AW, Aldick T, Schurk-Bulgrin R, Karch H. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin Infect Dis. 2006b;43:1160–1167. doi: 10.1086/508195. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Blumer C, Kleefeld A, Lehnen D, Heintz M, Dobrindt U, Nagy G, et al. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology. 2005;151:3287–3298. doi: 10.1099/mic.0.28098-0. [DOI] [PubMed] [Google Scholar]
- Brzuszkiewicz E, Brüggemann H, Liesegang H, Emmerth M, Olschläger T, Nagy G, et al. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci USA. 2006;103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno SM, Santiviago CA, Murillo AA, Fuentes JA, Trombert AN, Rodas PI, et al. Precise excision of the large pathogenicity island, SPI7, in Salmonella enterica serovar Typhi. J Bacteriol. 2004;186:3202–3213. doi: 10.1128/JB.186.10.3202-3213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Huang K-L, Xu W-T, Li Y, Luo Y-B. Real-time quantitative PCR detection of Escherichia coli O157:H7. Chin J Agric Biotechol. 2007;4:15–19. [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIN 38406-32. Deutsche Einheitsverfahren zur Wasser-, Abwasser- und Schlammuntersuchung – Kationen (Gruppe E) – Teil 32: Bestimmung Von Eisen Mittels Atomabsorptionsspektrometrie (E 32) Berlin: Beuth Verlag GmbH; 2000. pp. 1–15. [Google Scholar]
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- Donnenberg MS, Tzipori S, McKee ML, O'Brien AD, Alroy J, Kaper JB. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Invest. 1993;92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund M, Bielaszewska M, Nakari UM, Karch H, Siitonen A. Molecular and phenotypic profiling of sorbitol-fermenting Escherichia coli O157:H- human isolates from Finland. Clin Microbiol Infect. 2006;12:634–641. doi: 10.1111/j.1469-0691.2006.01478.x. [DOI] [PubMed] [Google Scholar]
- Feng P, Lampel KA, Karch H, Whittam TS. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–1753. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- Feng P, Dey M, Abe A, Takeda T. Isogenic strain of Escherichia coli O157:H7 that has lost both Shiga toxin 1 and 2 genes. Clin Diagn Lab Immunol. 2001;8:711–717. doi: 10.1128/CDLI.8.4.711-717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich AW, Bielaszewska M, Zhang WL, Pulz M, Kuczius T, Ammon A, Karch H. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J Infect Dis. 2002;185:74–84. doi: 10.1086/338115. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, et al. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol. 2005;43:1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janka A, Becker G, Sonntag A-K, Bielaszewska M, Dobrindt U, Karch H. Presence and characterization of a mosaic genomic island which distinguishes sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H- from E. coli O157:H7. Appl Environ Microbiol. 2005;71:4875–4878. doi: 10.1128/AEM.71.8.4875-4878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Jelacic S, Schoening LM, Clabots C, Shaikh N, Mobley HL, Tarr PI. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect Immun. 2005;73:965–971. doi: 10.1128/IAI.73.2.965-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper JB, Karmali MA. The continuing evolution of a bacterial pathogen. Proc Natl Acad Sci USA. 2008;105:4535–4536. doi: 10.1073/pnas.0801435105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch H, Tarr PI, Bielaszewska M. Enterohaemorrhagic Escherichia coli in human medicine. Int J Med Microbiol. 2005;295:405–418. doi: 10.1016/j.ijmm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Kresse AU, Rienäcker I, Valle AM, Steinrück H, Claus H, Payne SM, et al. Enterohaemorrhagic Escherichia coli O157 and non-O157 serovars differ in their mechanisms for iron supply. Int J Med Microbiol. 2007;297:9–15. doi: 10.1016/j.ijmm.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Leopold SR, Magrini V, Holt NJ, Shaikh N, Mardis ER, Cagno J, et al. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc Natl Acad Sci USA. 2009;106:8713–8718. doi: 10.1073/pnas.0812949106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léveillé S, Caza M, Johnson JR, Clabots C, Sabri M, Dozois CM. Iha from an Escherichia coli urinary tract infection outbreak clonal group A strain is expressed in vivo in the mouse urinary tract and functions as a catecholate siderophore receptor. Infect Immun. 2006;74:3427–3436. doi: 10.1128/IAI.00107-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesegang A, Sachse U, Prager R, Claus H, Steinruck H, Aleksic S, et al. Clonal diversity of Shiga toxin-producing Escherichia coli O157:H7/H- in Germany-a ten-year study. Int J Med Microbiol. 2000;290:269–278. doi: 10.1016/S1438-4221(00)80125-3. [DOI] [PubMed] [Google Scholar]
- Luck SN, Turner SA, Rajakumar K, Sakellaris H, Adler B. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect Immun. 2001;69:6012–6021. doi: 10.1128/IAI.69.10.6012-6021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli AT, Fernandez RE, Bloch CA, Rode CK, Fasano A. ‘Black holes’ and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci USA. 1998;35:3943–3948. doi: 10.1073/pnas.95.7.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellmann A, Bielaszewska M, Zimmerhackl LB, Prager R, Harmsen D, Tschape H, Karch H. Enterohemorrhagic Escherichia coli in human infection: in vivo evolution of a bacterial pathogen. Clin Infect Dis. 2005;41:785–792. doi: 10.1086/432722. [DOI] [PubMed] [Google Scholar]
- Mellmann A, Lu S, Karch H, Xu JG, Harmsen D, Schmidt MA, Bielaszewska M. Recycling of Shiga toxin 2 genes in sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl Environ Microbiol. 2008;74:67–72. doi: 10.1128/AEM.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellmann A, Bielaszewska M, Karch H. Intra-host genome alterations in enterohemorrhagic Escherichia coli. Gastroenterology. 2009;136:1925–1938. doi: 10.1053/j.gastro.2008.12.072. [DOI] [PubMed] [Google Scholar]
- Middendorf B, Hochhut B, Leipold K, Dobrindt U, Blum-Oehler G, Hacker J. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J Bacteriol. 2004;186:3086–3096. doi: 10.1128/JB.186.10.3086-3096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase T, Yamai S, Watanabe H. Changes in pulsed-field gel electrophoresis patterns in clinical isolates of enterohemorrhagic Escherichia coli O157:H7 associated with loss of Shiga toxin genes. Curr Microbiol. 1999;38:48–50. doi: 10.1007/pl00006771. [DOI] [PubMed] [Google Scholar]
- Nagano I, Kunishima M, Itoh Y, Wu Z, Takahashi Y. Detection of verotoxin-producing Escherichia coli O157:H7 by multiplex polymerase chain reaction. Microbiol Immunol. 1998;42:371–376. doi: 10.1111/j.1348-0421.1998.tb02297.x. [DOI] [PubMed] [Google Scholar]
- Onoue Y, Konuma H, Nakagawa H, Hara-Kudo Y, Fujita T, Kumagai S. Collaborative evaluation of detection methods for Escherichia coli O157:H7 from radish sprouts and ground beef. Int J Food Microbiol. 1999;46:27–36. doi: 10.1016/s0168-1605(98)00174-3. [DOI] [PubMed] [Google Scholar]
- Orth D, Grif K, Dierich MP, Würzner R. Variability in tellurite resistance and the ter gene cluster among Shiga toxin-producing Escherichia coli isolated from humans, animals and food. Res Microbiol. 2007;158:105–111. doi: 10.1016/j.resmic.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Parreira VR, Liao JH, Kim SH, Gyles CL. A homolog of the O157 urease-encoding O island 48 is present in porcine O149:H10 enterotoxigenic Escherichia coli. Vet Res. 2008;39:38. doi: 10.1051/vetres:2008015. DOI: 10.1051/vetres:2008015. [DOI] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Prager R, Strutz U, Fruth A, Tschape H. Subtyping of pathogenic Escherichia coli strains using flagellar (H)-antigens: serotyping versus fliC polymorphisms. Int J Med Microbiol. 2003;292:477–486. doi: 10.1078/1438-4221-00226. [DOI] [PubMed] [Google Scholar]
- Rajanna C, Wang J, Zhang D, Xu Z, Ali A, Hou YM, Karaolis DK. The vibrio pathogenicity island of epidemic Vibrio cholerae forms precise extrachromosomal circular excision products. J Bacteriol. 2003;185:6893–6901. doi: 10.1128/JB.185.23.6893-6901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahm DF, Washington JA. Antibacterial susceptibility tests: dilution methods. In: Ballows A, Hausler WJ Jr, Hermann KL, Isenberg HD, Shadomy HJ, editors. Manual of Clinical Microbiology. Washington, DC: American Society for Microbiology; 1991. pp. 1105–1116. [Google Scholar]
- Sakellaris H, Luck SN, Al-Hasani K, Rajakumar K, Turner SA, Adler B. Regulated site-specific recombination of the she pathogenicity island of Shigella flexneri. Mol Microbiol. 2004;52:1329–1336. doi: 10.1111/j.1365-2958.2004.04048.x. [DOI] [PubMed] [Google Scholar]
- Scaletsky IC, Pedroso MZ, Oliva CA, Carvalho RL, Morais MB, Fagundes-Neto U. A localized adherence-like pattern as a second pattern of adherence of classic enteropathogenic Escherichia coli to HEp-2 cells that is associated with infantile diarrhea. Infect Immun. 1999;67:3410–3415. doi: 10.1128/iai.67.7.3410-3415.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübbe S, Kube M, Scheffel A, Wawer C, Heyen U, Meyerdierks A, et al. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J Bacteriol. 2003;185:5779–5790. doi: 10.1128/JB.185.19.5779-5790.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Shen S, Mascarenhas M, Rahn K, Kaper JB, Karmali MA. Evidence for a hybrid genomic island in verocytotoxin-producing Escherichia coli CL3 (serotype O113:H21) containing segments of EDL933 (serotype O157:H7) O islands 122 and 48. Infect Immun. 2004;72:1496–1503. doi: 10.1128/IAI.72.3.1496-1503.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag A-K, Prager R, Bielaszewska M, Zhang W, Fruth A, Tschäpe H, Karch H. Phenotypic and genotypic analyses of enterohemorrhagic Escherichia coli O145 strains from patients in Germany. J Clin Microbiol. 2004;42:954–962. doi: 10.1128/JCM.42.3.954-962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag A-K, Bielaszewska M, Mellmann A, Dierksen N, Schierack P, Wieler LH, et al. Shiga toxin 2e-producing Escherichia coli isolates from humans and pigs differ in their virulence profiles and interactions with intestinal epithelial cells. Appl Environ Microbiol. 2005;71:8855–8863. doi: 10.1128/AEM.71.12.8855-8863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr PI, Bilge SS, Vary JC, Jr, Jelacic S, Habeeb RL, Ward TR, et al. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun. 2000;68:1400–1407. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- Tauschek M, Strugnell RA, Robins-Browne RM. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol Microbiol. 2002;44:1533–1550. doi: 10.1046/j.1365-2958.2002.02968.x. [DOI] [PubMed] [Google Scholar]
- Taylor DE, Rooker M, Keelan M, Ng LK, Martin I, Perna NT, et al. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J Bacteriol. 2002;184:4690–4698. doi: 10.1128/JB.184.17.4690-4698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Payne SM. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1997;23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- Torres AG, Giron JA, Perna NT, Burland V, Blattner FR, Avelino-Flores F, Kaper JB. Identification and characterization of lpfABCC'DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2002;70:5416–5427. doi: 10.1128/IAI.70.10.5416-5427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Kanack KJ, Tutt CB, Popov V, Kaper JB. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol Lett. 2004;238:333–344. doi: 10.1016/j.femsle.2004.07.053. [DOI] [PubMed] [Google Scholar]
- Torres AG, Blanco M, Valenzuela P, Slater TM, Patel SD, Dahbi G, et al. Genes related to long polar fimbriae of pathogenic Escherichia coli strains as reliable markers to identify virulent isolates. J Clin Microbiol. 2009;47:2442–2451. doi: 10.1128/JCM.00566-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SA, Luck SN, Sakellaris H, Rajakumar K, Adler B. Nested deletions of the SRL pathogenicity island of Shigella flexneri 2a. J Bacteriol. 2001;183:5535–5543. doi: 10.1128/JB.183.19.5535-5543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SA, Luck SN, Sakellaris H, Rajakumar K, Adler B. Role of attP in integrase-mediated integration of the Shigella Resistance Locus pathogenicity island of Shigella flexneri. Antimicrob Agents Chemother. 2004;48:1028–1031. doi: 10.1128/AAC.48.3.1028-1031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S, Kube M, Schubbe S, Reinhardt R, Schuler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacteriol. 2005;187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duynhoven YT, De Jager CM, Heuvelink AE, Van Der Zwaluw WK, Maas HM, Van Pelt W, Wannet WJ. Enhanced laboratory-based surveillance of Shiga-toxin-producing Escherichia coli O157 in The Netherlands. Eur J Clin Microbiol Infect Dis. 2002;21:513–522. doi: 10.1007/s10096-002-0756-7. [DOI] [PubMed] [Google Scholar]
- Waldor MK. Mobilizable genomic islands: going mobile with oriT mimicry. Mol Microbiol. 2010;78:537–540. doi: 10.1111/j.1365-2958.2010.07365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, et al. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ Microbiol. 2008;10:589–604. doi: 10.1111/j.1462-2920.2007.01479.x. [DOI] [PubMed] [Google Scholar]
- Whelan KF, Colleran E, Taylor DE. Phage inhibition, colicin resistance, and tellurite resistance are encoded by a single cluster of genes on the IncHI2 plasmid R478. J Bacteriol. 1995;177:5016–5027. doi: 10.1128/jb.177.17.5016-5027.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Wheatcroft R, Chambers JR, Liu B, Zhu J, Gyles CL. Contributions of O island 48 to adherence of enterohemorrhagic Escherichia coli O157:H7 to epithelial cells in vitro and in ligated pig ileal loops. Appl Environ Microbiol. 2009;75:5779–5786. doi: 10.1128/AEM.00507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadik PM, Chapman PA, Siddons CA. Use of tellurite for the selection of verocytotoxigenic Escherichia coli O157. J Med Microbiol. 1993;39:155–158. doi: 10.1099/00222615-39-2-155. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bielaszewska M, Friedrich AW, Kuczius T, Karch H. Transcriptional analysis of genes encoding Shiga toxin 2 and its variants in Escherichia coli. Appl Environ Microbiol. 2005;71:558–561. doi: 10.1128/AEM.71.1.558-561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


